

You can:
| Name | Alpha-2A adrenergic receptor |
|---|---|
| Species | Bos taurus (Bovine) |
| Gene | ADRA2A |
| Synonym | Alpha-2A adrenoceptor Alpha-2A adrenoreceptor Alpha-2AAR Alpha-2D adrenergic receptor |
| Disease | N/A for non-human GPCRs |
| Length | 452 |
| Amino acid sequence | MGSLQPDAGNASWNGTEAPGGGARATPYSLQVTLTLVCLAGLLMLFTVFGNVLVIIAVFTSRALKAPQNLFLVSLASADILVATLVIPFSLANEVMGYWYFGKAWCEIYLALDVLFCTSSIVHLCAISLDRYWSITQAIEYNLKRTPRRIKAIIVTVWVISAVISFPPLISFEKKRGRSGQPSAEPRCEINDQKWYVISSSIGSFFAPCLIMILVYVRIYQIAKRRTRVPPSRRGPDATAAELPGSAERRPNGLGPERGGVGPVGAEVESLQVQLNGAPGEPAPAGAGADALDLEESSSSEHAERPPGSRRSERGPRAKGKARASQVKPGDSLPRRGPGATGLGAPTAGPAEERSGGGAKASRWRGRQNREKRFTFVLAVVIGVFVVCWFPFFFTYTLTAIGCPVPPTLFKFFFWFGYCNSSLNPVIYTIFNHDFRRAFKKILCRGDRKRIV |
| UniProt | Q28838 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4744 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 4220 | 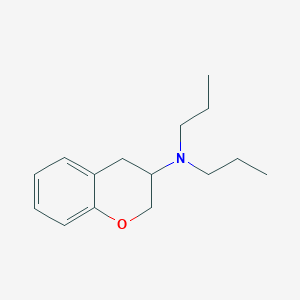 CHEMBL25984 CHEMBL25984 | C15H23NO | 233.355 | 2 / 0 | 3.8 | Yes |
| 4974 | 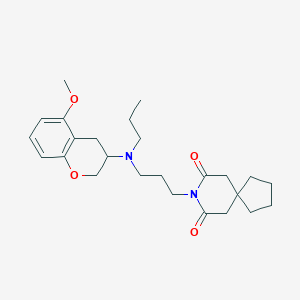 CHEMBL50188 CHEMBL50188 | C25H36N2O4 | 428.573 | 5 / 0 | 4.2 | Yes |
| 5544 | 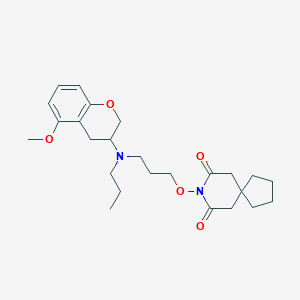 CHEMBL50722 CHEMBL50722 | C25H36N2O5 | 444.572 | 6 / 0 | 4.3 | Yes |
| 5917 | 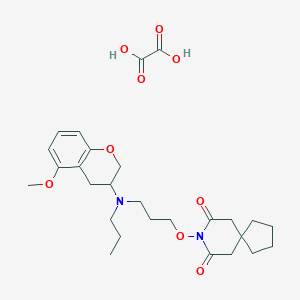 CHEMBL50722 CHEMBL50722 | C27H38N2O9 | 534.606 | 10 / 2 | N/A | No |
| 6362 | 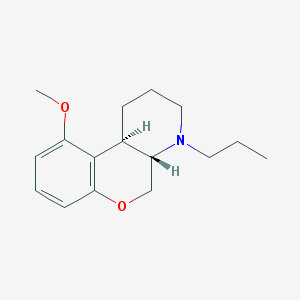 CHEMBL163281 CHEMBL163281 | C16H23NO2 | 261.365 | 3 / 0 | 3.2 | Yes |
| 11808 | 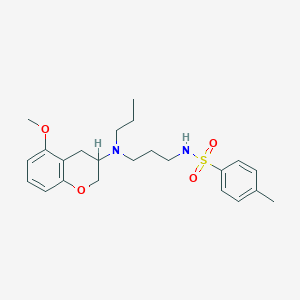 CHEMBL54266 CHEMBL54266 | C23H32N2O4S | 432.579 | 6 / 1 | 4.3 | Yes |
