

You can:
| Name | Platelet-activating factor receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Ptafr |
| Synonym | AGEPC receptor PAF receptor PAF-R PAFr |
| Disease | N/A for non-human GPCRs |
| Length | 341 |
| Amino acid sequence | MEQNGSFRVDSEFRYTLFPIVYSVIFVLGVVANGYVLWVFATLYPSKKLNEIKIFMVNLTVADLLFLMTLPLWIVYYSNEGDWIVHKFLCNLAGCLFFINTYCSVAFLGVITYNRYQAVAYPIKTAQATTRKRGITLSLVIWISIAATASYFLATDSTNVVPKKDGSGNITRCFEHYEPYSVPILVVHIFITSCFFLVFFLIFYCNMVIIHTLLTRPVRQQRKPEVKRRALWMVCTVLAVFVICFVPHHVVQLPWTLAELGYQTNFHQAINDAHQITLCLLSTNCVLDPVIYCFLTKKFRKHLSEKFYSMRSSRKCSRATSDTCTEVMMPANQTPVLPLKN |
| UniProt | P46002 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4127 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 4049 |  CHEMBL51798 CHEMBL51798 | C31H39NO8S2 | 617.772 | 10 / 1 | 5.1 | No |
| 5414 |  CHEMBL418989 CHEMBL418989 | C28H31N5O3S | 517.648 | 7 / 0 | 2.9 | No |
| 6168 |  CHEMBL134025 CHEMBL134025 | C16H22N4O | 286.379 | 4 / 0 | 1.3 | Yes |
| 6933 |  CHEMBL127766 CHEMBL127766 | C36H61NO6S2 | 668.005 | 7 / 0 | N/A | No |
| 7575 |  CHEMBL339676 CHEMBL339676 | C36H59NO6S2 | 665.989 | 7 / 0 | N/A | No |
| 10832 |  CHEMBL3216875 CHEMBL3216875 | C27H29Cl2N3O2 | 498.448 | 5 / 2 | N/A | N/A |
| 11375 |  CV-6209 CV-6209 | C34H60ClN3O6 | 642.319 | 7 / 1 | N/A | No |
| 12794 |  CHEMBL174262 CHEMBL174262 | C25H20N4O3 | 424.46 | 5 / 1 | 3.7 | Yes |
