

You can:
| Name | D(1B) dopamine receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | DRD5 |
| Synonym | Gpcr1 DRD1L2 DRD1B dopamine receptor 5 Dopamine D5 receptor [ Show all ] |
| Disease | Solid tumours Schizophrenia |
| Length | 477 |
| Amino acid sequence | MLPPGSNGTAYPGQFALYQQLAQGNAVGGSAGAPPLGPSQVVTACLLTLLIIWTLLGNVLVCAAIVRSRHLRANMTNVFIVSLAVSDLFVALLVMPWKAVAEVAGYWPFGAFCDVWVAFDIMCSTASILNLCVISVDRYWAISRPFRYKRKMTQRMALVMVGLAWTLSILISFIPVQLNWHRDQAASWGGLDLPNNLANWTPWEEDFWEPDVNAENCDSSLNRTYAISSSLISFYIPVAIMIVTYTRIYRIAQVQIRRISSLERAAEHAQSCRSSAACAPDTSLRASIKKETKVLKTLSVIMGVFVCCWLPFFILNCMVPFCSGHPEGPPAGFPCVSETTFDVFVWFGWANSSLNPVIYAFNADFQKVFAQLLGCSHFCSRTPVETVNISNELISYNQDIVFHKEIAAAYIHMMPNAVTPGNREVDNDEEEGPFDRMFQIYQTSPDGDPVAESVWELDCEGEISLDKITPFTPNGFH |
| UniProt | P21918 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P21918 |
| 3D structure model | This predicted structure model is from GPCR-EXP P21918. |
| BioLiP | N/A |
| Therapeutic Target Database | T46828 |
| ChEMBL | CHEMBL1850 |
| IUPHAR | 218 |
| DrugBank | BE0000145, BE0004889 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 81 | 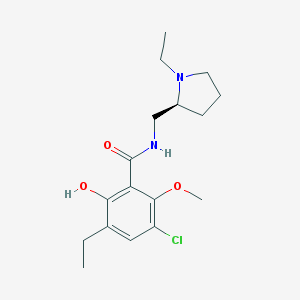 Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 99 | 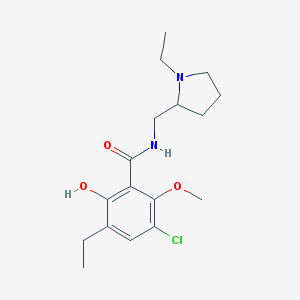 AC1O8PYY AC1O8PYY | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 1016 | 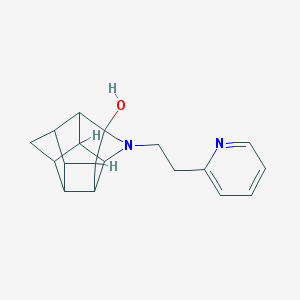 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1060 | 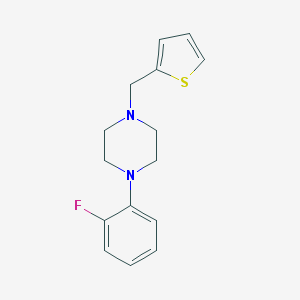 CHEMBL2420770 CHEMBL2420770 | C15H17FN2S | 276.373 | 4 / 0 | 3.2 | Yes |
| 2257 |  1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2409 |  CHEMBL425069 CHEMBL425069 | C27H28ClN5O3 | 506.003 | 6 / 1 | 4.3 | No |
| 3154 |  AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3460 |  CHEMBL201177 CHEMBL201177 | C21H24N2 | 304.437 | 1 / 1 | 4.6 | Yes |