| 13685 | 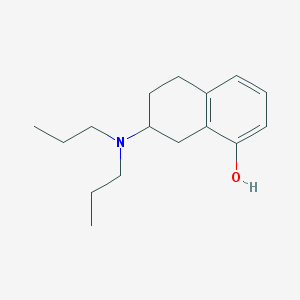 8-OH-Dpat 8-OH-Dpat | C16H25NO | 247.382 | 2 / 1 | 4.1 | Yes |
| 20072 | 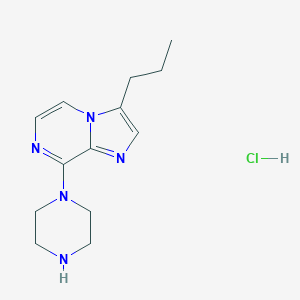 CHEMBL545293 CHEMBL545293 | C13H20ClN5 | 281.788 | 4 / 2 | N/A | N/A |
| 22833 | 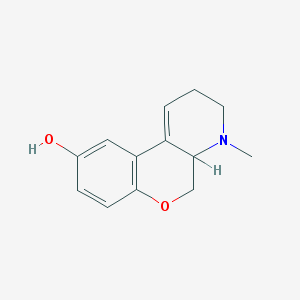 CHEMBL163167 CHEMBL163167 | C13H15NO2 | 217.268 | 3 / 1 | 1.6 | Yes |
| 25279 | 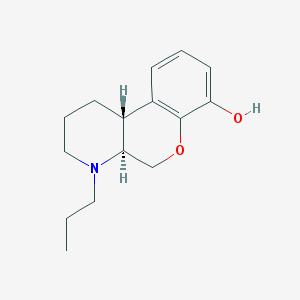 CHEMBL162170 CHEMBL162170 | C15H21NO2 | 247.338 | 3 / 1 | 2.9 | Yes |
| 26549 | 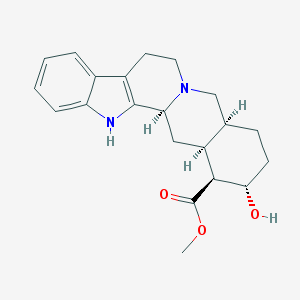 Rauwolscine Rauwolscine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26619 | 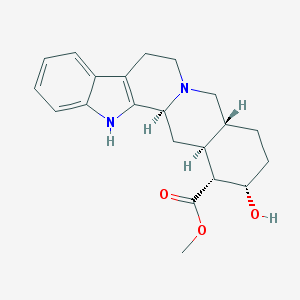 Yohimbine Yohimbine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 28845 | 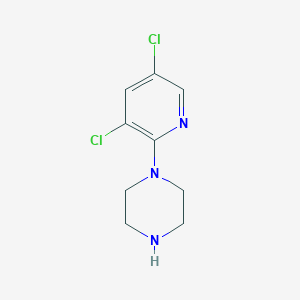 1-(3,5-dichloropyridin-2-yl)piperazine 1-(3,5-dichloropyridin-2-yl)piperazine | C9H11Cl2N3 | 232.108 | 3 / 1 | 1.9 | Yes |
| 32934 | 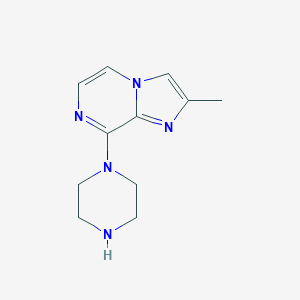 CHEMBL544354 CHEMBL544354 | C11H15N5 | 217.276 | 4 / 1 | 0.9 | Yes |
| 33286 | 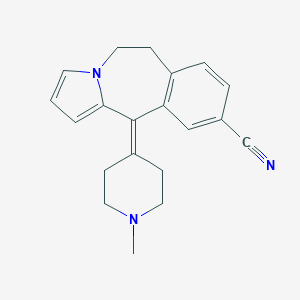 CHEMBL176365 CHEMBL176365 | C20H21N3 | 303.409 | 2 / 0 | 2.2 | Yes |
| 39332 | 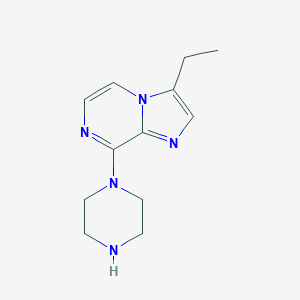 CHEMBL538537 CHEMBL538537 | C12H17N5 | 231.303 | 4 / 1 | 1.3 | Yes |
| 39945 |  CHEMBL124926 CHEMBL124926 | C10H11Br2N5 | 361.041 | 4 / 1 | 2.5 | Yes |
| 40769 |  3-chloro-8-(1-piperazinyl)imidazo[1,2-a]pyrazine 3-chloro-8-(1-piperazinyl)imidazo[1,2-a]pyrazine | C10H12ClN5 | 237.691 | 4 / 1 | 1.5 | Yes |
| 42778 |  BDBM50002148 BDBM50002148 | C10H15N5 | 205.265 | 3 / 1 | -1.5 | Yes |
| 47977 |  CHEMBL162282 CHEMBL162282 | C15H21NO2 | 247.338 | 3 / 1 | 2.9 | Yes |