| 14654 |  CHEMBL125576 CHEMBL125576 | C33H56ClN3O6 | 626.276 | 8 / 1 | N/A | No |
| 14863 |  CHEMBL318908 CHEMBL318908 | C27H27N5O2 | 453.546 | 6 / 0 | 4.0 | Yes |
| 18140 |  CHEMBL103571 CHEMBL103571 | C28H29N5O2 | 467.573 | 5 / 0 | 3.1 | Yes |
| 20160 |  CHEMBL443811 CHEMBL443811 | C27H44N4O | 440.676 | 4 / 0 | 7.3 | No |
| 21992 |  CHEMBL367418 CHEMBL367418 | C30H30FN5O2 | 511.601 | 5 / 0 | 4.8 | No |
| 22019 |  CHEMBL132028 CHEMBL132028 | C26H34N2O8 | 502.564 | 8 / 0 | 2.4 | No |
| 25839 |  CHEMBL3215984 CHEMBL3215984 | C18H20Cl2N4O | 379.285 | 4 / 2 | N/A | N/A |
| 26734 |  CHEMBL103903 CHEMBL103903 | C27H26N4O | 422.532 | 4 / 0 | 4.3 | Yes |
| 27160 |  CHEMBL545072 CHEMBL545072 | C27H30ClN5O | 476.021 | 5 / 1 | N/A | N/A |
| 27881 |  CHEMBL338689 CHEMBL338689 | C32H55IN2O5 | 674.705 | 6 / 0 | N/A | No |
| 31796 |  CHEMBL103561 CHEMBL103561 | C28H31N5O | 453.59 | 5 / 1 | 3.3 | Yes |
| 33163 |  CHEMBL127531 CHEMBL127531 | C24H50NO5P | 463.64 | 5 / 0 | 6.7 | No |
| 33662 |  CHEMBL102070 CHEMBL102070 | C27H28N6O2 | 468.561 | 6 / 1 | 1.7 | Yes |
| 33881 |  CHEMBL173497 CHEMBL173497 | C28H24FN5O2 | 481.531 | 5 / 1 | 3.5 | Yes |
| 34323 |  CHEMBL367673 CHEMBL367673 | C25H27N5O4 | 461.522 | 6 / 0 | 2.6 | Yes |
| 34965 |  CHEMBL102918 CHEMBL102918 | C28H29N5O2 | 467.573 | 5 / 1 | 2.8 | Yes |
| 35541 |  CHEMBL293773 CHEMBL293773 | C31H38O10S2 | 634.755 | 10 / 0 | 4.5 | No |
