

You can:
| Name | Alpha-2C adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA2C |
| Synonym | alpha2-C4 Adra-2c ADRA2L2 ADRA2RL2 Adrenergic alpha2C- receptor class I [ Show all ] |
| Disease | Diabetic nephropathy; Fibromyalgia Hypertension Heart failure Glaucoma Alzheimer disease [ Show all ] |
| Length | 462 |
| Amino acid sequence | MASPALAAALAVAAAAGPNASGAGERGSGGVANASGASWGPPRGQYSAGAVAGLAAVVGFLIVFTVVGNVLVVIAVLTSRALRAPQNLFLVSLASADILVATLVMPFSLANELMAYWYFGQVWCGVYLALDVLFCTSSIVHLCAISLDRYWSVTQAVEYNLKRTPRRVKATIVAVWLISAVISFPPLVSLYRQPDGAAYPQCGLNDETWYILSSCIGSFFAPCLIMGLVYARIYRVAKLRTRTLSEKRAPVGPDGASPTTENGLGAAAGAGENGHCAPPPADVEPDESSAAAERRRRRGALRRGGRRRAGAEGGAGGADGQGAGPGAAESGALTASRSPGPGGRLSRASSRSVEFFLSRRRRARSSVCRRKVAQAREKRFTFVLAVVMGVFVLCWFPFFFSYSLYGICREACQVPGPLFKFFFWIGYCNSSLNPVIYTVFNQDFRRSFKHILFRRRRRGFRQ |
| UniProt | P18825 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P18825 |
| 3D structure model | This predicted structure model is from GPCR-EXP P18825. |
| BioLiP | N/A |
| Therapeutic Target Database | T01777 |
| ChEMBL | CHEMBL1916 |
| IUPHAR | 27 |
| DrugBank | BE0004864, BE0000342, BE0004888 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 122 | 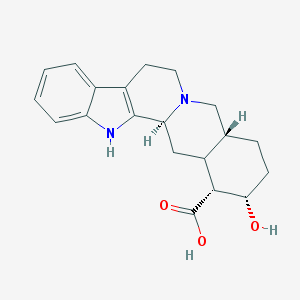 CHEMBL382310 CHEMBL382310 | C20H24N2O3 | 340.423 | 4 / 3 | 0.3 | Yes |
| 368 | 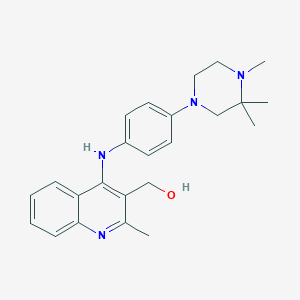 CHEMBL218675 CHEMBL218675 | C24H30N4O | 390.531 | 5 / 2 | 4.3 | Yes |