| 51662 | 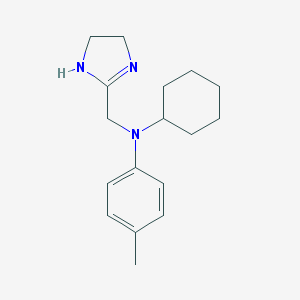 CHEMBL541208 CHEMBL541208 | C17H25N3 | 271.408 | 2 / 1 | 3.2 | Yes |
| 52778 | 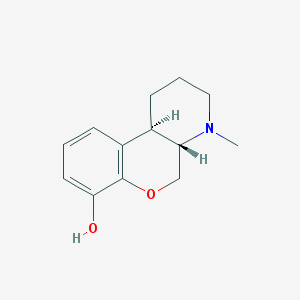 CHEMBL165290 CHEMBL165290 | C13H17NO2 | 219.284 | 3 / 1 | 2.0 | Yes |
| 54608 | 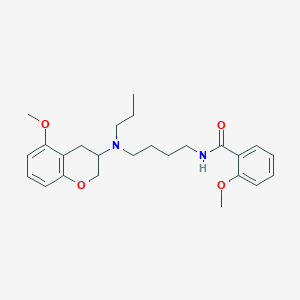 CHEMBL48925 CHEMBL48925 | C25H34N2O4 | 426.557 | 5 / 1 | 4.6 | Yes |
| 57187 | 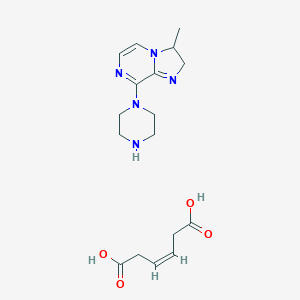 CID 44350012 CID 44350012 | C17H25N5O4 | 363.418 | 7 / 3 | N/A | N/A |
| 57404 |  CHEMBL298534 CHEMBL298534 | C24H31FN2O3 | 414.521 | 5 / 1 | 4.7 | Yes |
| 62318 |  CHEMBL51888 CHEMBL51888 | C22H30N2O4S | 418.552 | 6 / 1 | 3.9 | Yes |
| 62508 |  CHEMBL52438 CHEMBL52438 | C19H25NO | 283.415 | 2 / 0 | 5.1 | No |
| 63073 |  CHEMBL11592 CHEMBL11592 | C26H38N2O4 | 442.6 | 5 / 0 | 4.6 | Yes |
| 68902 | 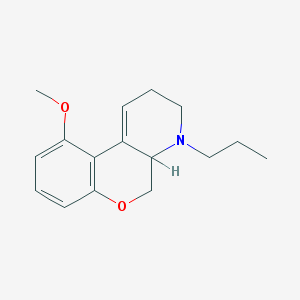 CHEMBL349824 CHEMBL349824 | C16H21NO2 | 259.349 | 3 / 0 | 2.8 | Yes |
| 68947 | 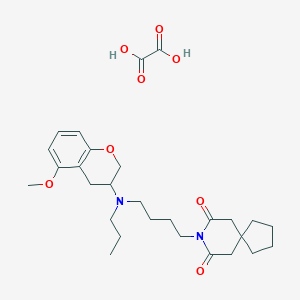 CHEMBL301060 CHEMBL301060 | C28H40N2O8 | 532.634 | 9 / 2 | N/A | No |
| 69547 | 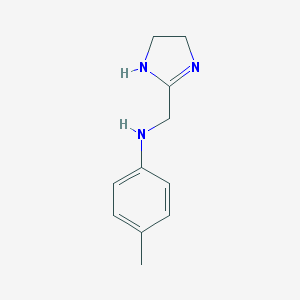 CHEMBL300053 CHEMBL300053 | C11H15N3 | 189.262 | 2 / 2 | 1.3 | Yes |
| 71133 | 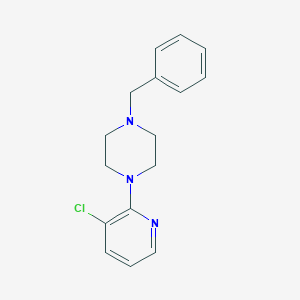 CHEMBL47471 CHEMBL47471 | C16H18ClN3 | 287.791 | 3 / 0 | 3.2 | Yes |
| 71959 | 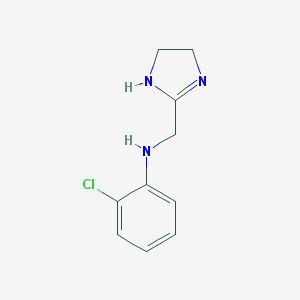 CHEMBL51358 CHEMBL51358 | C10H12ClN3 | 209.677 | 2 / 2 | 1.6 | Yes |
| 73652 | 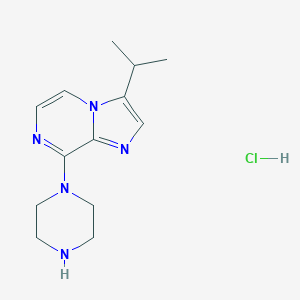 CHEMBL555391 CHEMBL555391 | C13H20ClN5 | 281.788 | 4 / 2 | N/A | N/A |
| 75638 | 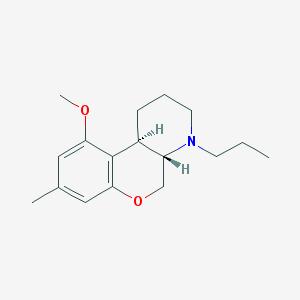 CHEMBL162510 CHEMBL162510 | C17H25NO2 | 275.392 | 3 / 0 | 3.5 | Yes |