| 36382 |  CHEMBL1203313 CHEMBL1203313 | C22H28ClN5O | 413.95 | 5 / 2 | N/A | N/A |
| 36645 |  CHEMBL125078 CHEMBL125078 | C36H61NO5S2 | 652.006 | 6 / 0 | N/A | No |
| 37975 |  CHEMBL542731 CHEMBL542731 | C26H29ClN4 | 432.996 | 4 / 1 | N/A | N/A |
| 38160 | 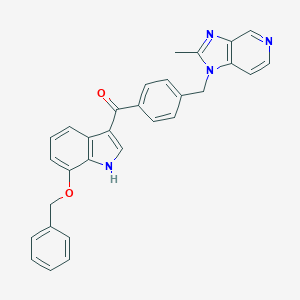 CHEMBL368292 CHEMBL368292 | C30H24N4O2 | 472.548 | 4 / 1 | 5.3 | No |
| 40244 | 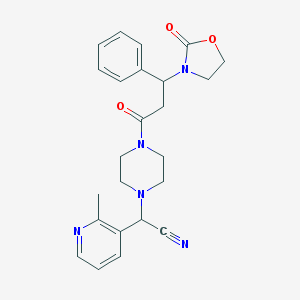 CHEMBL318163 CHEMBL318163 | C24H27N5O3 | 433.512 | 6 / 0 | 1.5 | Yes |
| 41305 | 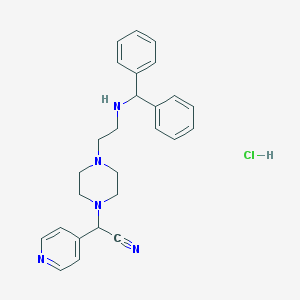 CHEMBL555567 CHEMBL555567 | C26H30ClN5 | 448.011 | 5 / 2 | N/A | N/A |
| 41647 | 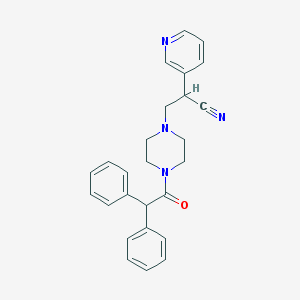 CHEMBL132824 CHEMBL132824 | C26H26N4O | 410.521 | 4 / 0 | 3.1 | Yes |
| 43852 | 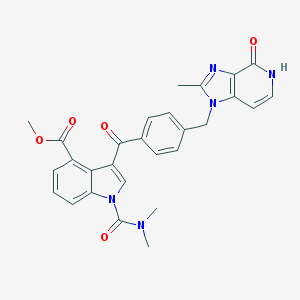 CHEMBL170398 CHEMBL170398 | C28H25N5O5 | 511.538 | 6 / 1 | 3.0 | No |
| 45343 | 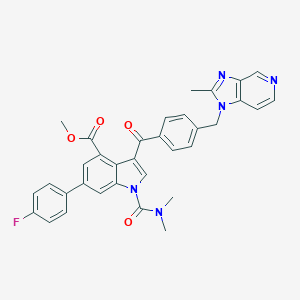 CHEMBL174592 CHEMBL174592 | C34H28FN5O4 | 589.627 | 7 / 0 | 5.3 | No |
| 45692 | 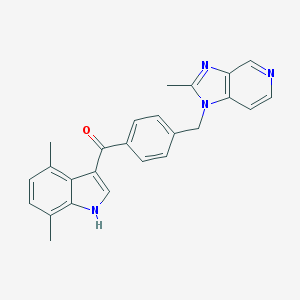 CHEMBL176797 CHEMBL176797 | C25H22N4O | 394.478 | 3 / 1 | 4.6 | Yes |
| 48800 | 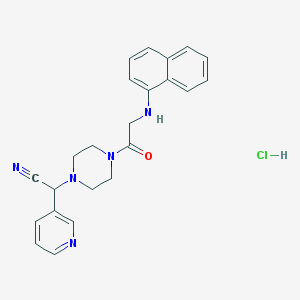 CHEMBL540829 CHEMBL540829 | C23H24ClN5O | 421.929 | 5 / 2 | N/A | N/A |
| 50764 |  CHEMBL52369 CHEMBL52369 | C31H38O8S2 | 602.757 | 9 / 0 | 5.8 | No |