| 1005 | 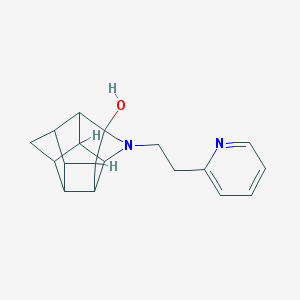 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1495 | 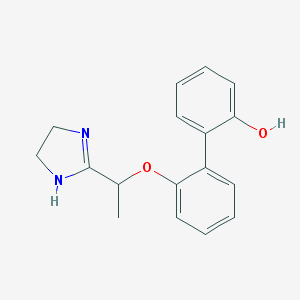 CHEMBL373830 CHEMBL373830 | C17H18N2O2 | 282.343 | 3 / 2 | 2.6 | Yes |
| 1558 | 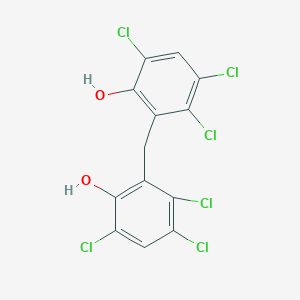 hexachlorophene hexachlorophene | C13H6Cl6O2 | 406.889 | 2 / 2 | 7.5 | No |
| 1784 | 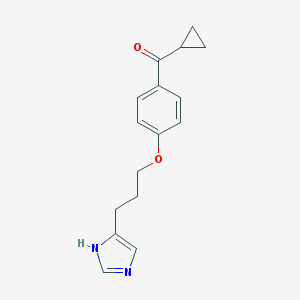 ciproxifan ciproxifan | C16H18N2O2 | 270.332 | 3 / 1 | 2.5 | Yes |
| 441747 | 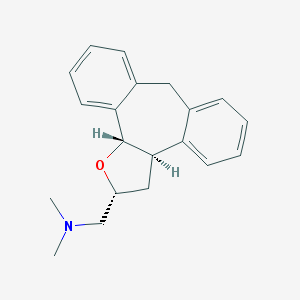 CHEMBL329566 CHEMBL329566 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1926 | 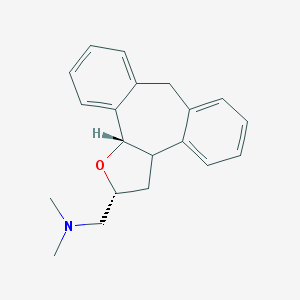 CHEMBL371352 CHEMBL371352 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 2287 | 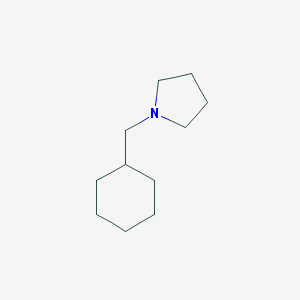 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 3127 | 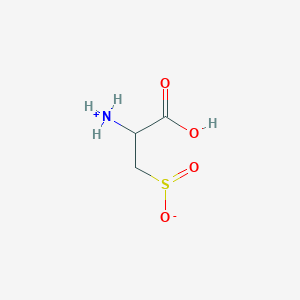 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3769 | 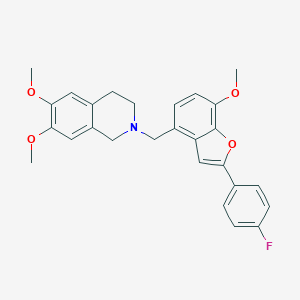 CHEMBL246851 CHEMBL246851 | C27H26FNO4 | 447.506 | 6 / 0 | 5.4 | No |
| 3912 | 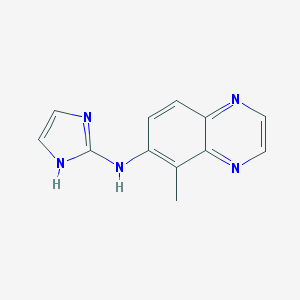 CHEMBL75491 CHEMBL75491 | C12H11N5 | 225.255 | 4 / 2 | 1.6 | Yes |
| 4320 |  sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 4415 |  CHEMBL491419 CHEMBL491419 | C28H33N3O4 | 475.589 | 7 / 0 | 4.0 | Yes |
| 4567 |  CHEMBL373535 CHEMBL373535 | C16H17N3O | 267.332 | 3 / 1 | 1.9 | Yes |
| 4658 |  120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 5056 | 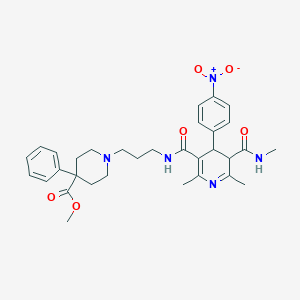 BDBM50068818 BDBM50068818 | C32H39N5O6 | 589.693 | 8 / 2 | 2.9 | No |