| 3576 |  CHEMBL238915 CHEMBL238915 | C21H24ClN3O3S | 433.951 | 5 / 0 | 3.6 | Yes |
| 3895 |  CHEMBL239917 CHEMBL239917 | C22H26ClN3O | 383.92 | 3 / 0 | 3.9 | Yes |
| 3999 |  MLS000118160 MLS000118160 | C20H18N4O3 | 362.389 | 6 / 1 | 3.2 | Yes |
| 4720 |  cid_654562 cid_654562 | C19H20N4O2S | 368.455 | 6 / 1 | 2.6 | Yes |
| 5364 |  CHEMBL396560 CHEMBL396560 | C21H24N2O | 320.436 | 2 / 1 | 4.3 | Yes |
| 5380 |  Indolo[2,3-b]quinoxalin-6-yl-acetic acid ethyl ester Indolo[2,3-b]quinoxalin-6-yl-acetic acid ethyl ester | C18H15N3O2 | 305.337 | 4 / 0 | 3.2 | Yes |
| 5676 |  AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 463718 |  CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7128 |  CHEMBL398813 CHEMBL398813 | C23H25F3N4O | 430.475 | 7 / 0 | 4.1 | Yes |
| 8459 |  CHEMBL598105 CHEMBL598105 | C26H23ClN2O | 414.933 | 3 / 1 | 5.8 | No |
| 9708 |  MLS000102793 MLS000102793 | C20H15N3O4S2 | 425.477 | 8 / 1 | 3.5 | Yes |
| 10175 |  CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10740 | 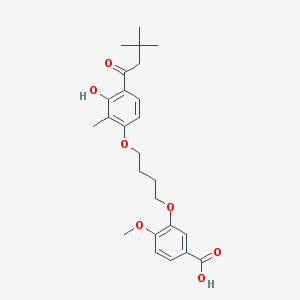 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10877 | 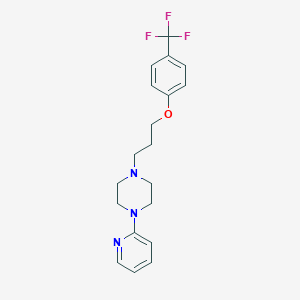 CHEMBL3289645 CHEMBL3289645 | C19H22F3N3O | 365.4 | 7 / 0 | 4.0 | Yes |
| 11016 | 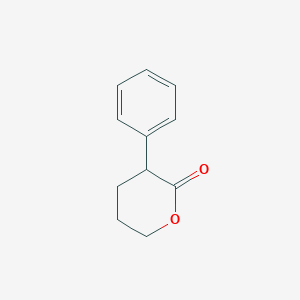 CHEMBL201086 CHEMBL201086 | C11H12O2 | 176.215 | 2 / 0 | 2.2 | Yes |
| 11139 | 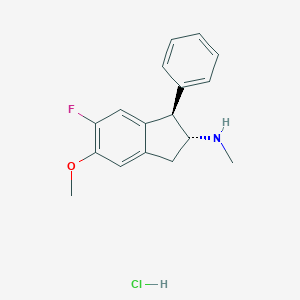 CHEMBL558364 CHEMBL558364 | C17H19ClFNO | 307.793 | 3 / 2 | N/A | N/A |
| 12022 | 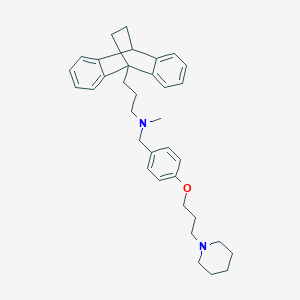 CHEMBL517244 CHEMBL517244 | C35H44N2O | 508.75 | 3 / 0 | 7.8 | No |
| 12832 | 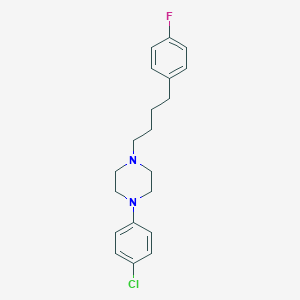 CHEMBL1940408 CHEMBL1940408 | C20H24ClFN2 | 346.874 | 3 / 0 | 5.3 | No |