| 52004 |  CHEMBL3216197 CHEMBL3216197 | C26H28Cl2N4O | 483.437 | 4 / 2 | N/A | N/A |
| 53027 |  CHEMBL427307 CHEMBL427307 | C34H29N5O4 | 571.637 | 6 / 0 | 5.0 | No |
| 54180 |  CHEMBL3215754 CHEMBL3215754 | C22H22Cl2N4O | 429.345 | 4 / 2 | N/A | N/A |
| 55773 | 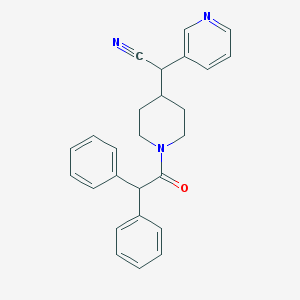 CHEMBL340544 CHEMBL340544 | C26H25N3O | 395.506 | 3 / 0 | 4.1 | Yes |
| 57206 | 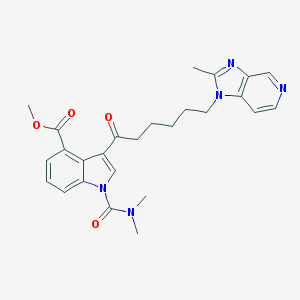 CHEMBL173304 CHEMBL173304 | C26H29N5O4 | 475.549 | 6 / 0 | 2.9 | Yes |
| 59840 | 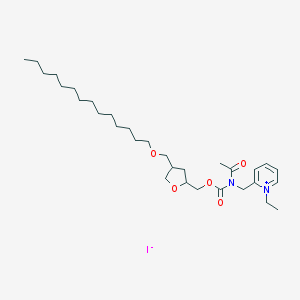 CHEMBL127010 CHEMBL127010 | C31H53IN2O5 | 660.678 | 6 / 0 | N/A | No |
| 61412 | 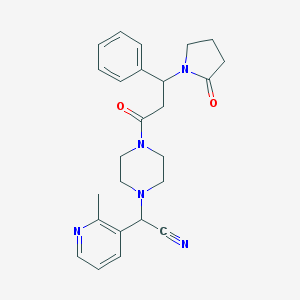 CHEMBL100190 CHEMBL100190 | C25H29N5O2 | 431.54 | 5 / 0 | 1.4 | Yes |
| 61851 | 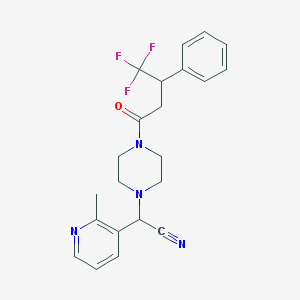 CHEMBL103636 CHEMBL103636 | C22H23F3N4O | 416.448 | 7 / 0 | 3.1 | Yes |
| 63931 | 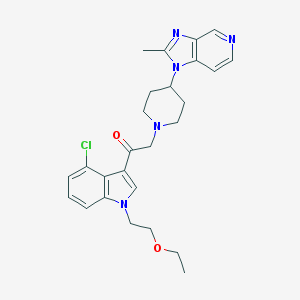 CHEMBL173531 CHEMBL173531 | C26H30ClN5O2 | 480.009 | 5 / 0 | 3.5 | Yes |
| 64515 | 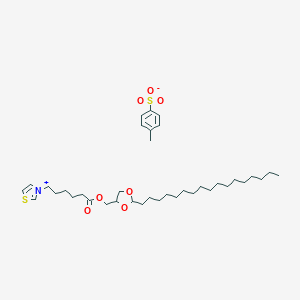 CHEMBL125419 CHEMBL125419 | C37H61NO7S2 | 696.015 | 8 / 0 | N/A | No |
| 66677 | 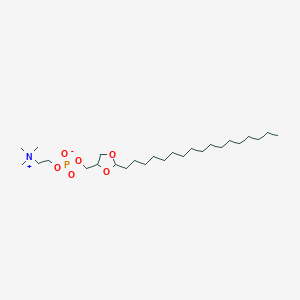 CHEMBL127183 CHEMBL127183 | C26H54NO6P | 507.693 | 6 / 0 | 7.5 | No |
| 66833 |  CHEMBL322184 CHEMBL322184 | C26H27N5O | 425.536 | 5 / 1 | 2.7 | Yes |
| 71191 |  CHEMBL335853 CHEMBL335853 | C25H24N4S | 412.555 | 4 / 0 | 3.8 | Yes |