| 5124 | 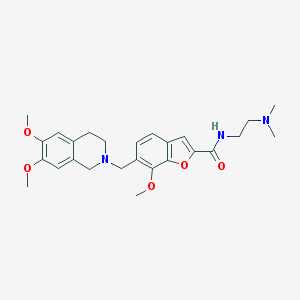 CHEMBL248272 CHEMBL248272 | C26H33N3O5 | 467.566 | 7 / 1 | 3.3 | Yes |
| 5684 | 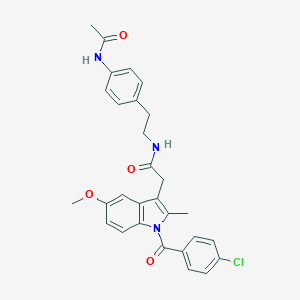 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 557468 | 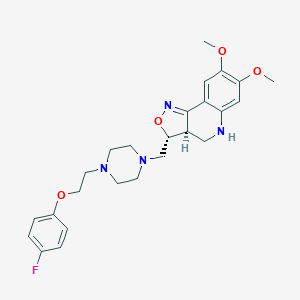 CHEMBL2112969 CHEMBL2112969 | C25H31FN4O4 | 470.545 | 9 / 1 | 3.2 | Yes |
| 463712 |  CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7347 |  CHEMBL179648 CHEMBL179648 | C21H26N2O | 322.452 | 3 / 0 | 3.7 | Yes |
| 7404 |  CHEMBL1256610 CHEMBL1256610 | C15H16N2 | 224.307 | 1 / 1 | 2.9 | Yes |
| 8054 |  CHEMBL163247 CHEMBL163247 | C22H19Cl2N3O2 | 428.313 | 4 / 0 | 4.0 | Yes |
| 521680 |  CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8715 |  CHEMBL308863 CHEMBL308863 | C12H15N3O2 | 233.271 | 4 / 2 | 2.2 | Yes |
| 10525 |  CHEMBL30739 CHEMBL30739 | C13H16N2S | 232.345 | 2 / 1 | 3.2 | Yes |
| 10738 |  CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 11150 | 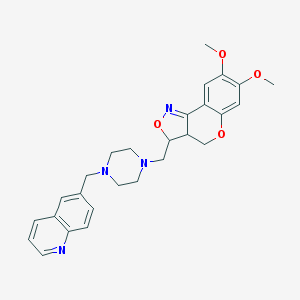 CHEMBL90966 CHEMBL90966 | C27H30N4O4 | 474.561 | 8 / 0 | 3.2 | Yes |
| 11586 | 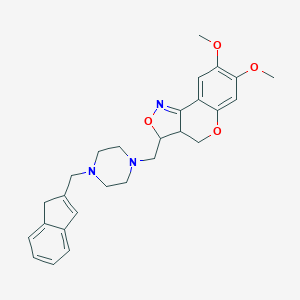 CHEMBL328377 CHEMBL328377 | C27H31N3O4 | 461.562 | 7 / 0 | 3.3 | Yes |
| 521783 | 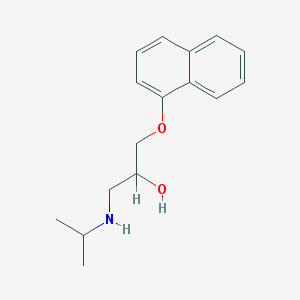 propranolol propranolol | C16H21NO2 | 259.349 | 3 / 2 | 3.0 | Yes |
| 12456 | 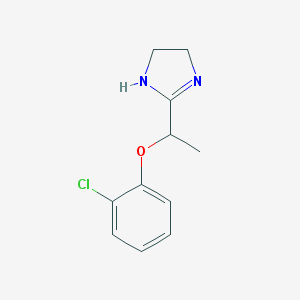 CHEMBL1956191 CHEMBL1956191 | C11H13ClN2O | 224.688 | 2 / 1 | 2.0 | Yes |