| 553324 | 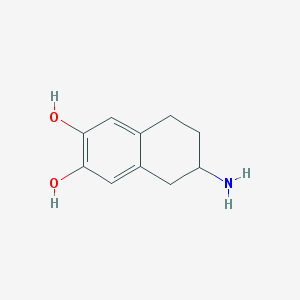 ADTN ADTN | C10H13NO2 | 179.219 | 3 / 3 | 1.4 | Yes |
| 13698 | 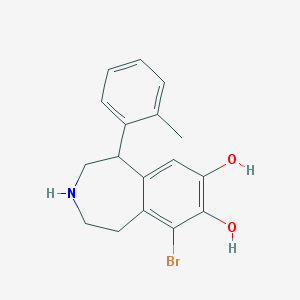 BDBM86280 BDBM86280 | C17H18BrNO2 | 348.24 | 3 / 3 | 3.2 | Yes |
| 13814 |  CHEMBL380330 CHEMBL380330 | C19H23NO | 281.399 | 2 / 0 | 4.1 | Yes |
| 14164 |  cid_652353 cid_652353 | C14H14N4OS | 286.353 | 5 / 2 | 2.8 | Yes |
| 15484 |  CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 442284 |  CHEMBL3321789 CHEMBL3321789 | C22H25ClFNO2 | 389.895 | 4 / 1 | 4.8 | Yes |
| 16886 |  Glemanserin Glemanserin | C20H25NO | 295.426 | 2 / 1 | 3.8 | Yes |
| 19203 |  CHEMBL559164 CHEMBL559164 | C20H22FNO | 311.4 | 3 / 0 | 4.5 | Yes |
| 20195 |  imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 522149 |  CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522209 |  CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 22483 |  CHEMBL238916 CHEMBL238916 | C22H24F3N3O3S | 467.507 | 8 / 0 | 3.8 | Yes |
| 23355 |  sulpiride sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 24098 |  CHEMBL389559 CHEMBL389559 | C20H25NO | 295.426 | 2 / 1 | 4.5 | Yes |
| 24219 |  CHEMBL382780 CHEMBL382780 | C21H22N2O | 318.42 | 1 / 1 | 3.9 | Yes |
| 26046 |  LISURIDE LISURIDE | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26070 |  S-(-)-Lisuride S-(-)-Lisuride | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26088 |  AC1L1H1T AC1L1H1T | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26157 |  SMR000093497 SMR000093497 | C18H21NO4 | 315.369 | 4 / 1 | 3.2 | Yes |
| 26273 |  SMR000028039 SMR000028039 | C20H18N2O4S | 382.434 | 5 / 1 | 2.8 | Yes |
| 27016 |  CHEMBL2420891 CHEMBL2420891 | C19H18Cl2N2S | 377.327 | 3 / 0 | 5.6 | No |