| 77264 | 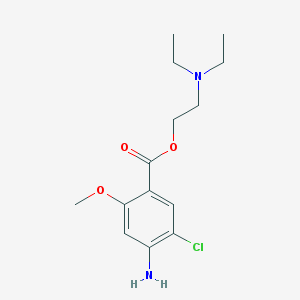 SDZ-205557 SDZ-205557 | C14H21ClN2O3 | 300.783 | 5 / 1 | 3.2 | Yes |
| 79461 | 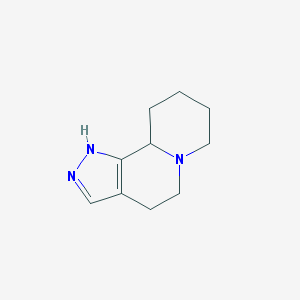 CHEMBL156283 CHEMBL156283 | C10H15N3 | 177.251 | 2 / 1 | 1.0 | Yes |
| 79511 | 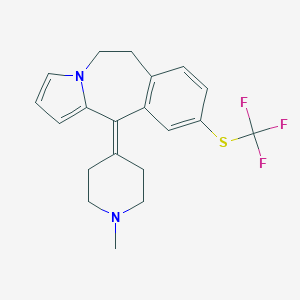 CHEMBL368475 CHEMBL368475 | C20H21F3N2S | 378.457 | 5 / 0 | 4.2 | Yes |
| 82721 | 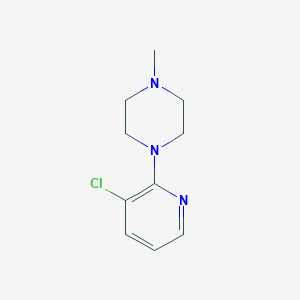 87394-57-8 87394-57-8 | C10H14ClN3 | 211.693 | 3 / 0 | 1.7 | Yes |
| 83013 | 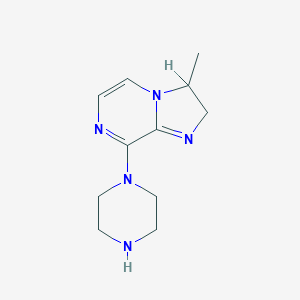 BDBM50002141 BDBM50002141 | C11H17N5 | 219.292 | 3 / 1 | -1.1 | Yes |
| 85377 | 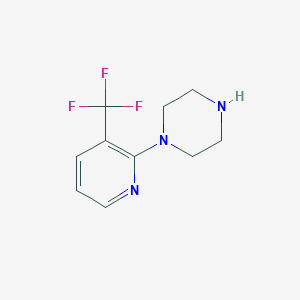 87394-63-6 87394-63-6 | C10H12F3N3 | 231.222 | 6 / 1 | 1.5 | Yes |
| 88082 | 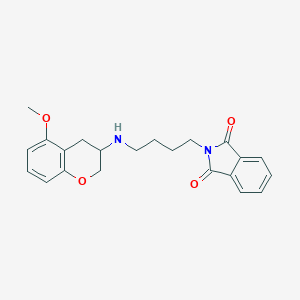 CHEMBL296395 CHEMBL296395 | C22H24N2O4 | 380.444 | 5 / 1 | 2.9 | Yes |
| 88271 | 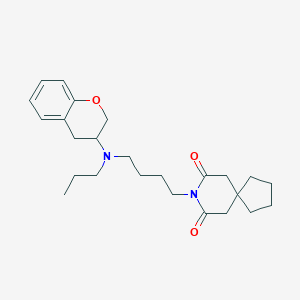 CHEMBL50993 CHEMBL50993 | C25H36N2O3 | 412.574 | 4 / 0 | 4.6 | Yes |
| 92988 | 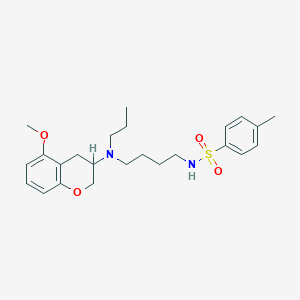 CHEMBL300735 CHEMBL300735 | C24H34N2O4S | 446.606 | 6 / 1 | 4.6 | Yes |
| 94939 | 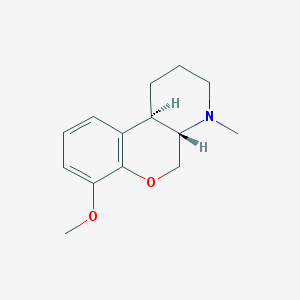 CHEMBL349776 CHEMBL349776 | C14H19NO2 | 233.311 | 3 / 0 | 2.3 | Yes |
| 95814 | 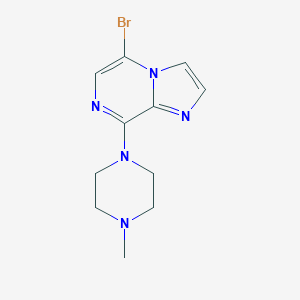 CHEMBL338704 CHEMBL338704 | C11H14BrN5 | 296.172 | 4 / 0 | 2.0 | Yes |
| 95875 | 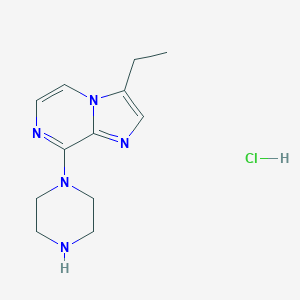 CHEMBL538537 CHEMBL538537 | C12H18ClN5 | 267.761 | 4 / 2 | N/A | N/A |
| 98236 | 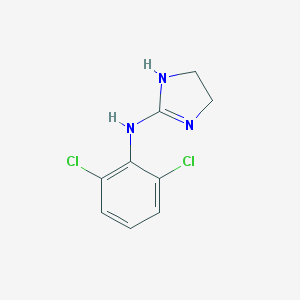 clonidine clonidine | C9H9Cl2N3 | 230.092 | 1 / 2 | 1.6 | Yes |
| 99330 | 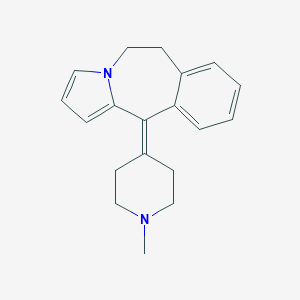 CHEMBL368413 CHEMBL368413 | C19H22N2 | 278.399 | 1 / 0 | 2.5 | Yes |