| 71524 |  CHEMBL176678 CHEMBL176678 | C30H31FN6O2 | 526.616 | 6 / 0 | 4.2 | No |
| 71705 |  CHEMBL131597 CHEMBL131597 | C25H25N5O | 411.509 | 4 / 1 | 3.0 | Yes |
| 72244 |  CHEMBL423711 CHEMBL423711 | C28H26N4O | 434.543 | 4 / 0 | 4.1 | Yes |
| 73129 |  CHEMBL300910 CHEMBL300910 | C32H37NO9S2 | 643.766 | 11 / 0 | 4.1 | No |
| 74681 |  CHEMBL3216401 CHEMBL3216401 | C27H30Cl2N4O | 497.464 | 4 / 2 | N/A | N/A |
| 75904 |  CHEMBL368657 CHEMBL368657 | C30H29N5O4 | 523.593 | 6 / 0 | 4.3 | No |
| 76382 | 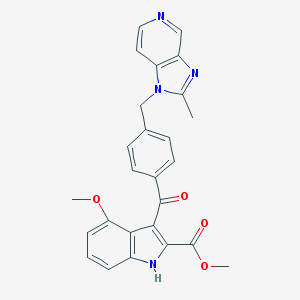 CHEMBL424947 CHEMBL424947 | C26H22N4O4 | 454.486 | 6 / 1 | 4.0 | Yes |
| 78314 | 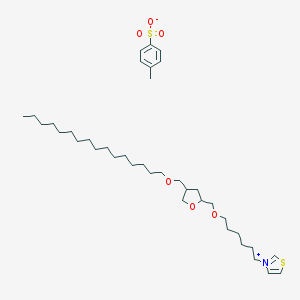 CHEMBL338977 CHEMBL338977 | C38H65NO6S2 | 696.059 | 7 / 0 | N/A | No |
| 79433 | 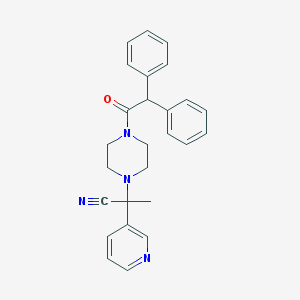 CHEMBL133921 CHEMBL133921 | C26H26N4O | 410.521 | 4 / 0 | 3.4 | Yes |
| 82261 | 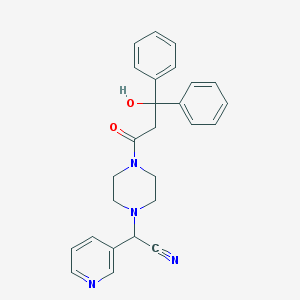 CHEMBL103753 CHEMBL103753 | C26H26N4O2 | 426.52 | 5 / 1 | 2.2 | Yes |
| 82464 |  CHEMBL124394 CHEMBL124394 | C33H56IN3O6 | 717.73 | 8 / 1 | N/A | No |
| 84290 |  CHEMBL427121 CHEMBL427121 | C26H23N5O2 | 437.503 | 4 / 0 | 3.7 | Yes |
| 84398 |  CHEMBL125315 CHEMBL125315 | C38H65NO6S2 | 696.059 | 7 / 0 | N/A | No |
| 84965 |  CHEMBL338940 CHEMBL338940 | C26H26N4O2 | 426.52 | 5 / 1 | 2.2 | Yes |
| 85406 |  CHEMBL124985 CHEMBL124985 | C35H61IN2O5 | 716.786 | 6 / 0 | N/A | No |