| 27190 |  CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27332 |  MLS000084541 MLS000084541 | C13H15N9OS | 345.385 | 10 / 2 | 1.5 | Yes |
| 27488 | 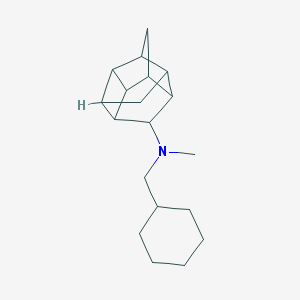 CHEMBL2432063 CHEMBL2432063 | C19H29N | 271.448 | 1 / 0 | 4.7 | Yes |
| 28533 | 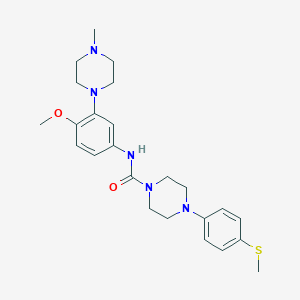 CHEMBL197796 CHEMBL197796 | C24H33N5O2S | 455.621 | 6 / 1 | 3.1 | Yes |
| 466501 | 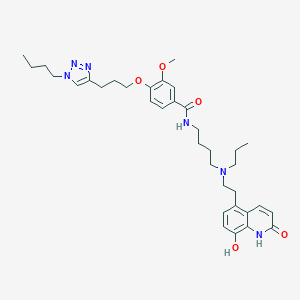 J3.522.165D J3.522.165D | C35H48N6O5 | 632.806 | 8 / 3 | 5.1 | No |
| 29169 | 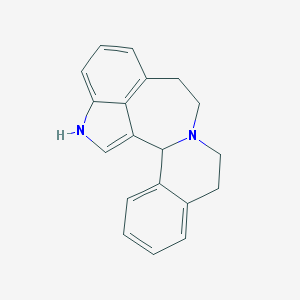 CHEMBL1086790 CHEMBL1086790 | C19H18N2 | 274.367 | 1 / 1 | 3.6 | Yes |
| 29413 |  CHEMBL1940420 CHEMBL1940420 | C18H22FN3O | 315.392 | 5 / 0 | 3.7 | Yes |
| 29571 |  CHEMBL3289653 CHEMBL3289653 | C21H23ClN2O | 354.878 | 3 / 0 | 5.2 | No |
| 29822 |  CHEMBL2420772 CHEMBL2420772 | C15H16Cl2N2S | 327.267 | 3 / 0 | 4.3 | Yes |
| 30687 |  MLS000101323 MLS000101323 | C15H17N3OS | 287.381 | 3 / 1 | 3.6 | Yes |
| 32118 | 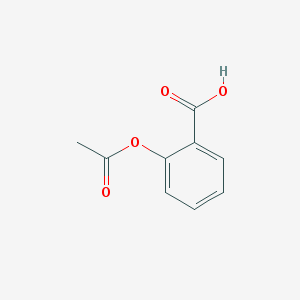 aspirin aspirin | C9H8O4 | 180.159 | 4 / 1 | 1.2 | Yes |
| 32315 | 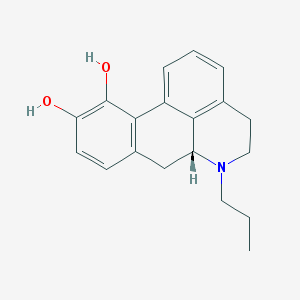 n-Propylapomorphine n-Propylapomorphine | C19H21NO2 | 295.382 | 3 / 2 | 3.2 | Yes |
| 32317 | 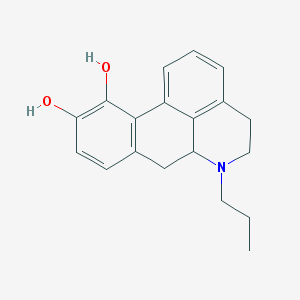 N-n-Propylnorapomorphine N-n-Propylnorapomorphine | C19H21NO2 | 295.382 | 3 / 2 | 3.2 | Yes |
| 33049 | 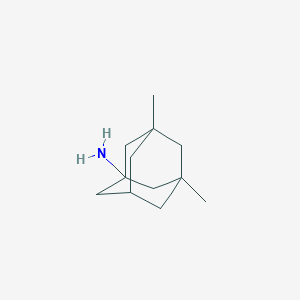 memantine memantine | C12H21N | 179.307 | 1 / 1 | 3.3 | Yes |
| 33893 |  CHEMBL556194 CHEMBL556194 | C16H17ClFNO | 293.766 | 3 / 2 | N/A | N/A |
| 35281 |  CHEMBL201525 CHEMBL201525 | C18H21N | 251.373 | 1 / 0 | 4.6 | Yes |
| 35343 |  CHEMBL600083 CHEMBL600083 | C19H19BrClNO | 392.721 | 2 / 1 | 5.1 | No |
| 38538 |  SMR000111657 SMR000111657 | C17H18N2O | 266.344 | 2 / 1 | 3.0 | Yes |
| 40285 |  Aripiprazole Aripiprazole | C23H27Cl2N3O2 | 448.388 | 4 / 1 | 4.6 | Yes |