| 101051 | 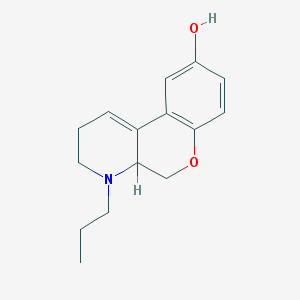 CHEMBL163061 CHEMBL163061 | C15H19NO2 | 245.322 | 3 / 1 | 2.5 | Yes |
| 101638 | 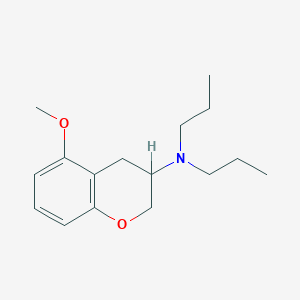 5-Meo-dpac 5-Meo-dpac | C16H25NO2 | 263.381 | 3 / 0 | 3.8 | Yes |
| 460151 | 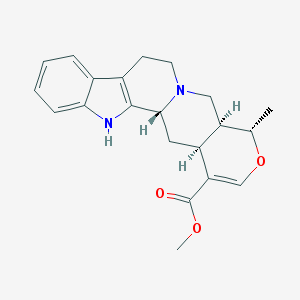 akuammigine akuammigine | C21H24N2O3 | 352.434 | 4 / 1 | 2.7 | Yes |
| 460152 | 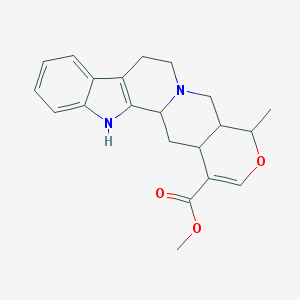 .delta.-Yohimbine .delta.-Yohimbine | C21H24N2O3 | 352.434 | 4 / 1 | 2.7 | Yes |
| 104245 | 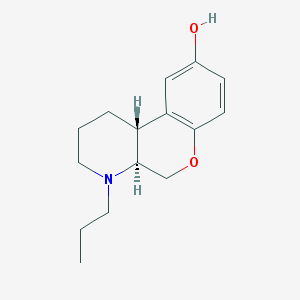 CHEMBL162955 CHEMBL162955 | C15H21NO2 | 247.338 | 3 / 1 | 2.9 | Yes |
| 108399 | 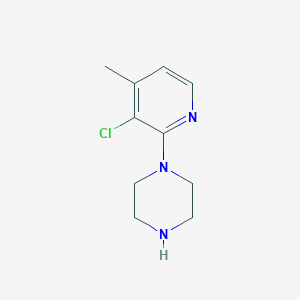 CHEMBL299007 CHEMBL299007 | C10H14ClN3 | 211.693 | 3 / 1 | 1.6 | Yes |
| 109376 | 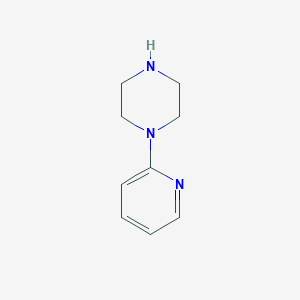 1-(2-Pyridyl)piperazine 1-(2-Pyridyl)piperazine | C9H13N3 | 163.224 | 3 / 1 | 0.7 | Yes |
| 110920 | 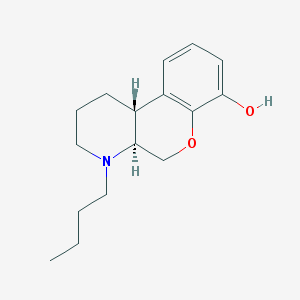 CHEMBL263574 CHEMBL263574 | C16H23NO2 | 261.365 | 3 / 1 | 3.2 | Yes |
| 112352 | 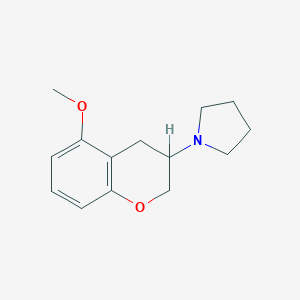 CHEMBL285010 CHEMBL285010 | C14H19NO2 | 233.311 | 3 / 0 | 2.5 | Yes |
| 117771 | 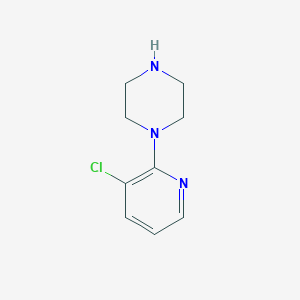 1-(3-chloropyridin-2-yl)piperazine 1-(3-chloropyridin-2-yl)piperazine | C9H12ClN3 | 197.666 | 3 / 1 | 1.3 | Yes |
| 120667 | 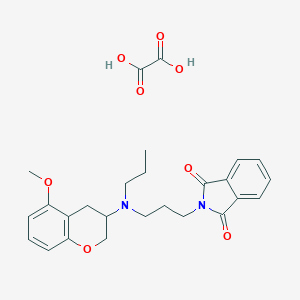 CHEMBL54089 CHEMBL54089 | C26H30N2O8 | 498.532 | 9 / 2 | N/A | N/A |
| 120820 | 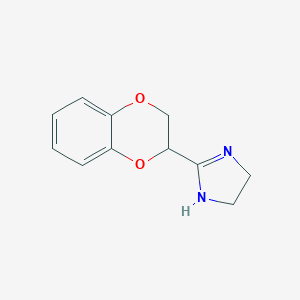 IDAZOXAN IDAZOXAN | C11H12N2O2 | 204.229 | 3 / 1 | 0.7 | Yes |