| 12491 |  CHEMBL1255723 CHEMBL1255723 | C15H16N2 | 224.307 | 1 / 1 | 3.1 | Yes |
| 13156 |  CHEMBL1256565 CHEMBL1256565 | C15H16N2O | 240.306 | 2 / 1 | 1.9 | Yes |
| 15012 |  EMETINE EMETINE | C29H40N2O4 | 480.649 | 6 / 1 | 4.7 | Yes |
| 464736 |  CHEMBL3588910 CHEMBL3588910 | C15H26N2OS | 282.446 | 3 / 1 | 2.7 | Yes |
| 15487 | 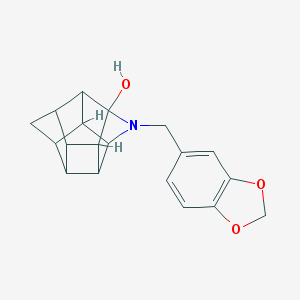 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15682 | 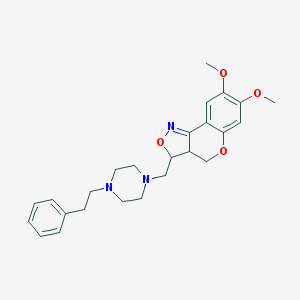 CHEMBL92496 CHEMBL92496 | C25H31N3O4 | 437.54 | 7 / 0 | 3.4 | Yes |
| 16253 | 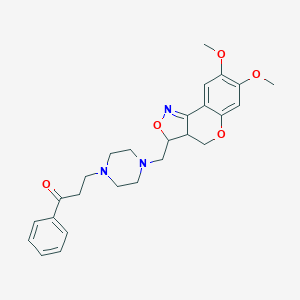 CHEMBL328735 CHEMBL328735 | C26H31N3O5 | 465.55 | 8 / 0 | 2.7 | Yes |
| 16309 | 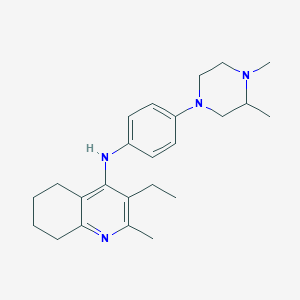 CHEMBL217768 CHEMBL217768 | C24H34N4 | 378.564 | 4 / 1 | 5.2 | No |
| 16786 |  Pipamperone Pipamperone | C21H30FN3O2 | 375.488 | 5 / 1 | 2.0 | Yes |
| 16877 |  Glemanserin Glemanserin | C20H25NO | 295.426 | 2 / 1 | 3.8 | Yes |
| 17579 |  CHEMBL176116 CHEMBL176116 | C27H31N3O4 | 461.562 | 7 / 0 | 4.0 | Yes |
| 17943 |  CHEMBL75257 CHEMBL75257 | C16H23N3O | 273.38 | 3 / 2 | 4.3 | Yes |
| 18689 |  SK&F-106686 SK&F-106686 | C14H16ClNS | 265.799 | 2 / 0 | 4.3 | Yes |
| 20051 |  CHEMBL72768 CHEMBL72768 | C11H14FN3S | 239.312 | 4 / 2 | 1.5 | Yes |
| 21115 |  CHEMBL217180 CHEMBL217180 | C18H24N4 | 296.418 | 4 / 1 | 3.1 | Yes |
| 21392 |  CHEMBL77913 CHEMBL77913 | C11H13N3O | 203.245 | 3 / 2 | 2.3 | Yes |