| 87137 |  CHEMBL127464 CHEMBL127464 | C33H53NO7S2 | 639.907 | 8 / 0 | N/A | No |
| 88467 |  CHEMBL52419 CHEMBL52419 | C33H43NO8S2 | 645.826 | 10 / 0 | 5.9 | No |
| 90195 |  CHEMBL103287 CHEMBL103287 | C25H31N5O | 417.557 | 5 / 0 | 2.3 | Yes |
| 90689 |  CHEMBL132655 CHEMBL132655 | C27H26ClN3O | 443.975 | 3 / 0 | 5.0 | Yes |
| 92512 |  CHEMBL338345 CHEMBL338345 | C35H60IN3O6 | 745.784 | 8 / 1 | N/A | No |
| 92638 |  CHEMBL172949 CHEMBL172949 | C30H25N5O3 | 503.562 | 5 / 0 | 4.4 | No |
| 93748 |  ABT-491 free base ABT-491 free base | C28H22FN5O2 | 479.515 | 5 / 0 | 4.0 | Yes |
| 97731 | 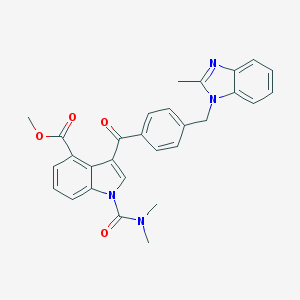 CHEMBL174198 CHEMBL174198 | C29H26N4O4 | 494.551 | 5 / 0 | 4.6 | Yes |
| 97816 | 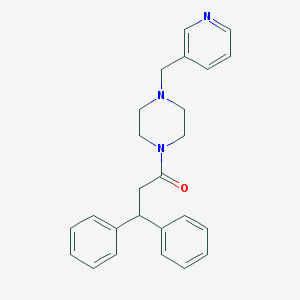 CHEMBL131690 CHEMBL131690 | C25H27N3O | 385.511 | 3 / 0 | 3.3 | Yes |
| 97964 | 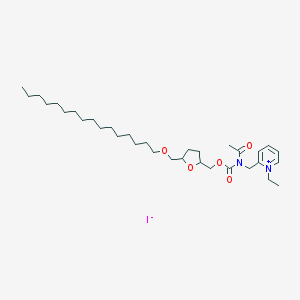 CHEMBL125080 CHEMBL125080 | C33H57IN2O5 | 688.732 | 6 / 0 | N/A | No |
| 99293 | 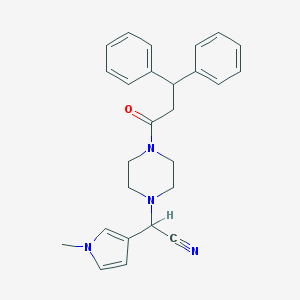 CHEMBL134877 CHEMBL134877 | C26H28N4O | 412.537 | 3 / 0 | 3.1 | Yes |
| 99564 | 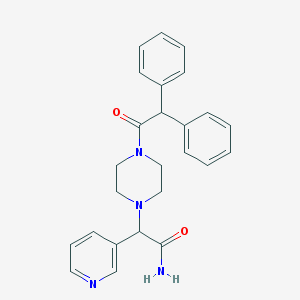 CHEMBL423717 CHEMBL423717 | C25H26N4O2 | 414.509 | 4 / 1 | 2.4 | Yes |
| 100180 | 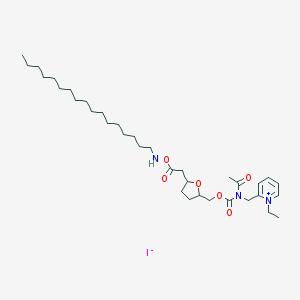 CHEMBL124906 CHEMBL124906 | C35H60IN3O6 | 745.784 | 8 / 1 | N/A | No |