| 40729 |  SMR000103840 SMR000103840 | C20H19F3N4O2 | 404.393 | 7 / 0 | 3.5 | Yes |
| 548359 |  CHEMBL3957865 CHEMBL3957865 | C21H23F2N3 | 355.433 | 4 / 0 | 5.1 | No |
| 42472 |  CHEMBL2432039 CHEMBL2432039 | C20H23N | 277.411 | 1 / 1 | 2.7 | Yes |
| 558588 |  CHEMBL464811 CHEMBL464811 | C32H38ClN5O | 544.14 | 5 / 1 | 5.8 | No |
| 45771 |  23981-80-8 23981-80-8 | C14H14O3 | 230.263 | 3 / 1 | 3.3 | Yes |
| 45996 |  cid_657898 cid_657898 | C16H16N4O2S | 328.39 | 7 / 2 | 2.3 | Yes |
| 46005 |  CHEMBL2420779 CHEMBL2420779 | C18H23N3O | 297.402 | 4 / 0 | 2.6 | Yes |
| 46071 |  CHEMBL437888 CHEMBL437888 | C19H26N2 | 282.431 | 1 / 1 | 4.2 | Yes |
| 46112 |  CHEMBL389357 CHEMBL389357 | C20H25NO | 295.426 | 2 / 0 | 4.5 | Yes |
| 47276 |  CHEMBL3289643 CHEMBL3289643 | C21H23ClN2O | 354.878 | 3 / 0 | 4.3 | Yes |
| 47491 |  CHEMBL2432051 CHEMBL2432051 | C21H27NO3 | 341.451 | 4 / 1 | 3.3 | Yes |
| 548440 |  CHEMBL3983633 CHEMBL3983633 | C23H26F2N2O | 384.471 | 4 / 1 | 5.2 | No |
| 47745 |  CHEMBL328866 CHEMBL328866 | C23H26ClN3O3 | 427.929 | 4 / 2 | 3.2 | Yes |
| 48978 |  CHEMBL240996 CHEMBL240996 | C23H26F3N3O | 417.476 | 6 / 0 | 4.2 | Yes |
| 49562 |  CHEMBL394777 CHEMBL394777 | C19H20ClNO | 313.825 | 2 / 0 | 4.5 | Yes |
| 50037 | 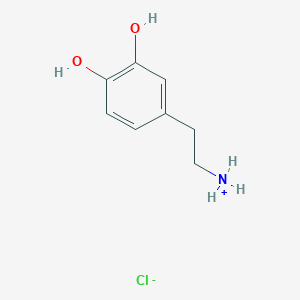 3-hydroxytyraminium chloride 3-hydroxytyraminium chloride | C8H12ClNO2 | 189.639 | 3 / 3 | N/A | N/A |
| 443755 | 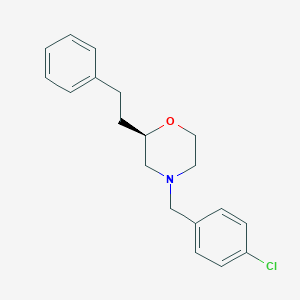 ML398 ML398 | C19H22ClNO | 315.841 | 2 / 0 | 4.4 | Yes |
| 443757 | 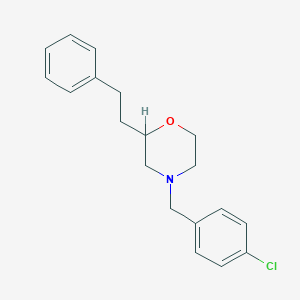 CHEMBL3335540 CHEMBL3335540 | C19H22ClNO | 315.841 | 2 / 0 | 4.4 | Yes |
| 52023 | 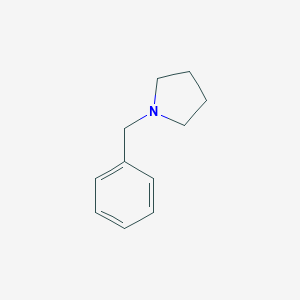 1-Benzylpyrrolidine 1-Benzylpyrrolidine | C11H15N | 161.248 | 1 / 0 | 2.2 | Yes |