| 21900 |  CHEMBL150161 CHEMBL150161 | C20H21ClFNO2 | 361.841 | 4 / 1 | 2.9 | Yes |
| 22176 |  suloctidil suloctidil | C20H35NOS | 337.566 | 3 / 2 | 5.8 | No |
| 22388 |  CHEMBL1956193 CHEMBL1956193 | C12H13F3N2O | 258.244 | 5 / 1 | 2.2 | Yes |
| 22607 |  CHEMBL284213 CHEMBL284213 | C11H12N2S | 204.291 | 2 / 1 | 2.3 | Yes |
| 23273 |  CHEMBL576623 CHEMBL576623 | C14H18N2O | 230.311 | 2 / 1 | 2.4 | Yes |
| 465812 | 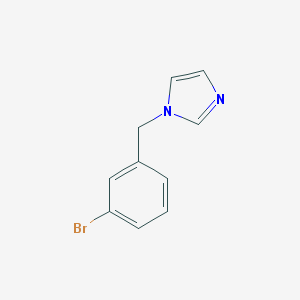 1-(3-Bromobenzyl)-1H-imidazole 1-(3-Bromobenzyl)-1H-imidazole | C10H9BrN2 | 237.1 | 1 / 0 | 2.3 | Yes |
| 26036 | 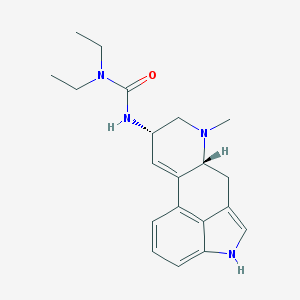 LISURIDE LISURIDE | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26284 | 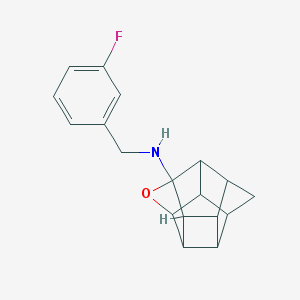 CHEMBL600053 CHEMBL600053 | C18H18FNO | 283.346 | 3 / 1 | 2.5 | Yes |
| 26353 | 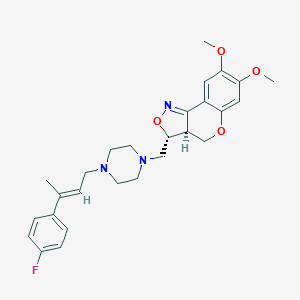 CHEMBL98541 CHEMBL98541 | C27H32FN3O4 | 481.568 | 8 / 0 | 4.3 | Yes |
| 26543 | 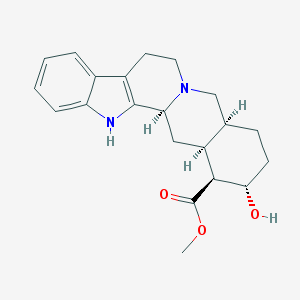 Rauwolscine Rauwolscine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26568 | 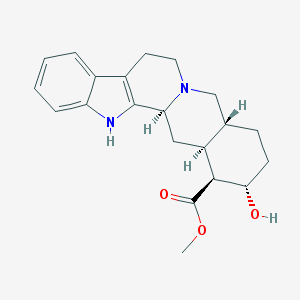 corynanthine corynanthine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26581 | 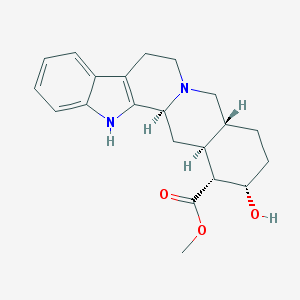 Yohimbine Yohimbine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26632 | 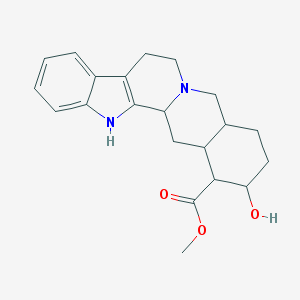 CHEBI:48565 CHEBI:48565 | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26654 | 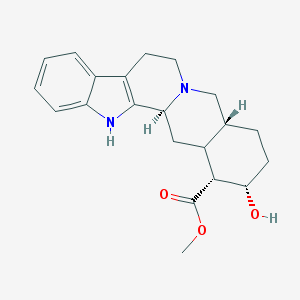 CHEMBL196400 CHEMBL196400 | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26935 | 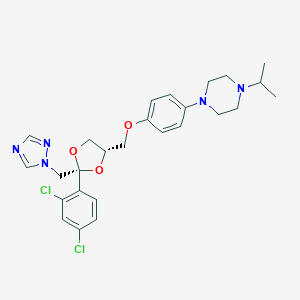 terconazole terconazole | C26H31Cl2N5O3 | 532.466 | 7 / 0 | 4.8 | No |