| 121142 | 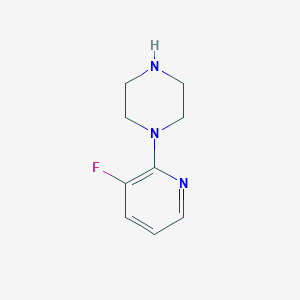 1-(3-fluoropyridin-2-yl)piperazine 1-(3-fluoropyridin-2-yl)piperazine | C9H12FN3 | 181.214 | 4 / 1 | 0.8 | Yes |
| 122582 | 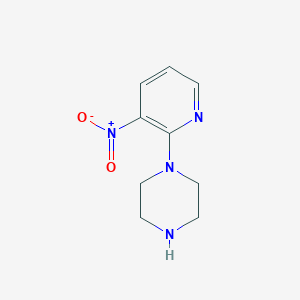 1-(3-nitropyridin-2-yl)piperazine 1-(3-nitropyridin-2-yl)piperazine | C9H12N4O2 | 208.221 | 5 / 1 | 0.5 | Yes |
| 125277 | 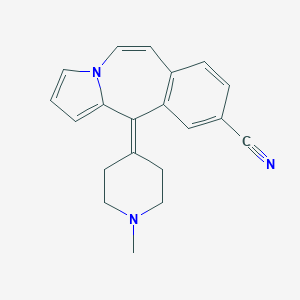 CHEMBL178376 CHEMBL178376 | C20H19N3 | 301.393 | 2 / 0 | 2.5 | Yes |
| 131351 | 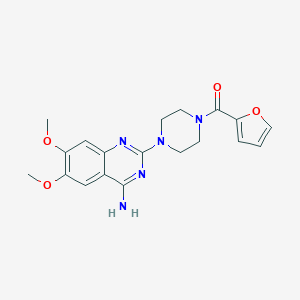 prazosin prazosin | C19H21N5O4 | 383.408 | 8 / 1 | 2.0 | Yes |
| 133951 | 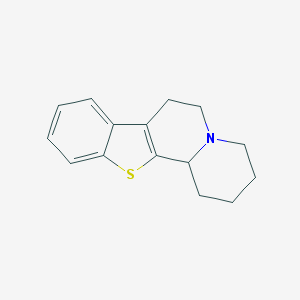 CHEMBL346492 CHEMBL346492 | C15H17NS | 243.368 | 2 / 0 | 3.7 | Yes |
| 139828 | 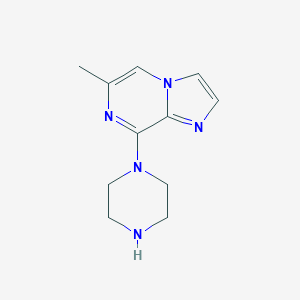 CHEMBL545289 CHEMBL545289 | C11H15N5 | 217.276 | 4 / 1 | 0.9 | Yes |
| 140737 | 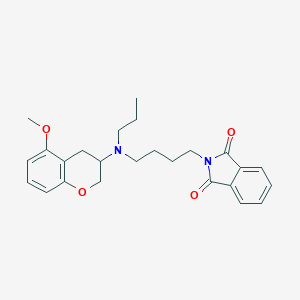 CHEMBL52396 CHEMBL52396 | C25H30N2O4 | 422.525 | 5 / 0 | 4.3 | Yes |
| 143148 | 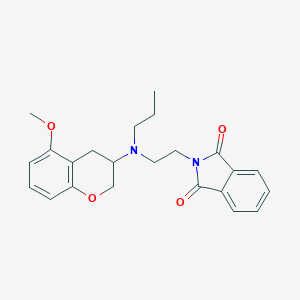 CHEMBL545198 CHEMBL545198 | C23H26N2O4 | 394.471 | 5 / 0 | 3.6 | Yes |
| 148419 | 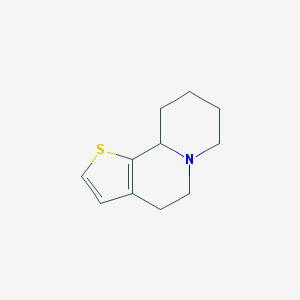 CHEMBL346016 CHEMBL346016 | C11H15NS | 193.308 | 2 / 0 | 2.4 | Yes |
| 152918 | 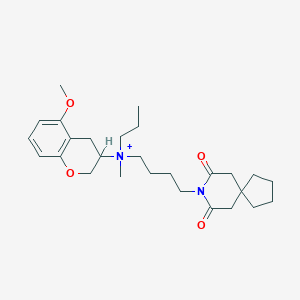 CHEMBL51985 CHEMBL51985 | C27H41N2O4+ | 457.635 | 4 / 0 | 4.6 | Yes |
| 153121 | 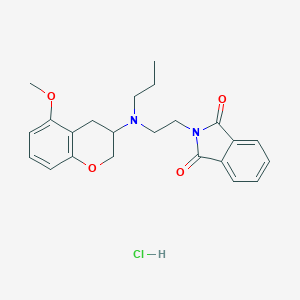 CHEMBL545198 CHEMBL545198 | C23H27ClN2O4 | 430.929 | 5 / 1 | N/A | N/A |
| 154922 | 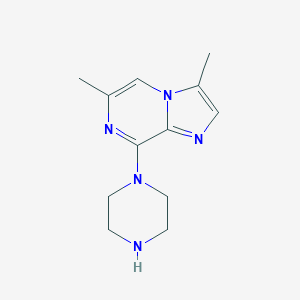 CHEMBL554785 CHEMBL554785 | C12H17N5 | 231.303 | 4 / 1 | 1.3 | Yes |
| 160851 | 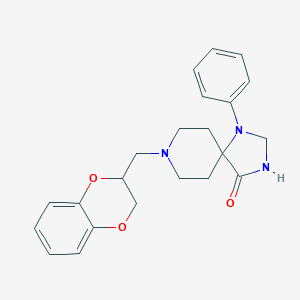 Spiroxatrine Spiroxatrine | C22H25N3O3 | 379.46 | 5 / 1 | 3.1 | Yes |