| 100776 | 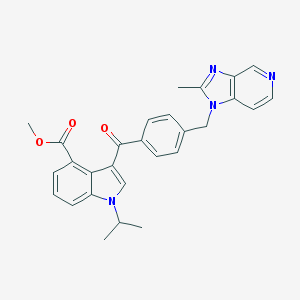 CHEMBL172819 CHEMBL172819 | C28H26N4O3 | 466.541 | 5 / 0 | 4.3 | Yes |
| 100920 | 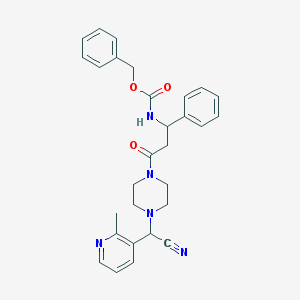 CHEMBL431049 CHEMBL431049 | C29H31N5O3 | 497.599 | 6 / 1 | 3.0 | Yes |
| 103624 | 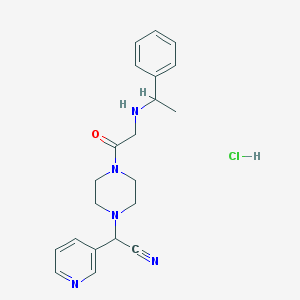 CHEMBL554463 CHEMBL554463 | C21H26ClN5O | 399.923 | 5 / 2 | N/A | N/A |
| 103827 | 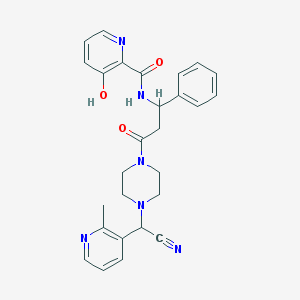 CHEMBL101342 CHEMBL101342 | C27H28N6O3 | 484.56 | 7 / 2 | 2.2 | Yes |
| 104291 | 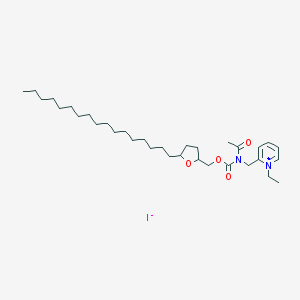 CHEMBL338519 CHEMBL338519 | C33H57IN2O4 | 672.733 | 5 / 0 | N/A | No |
| 105011 | 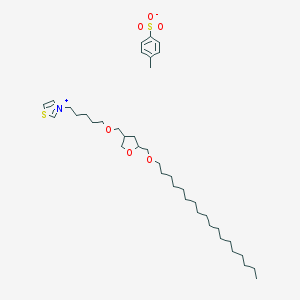 CHEMBL127009 CHEMBL127009 | C39H67NO6S2 | 710.086 | 7 / 0 | N/A | No |
| 105030 | 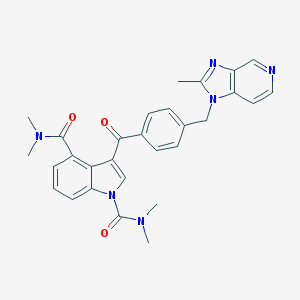 CHEMBL367530 CHEMBL367530 | C29H28N6O3 | 508.582 | 5 / 0 | 3.1 | No |
| 106475 | 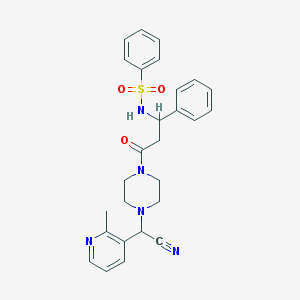 CHEMBL103231 CHEMBL103231 | C27H29N5O3S | 503.621 | 7 / 1 | 2.5 | No |
| 106484 | 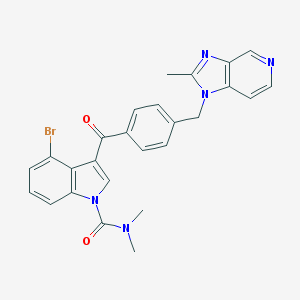 CHEMBL170370 CHEMBL170370 | C26H22BrN5O2 | 516.399 | 4 / 0 | 4.4 | No |
| 106593 | 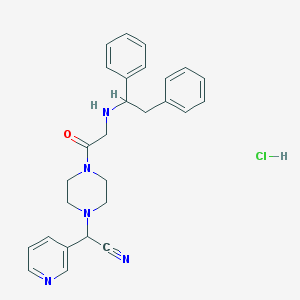 CHEMBL545773 CHEMBL545773 | C27H30ClN5O | 476.021 | 5 / 2 | N/A | N/A |
| 107199 | 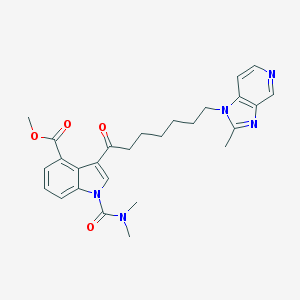 CHEMBL367425 CHEMBL367425 | C27H31N5O4 | 489.576 | 6 / 0 | 3.5 | Yes |