| 27171 | 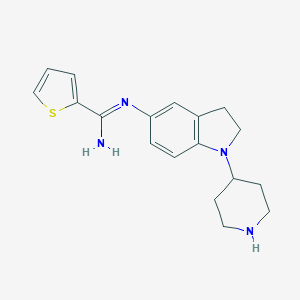 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27499 | 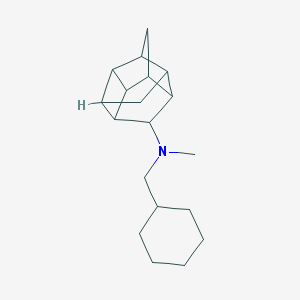 CHEMBL2432063 CHEMBL2432063 | C19H29N | 271.448 | 1 / 0 | 4.7 | Yes |
| 548195 | 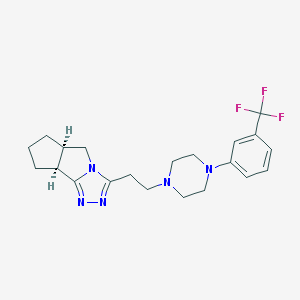 Lorpiprazole Lorpiprazole | C21H26F3N5 | 405.469 | 7 / 0 | 3.3 | Yes |
| 29410 | 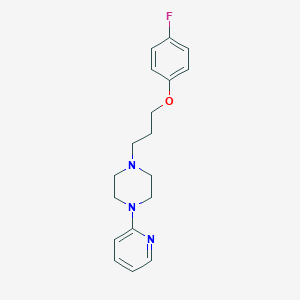 CHEMBL1940420 CHEMBL1940420 | C18H22FN3O | 315.392 | 5 / 0 | 3.7 | Yes |
| 29580 | 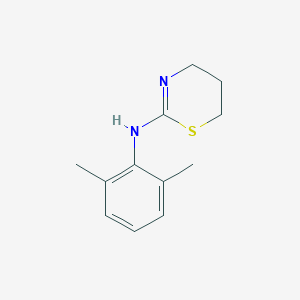 xylazine xylazine | C12H16N2S | 220.334 | 2 / 1 | 2.8 | Yes |
| 29662 | 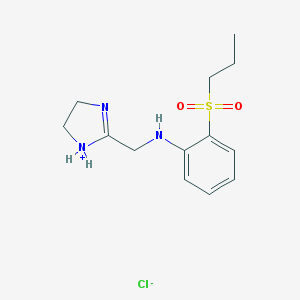 CID 10903225 CID 10903225 | C13H20ClN3O2S | 317.832 | 5 / 2 | N/A | N/A |
| 30195 |  SCHEMBL14542291 SCHEMBL14542291 | C22H21NO3 | 347.414 | 3 / 0 | 4.3 | Yes |
| 31085 |  CHEMBL165350 CHEMBL165350 | C22H20BrN3O2 | 438.325 | 4 / 0 | 3.2 | Yes |
| 31598 |  CHEMBL214996 CHEMBL214996 | C23H28N4O | 376.504 | 5 / 2 | 4.2 | Yes |
| 31838 |  CHEMBL328195 CHEMBL328195 | C25H31N3O4S | 469.6 | 8 / 0 | 3.9 | Yes |
| 31851 | 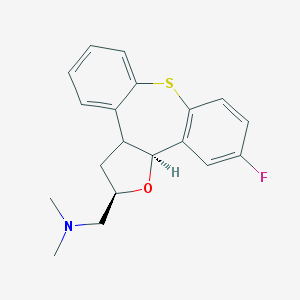 CHEMBL193639 CHEMBL193639 | C19H20FNOS | 329.433 | 4 / 0 | 3.8 | Yes |
| 31861 | 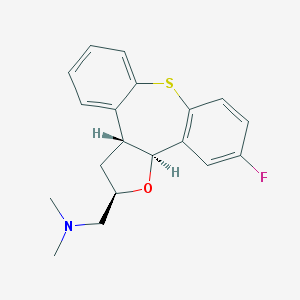 CHEMBL366164 CHEMBL366164 | C19H20FNOS | 329.433 | 4 / 0 | 3.8 | Yes |