| 161423 | 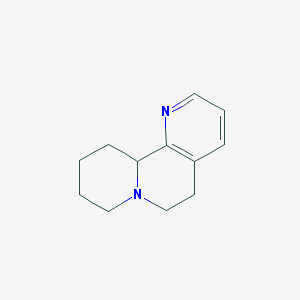 CHEMBL348538 CHEMBL348538 | C12H16N2 | 188.274 | 2 / 0 | 1.6 | Yes |
| 169121 | 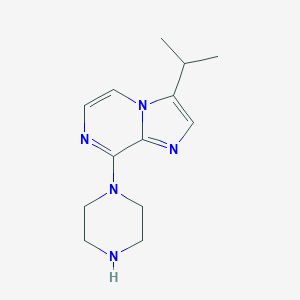 CHEMBL555391 CHEMBL555391 | C13H19N5 | 245.33 | 4 / 1 | 1.7 | Yes |
| 169281 | 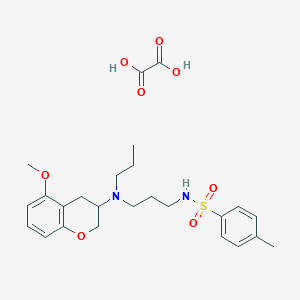 CHEMBL54266 CHEMBL54266 | C25H34N2O8S | 522.613 | 10 / 3 | N/A | No |
| 173364 | 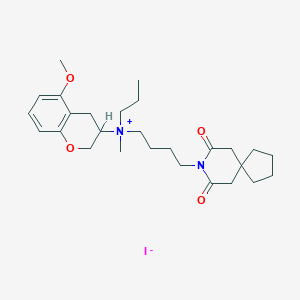 CHEMBL51985 CHEMBL51985 | C27H41IN2O4 | 584.539 | 5 / 0 | N/A | No |
| 175031 | 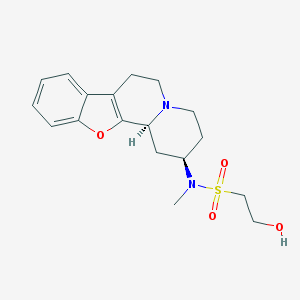 CHEMBL48603 CHEMBL48603 | C18H24N2O4S | 364.46 | 6 / 1 | 1.4 | Yes |
| 175033 | 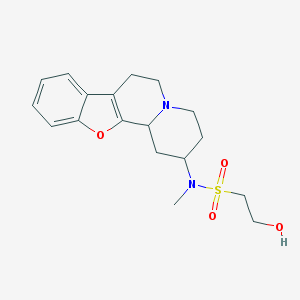 CHEMBL48558 CHEMBL48558 | C18H24N2O4S | 364.46 | 6 / 1 | 1.4 | Yes |
| 175035 | 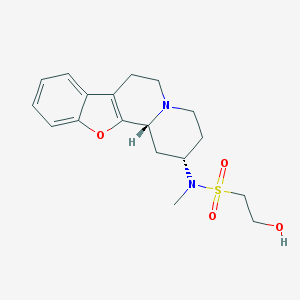 CHEMBL299595 CHEMBL299595 | C18H24N2O4S | 364.46 | 6 / 1 | 1.4 | Yes |
| 175482 | 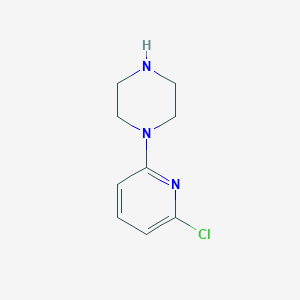 1-(6-chloropyridin-2-yl)piperazine 1-(6-chloropyridin-2-yl)piperazine | C9H12ClN3 | 197.666 | 3 / 1 | 1.6 | Yes |
| 181263 | 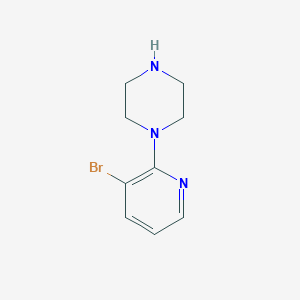 87394-56-7 87394-56-7 | C9H12BrN3 | 242.12 | 3 / 1 | 1.3 | Yes |
| 181920 | 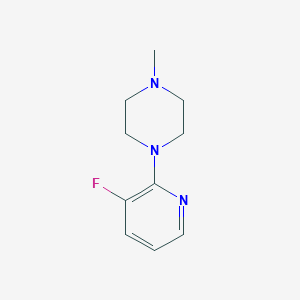 85386-90-9 85386-90-9 | C10H14FN3 | 195.241 | 4 / 0 | 1.2 | Yes |
| 182094 | 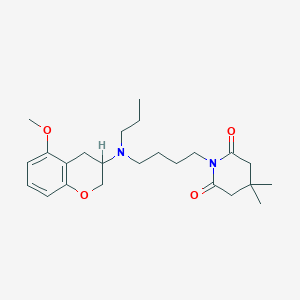 CHEMBL52485 CHEMBL52485 | C24H36N2O4 | 416.562 | 5 / 0 | 3.7 | Yes |
| 182301 | 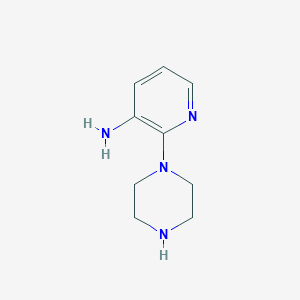 2-(Piperazin-1-yl)pyridin-3-amine 2-(Piperazin-1-yl)pyridin-3-amine | C9H14N4 | 178.239 | 4 / 2 | 0.0 | Yes |
| 186626 | 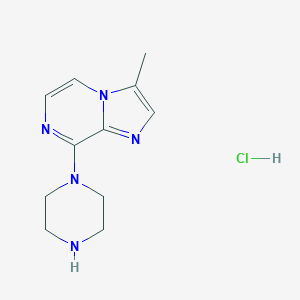 CHEMBL555637 CHEMBL555637 | C11H16ClN5 | 253.734 | 4 / 2 | N/A | N/A |