| 52908 | 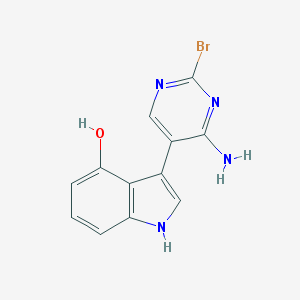 CHEMBL1829958 CHEMBL1829958 | C12H9BrN4O | 305.135 | 4 / 3 | 2.3 | Yes |
| 53173 | 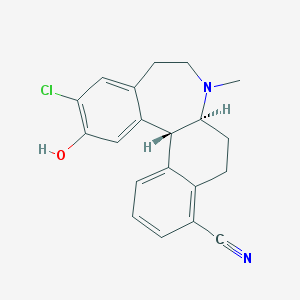 CHEMBL604127 CHEMBL604127 | C20H19ClN2O | 338.835 | 3 / 1 | 4.2 | Yes |
| 53668 | 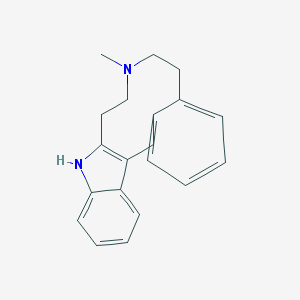 CHEMBL1087301 CHEMBL1087301 | C20H22N2 | 290.41 | 1 / 1 | 4.3 | Yes |
| 53778 | 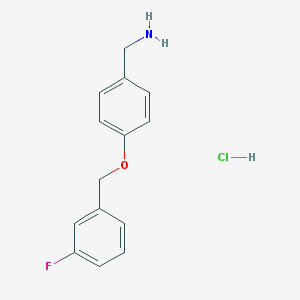 {4-[(3-fluorophenyl)methoxy]phenyl}methanamine hydrochloride {4-[(3-fluorophenyl)methoxy]phenyl}methanamine hydrochloride | C14H15ClFNO | 267.728 | 3 / 2 | N/A | N/A |
| 53824 |  CHEMBL596179 CHEMBL596179 | C18H21NO | 267.372 | 2 / 1 | 3.7 | Yes |
| 54056 |  ST-1535 ST-1535 | C12H16N8 | 272.316 | 6 / 1 | 1.8 | Yes |
| 54218 |  MLS000052086 MLS000052086 | C19H16N4S | 332.425 | 4 / 0 | 4.6 | Yes |
| 54484 |  CHEMBL540613 CHEMBL540613 | C19H20FNO | 297.373 | 3 / 1 | 4.2 | Yes |
| 55553 | 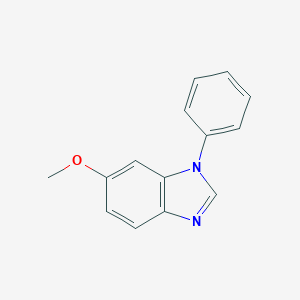 6-Methoxy-1-phenyl-1H-benzimidazole 6-Methoxy-1-phenyl-1H-benzimidazole | C14H12N2O | 224.263 | 2 / 0 | 3.1 | Yes |
| 55652 | 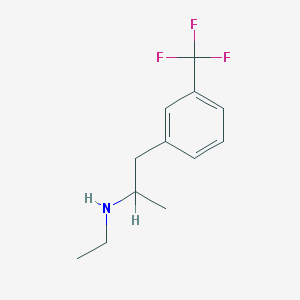 fenfluramine fenfluramine | C12H16F3N | 231.262 | 4 / 1 | 3.4 | Yes |
| 56316 | 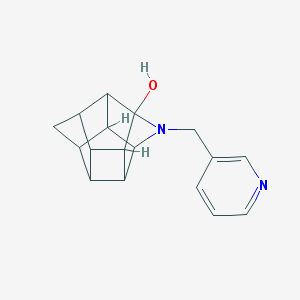 CHEMBL2205833 CHEMBL2205833 | C17H18N2O | 266.344 | 3 / 1 | 1.3 | Yes |
| 56861 | 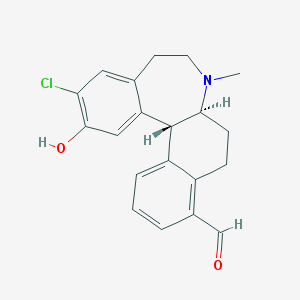 CHEMBL604316 CHEMBL604316 | C20H20ClNO2 | 341.835 | 3 / 1 | 3.9 | Yes |
| 548541 |  Aripiprazole lauroxil Aripiprazole lauroxil | C36H51Cl2N3O4 | 660.721 | 6 / 0 | 10.0 | No |
| 57214 |  CHEMBL212912 CHEMBL212912 | C25H23Cl2N5O | 480.393 | 4 / 1 | 5.0 | Yes |
| 58104 |  cinnarizine cinnarizine | C26H28N2 | 368.524 | 2 / 0 | 5.8 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417