| 31868 | 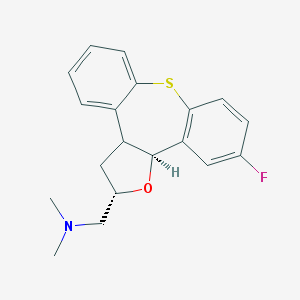 CHEMBL363581 CHEMBL363581 | C19H20FNOS | 329.433 | 4 / 0 | 3.8 | Yes |
| 32113 | 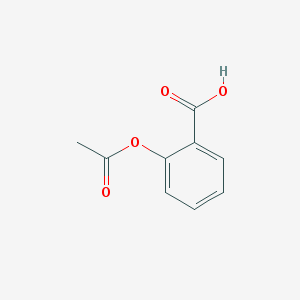 aspirin aspirin | C9H8O4 | 180.159 | 4 / 1 | 1.2 | Yes |
| 32540 | 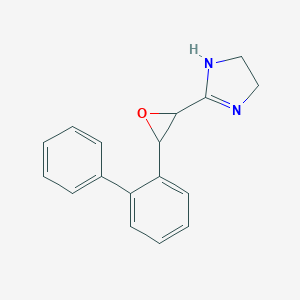 CHEMBL222933 CHEMBL222933 | C17H16N2O | 264.328 | 2 / 1 | 2.1 | Yes |
| 32632 | 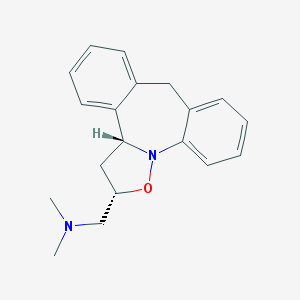 CHEMBL328573 CHEMBL328573 | C19H22N2O | 294.398 | 3 / 0 | 3.9 | Yes |
| 32653 | 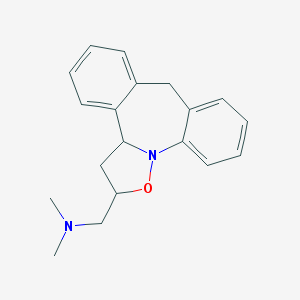 CHEMBL83658 CHEMBL83658 | C19H22N2O | 294.398 | 3 / 0 | 3.9 | Yes |
| 33052 | 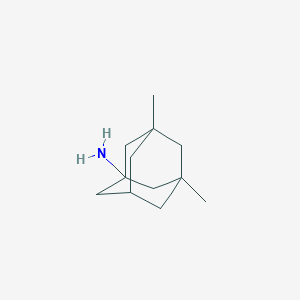 memantine memantine | C12H21N | 179.307 | 1 / 1 | 3.3 | Yes |
| 467026 |  CHEMBL3590089 CHEMBL3590089 | C27H34N4O3 | 462.594 | 5 / 1 | 3.7 | Yes |
| 33396 |  CHEMBL1956200 CHEMBL1956200 | C12H16N2O3S | 268.331 | 4 / 1 | 0.6 | Yes |
| 34136 |  CHEMBL492639 CHEMBL492639 | C14H19N3O2 | 261.325 | 4 / 1 | 1.7 | Yes |
| 35015 |  CHEMBL123769 CHEMBL123769 | C36H37N3O6 | 607.707 | 8 / 0 | 7.1 | No |
| 35244 | 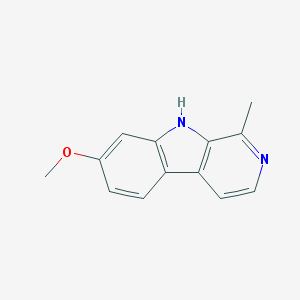 HARMINE HARMINE | C13H12N2O | 212.252 | 2 / 1 | 3.6 | Yes |
| 35773 | 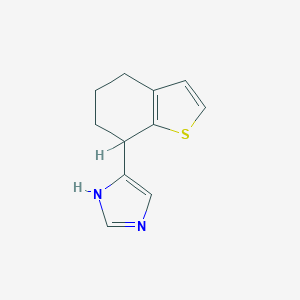 CHEMBL286269 CHEMBL286269 | C11H12N2S | 204.291 | 2 / 1 | 2.5 | Yes |
| 36995 | 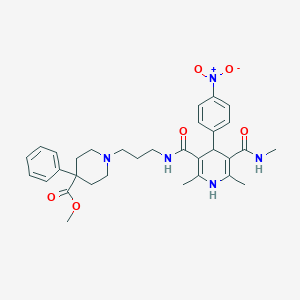 SNAP-5540 SNAP-5540 | C32H39N5O6 | 589.693 | 8 / 3 | 3.5 | No |