| 189323 | 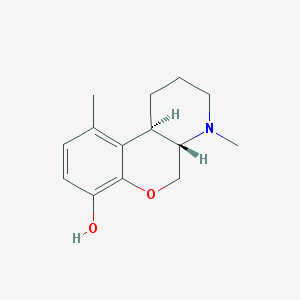 CHEMBL422024 CHEMBL422024 | C14H19NO2 | 233.311 | 3 / 1 | 2.3 | Yes |
| 189342 | 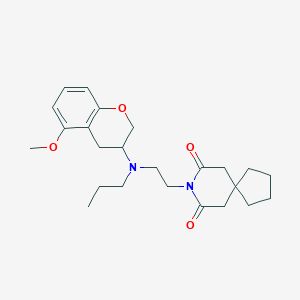 CHEMBL50841 CHEMBL50841 | C24H34N2O4 | 414.546 | 5 / 0 | 3.9 | Yes |
| 190616 | 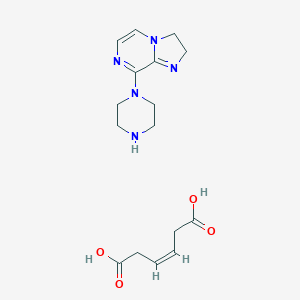 CID 44350063 CID 44350063 | C16H23N5O4 | 349.391 | 7 / 3 | N/A | N/A |
| 196337 | 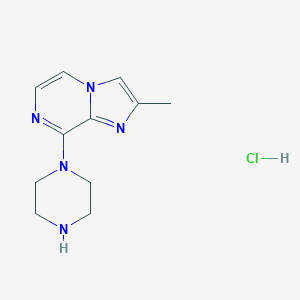 CHEMBL544354 CHEMBL544354 | C11H16ClN5 | 253.734 | 4 / 2 | N/A | N/A |
| 196478 | 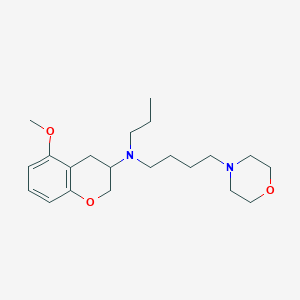 CHEMBL284912 CHEMBL284912 | C21H34N2O3 | 362.514 | 5 / 0 | 3.3 | Yes |
| 201555 | 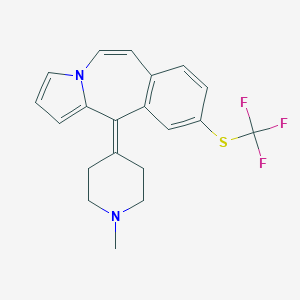 CHEMBL424967 CHEMBL424967 | C20H19F3N2S | 376.441 | 5 / 0 | 4.5 | Yes |
| 201791 | 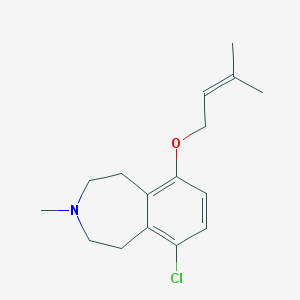 SK&F-104078 SK&F-104078 | C16H22ClNO | 279.808 | 2 / 0 | 4.4 | Yes |
| 203484 | 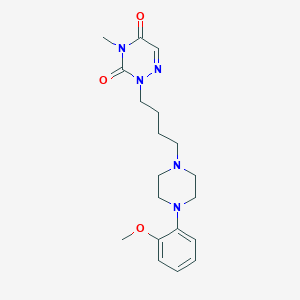 CUMI-101 CUMI-101 | C19H27N5O3 | 373.457 | 6 / 0 | 1.8 | Yes |
| 208398 | 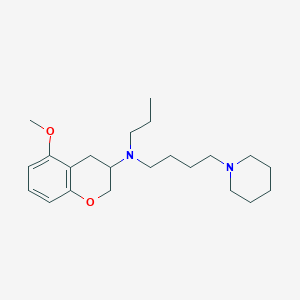 CHEMBL416747 CHEMBL416747 | C22H36N2O2 | 360.542 | 4 / 0 | 4.5 | Yes |
| 210574 | 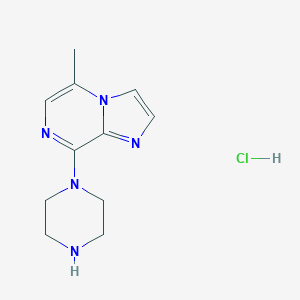 CHEMBL541088 CHEMBL541088 | C11H16ClN5 | 253.734 | 4 / 2 | N/A | N/A |
| 212198 | 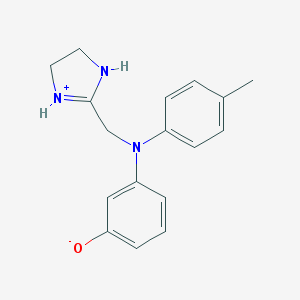 AC1NKXXY AC1NKXXY | C17H19N3O | 281.359 | 2 / 2 | 3.3 | Yes |
| 213837 | 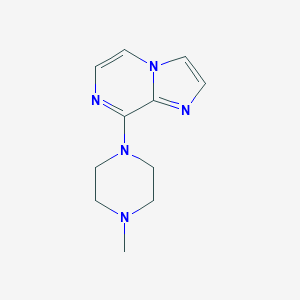 CHEMBL127303 CHEMBL127303 | C11H15N5 | 217.276 | 4 / 0 | 1.0 | Yes |
| 214499 | 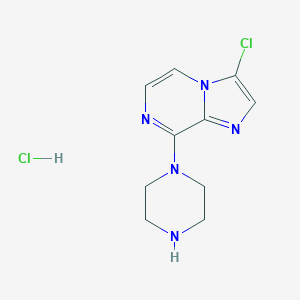 CHEMBL538788 CHEMBL538788 | C10H13Cl2N5 | 274.149 | 4 / 2 | N/A | N/A |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417