| 108487 | 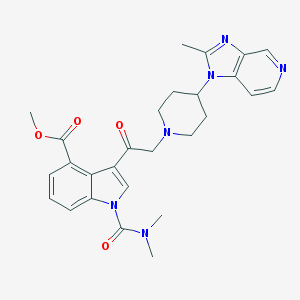 CHEMBL171294 CHEMBL171294 | C27H30N6O4 | 502.575 | 7 / 0 | 2.5 | No |
| 108490 | 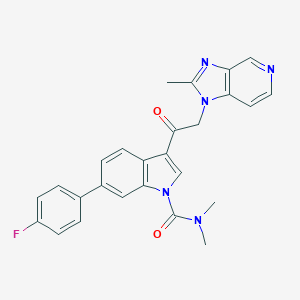 CHEMBL173104 CHEMBL173104 | C26H22FN5O2 | 455.493 | 5 / 0 | 4.0 | Yes |
| 110453 | 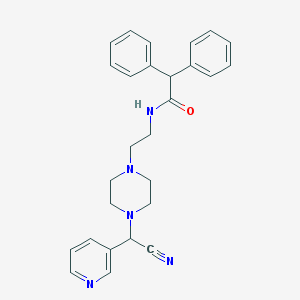 CHEMBL336816 CHEMBL336816 | C27H29N5O | 439.563 | 5 / 1 | 3.1 | Yes |
| 110468 | 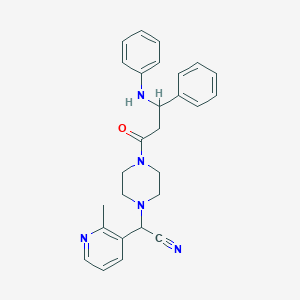 CHEMBL100547 CHEMBL100547 | C27H29N5O | 439.563 | 5 / 1 | 4.0 | Yes |
| 111413 | 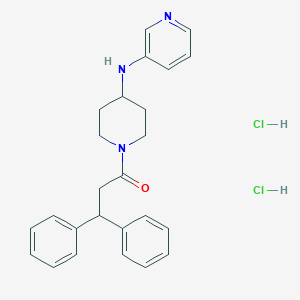 CHEMBL3216628 CHEMBL3216628 | C25H29Cl2N3O | 458.427 | 3 / 3 | N/A | N/A |
| 112370 | 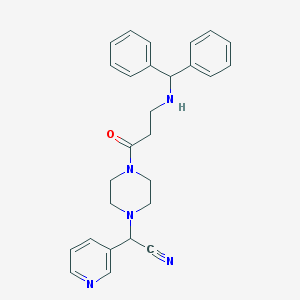 CHEMBL103801 CHEMBL103801 | C27H29N5O | 439.563 | 5 / 1 | 2.6 | Yes |
| 112741 | 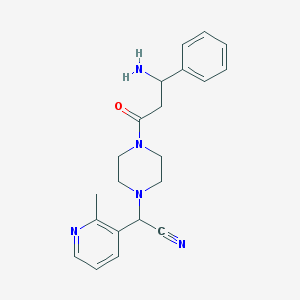 CHEMBL100853 CHEMBL100853 | C21H25N5O | 363.465 | 5 / 1 | 0.9 | Yes |
| 119323 | 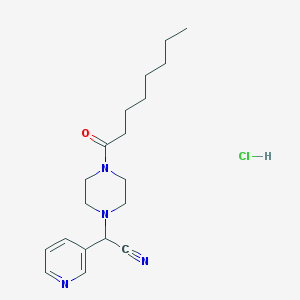 CHEMBL552771 CHEMBL552771 | C19H29ClN4O | 364.918 | 4 / 1 | N/A | N/A |
| 121251 | 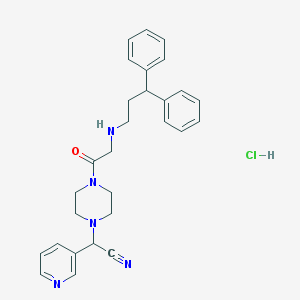 CHEMBL552805 CHEMBL552805 | C28H32ClN5O | 490.048 | 5 / 2 | N/A | N/A |
| 121988 | 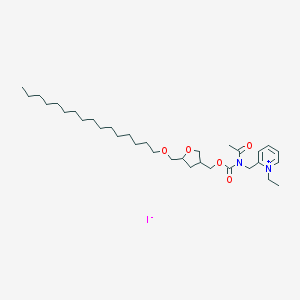 CHEMBL126016 CHEMBL126016 | C33H57IN2O5 | 688.732 | 6 / 0 | N/A | No |
| 122195 | 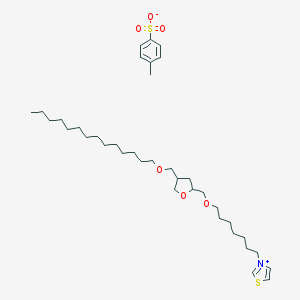 CHEMBL124581 CHEMBL124581 | C37H63NO6S2 | 682.032 | 7 / 0 | N/A | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417