| 522672 | 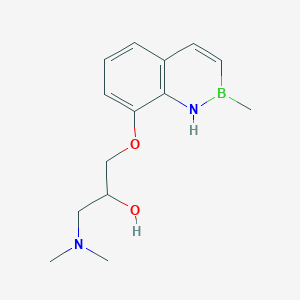 CHEMBL3736433 CHEMBL3736433 | C14H21BN2O2 | 260.144 | 4 / 2 | N/A | N/A |
| 39242 | 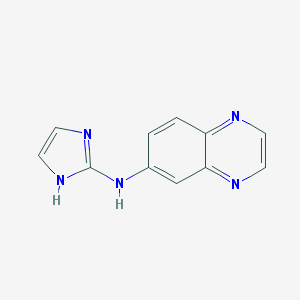 CHEMBL74449 CHEMBL74449 | C11H9N5 | 211.228 | 4 / 2 | 1.2 | Yes |
| 39462 | 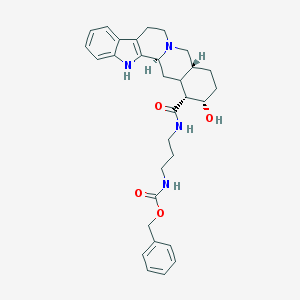 CHEMBL369938 CHEMBL369938 | C31H38N4O4 | 530.669 | 5 / 4 | 3.9 | No |
| 40068 | 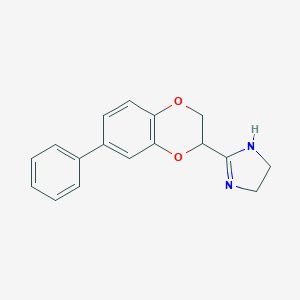 CHEMBL492445 CHEMBL492445 | C17H16N2O2 | 280.327 | 3 / 1 | 2.3 | Yes |
| 40250 | 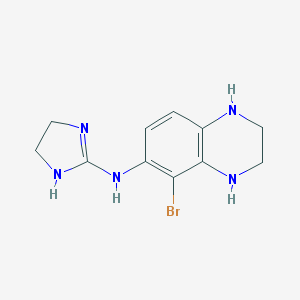 CHEMBL74283 CHEMBL74283 | C11H14BrN5 | 296.172 | 3 / 4 | 1.0 | Yes |
| 40289 | 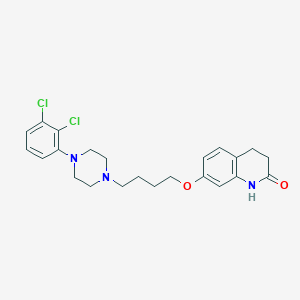 Aripiprazole Aripiprazole | C23H27Cl2N3O2 | 448.388 | 4 / 1 | 4.6 | Yes |
| 40619 | 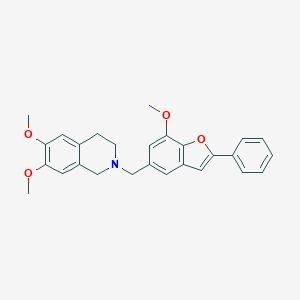 CHEMBL248064 CHEMBL248064 | C27H27NO4 | 429.516 | 5 / 0 | 5.3 | No |
| 40991 | 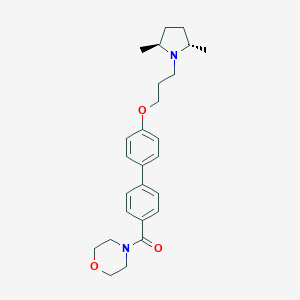 A-349821 A-349821 | C26H34N2O3 | 422.569 | 4 / 0 | 4.4 | Yes |
| 42048 | 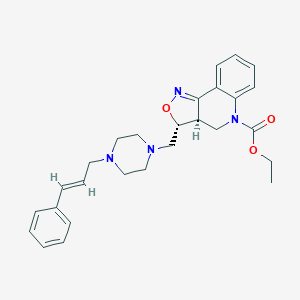 CHEMBL2112975 CHEMBL2112975 | C27H32N4O3 | 460.578 | 6 / 0 | 3.8 | Yes |
| 468117 | 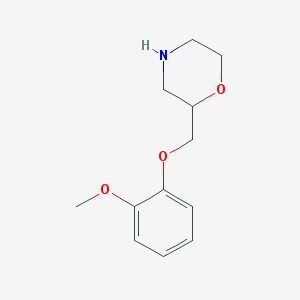 CHEMBL3588908 CHEMBL3588908 | C12H17NO3 | 223.272 | 4 / 1 | 1.1 | Yes |
| 42464 | 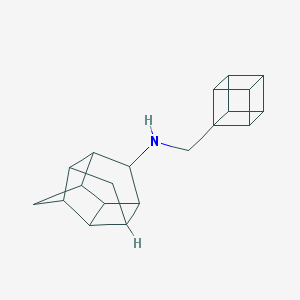 CHEMBL2432039 CHEMBL2432039 | C20H23N | 277.411 | 1 / 1 | 2.7 | Yes |
| 43324 | 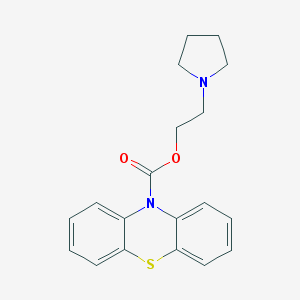 CHEMBL481153 CHEMBL481153 | C19H20N2O2S | 340.441 | 4 / 0 | 3.9 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417