

You can:
| Name | Trace amine-associated receptor 7b |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Taar7b |
| Synonym | TaR-12 TaR-7b Trace amine receptor 12 Trace amine receptor 7b |
| Disease | N/A for non-human GPCRs |
| Length | 358 |
| Amino acid sequence | MATDDDRFPWDQDSILSRDLLSASSMQLCYEKLNRSCVRSPYSPGPRLILYAVFGFGAVLAVCGNLLVMTSILHFRQLHSPANFLVASLACADFLVGLTVMPFSMVRSVEGCWYFGDIYCKFHSSFDGSFCYSSIFHLCFISADRYIAVSDPLIYPTRFTASVSGKCITFSWLLSIIYSFSLFYTGVNEAGLEDLVSALTCVGGCQIAVNQSWVFINFLLFLVPALVMMTVYSKIFLIAKQQAQNIEKMGKQTARASESYKDRVAKRERKAAKTLGIAVAAFLLSWLPYFIDSIIDAFLGFVTPTYVYEILVWIGYYNSAMNPLIYAFFYPWFRKAIKLIVTGKILRENSSATNLFPE |
| UniProt | Q923X8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2176813 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3316 | 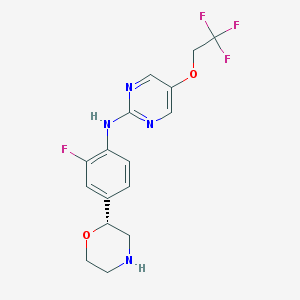 US8802673, 145 US8802673, 145 | C16H16F4N4O2 | 372.324 | 10 / 2 | 2.3 | Yes |
| 3319 | 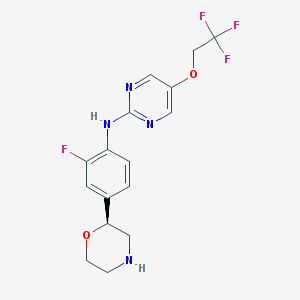 US8802673, 145 US8802673, 145 | C16H16F4N4O2 | 372.324 | 10 / 2 | 2.3 | Yes |
| 8421 | 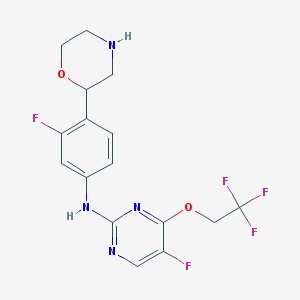 CHEMBL3641759 CHEMBL3641759 | C16H15F5N4O2 | 390.314 | 11 / 2 | 2.7 | No |
| 8951 | 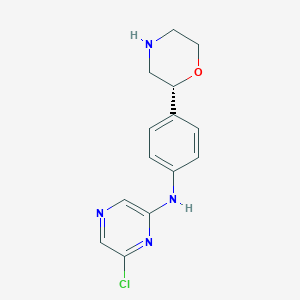 CHEMBL3641756 CHEMBL3641756 | C14H15ClN4O | 290.751 | 5 / 2 | 1.6 | Yes |
| 9803 | 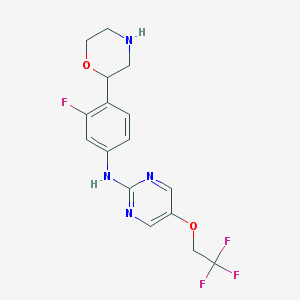 CHEMBL3641757 CHEMBL3641757 | C16H16F4N4O2 | 372.324 | 10 / 2 | 2.3 | Yes |
| 11757 | 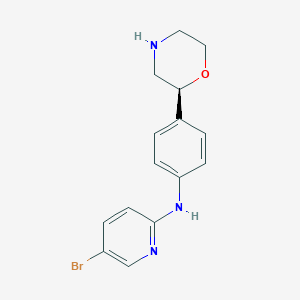 CHEMBL3701909 CHEMBL3701909 | C15H16BrN3O | 334.217 | 4 / 2 | 2.4 | Yes |
| 14388 | 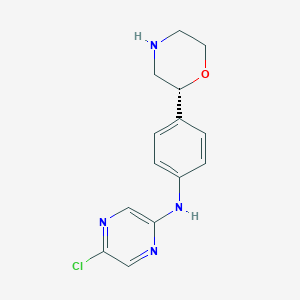 CHEMBL3641755 CHEMBL3641755 | C14H15ClN4O | 290.751 | 5 / 2 | 1.6 | Yes |
| 17472 | 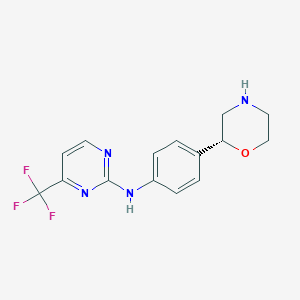 CHEMBL3641723 CHEMBL3641723 | C15H15F3N4O | 324.307 | 8 / 2 | 2.0 | Yes |
| 18473 | 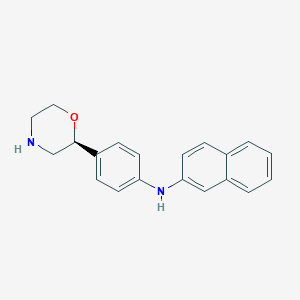 CHEMBL3701943 CHEMBL3701943 | C20H20N2O | 304.393 | 3 / 2 | 3.9 | Yes |
| 19029 | 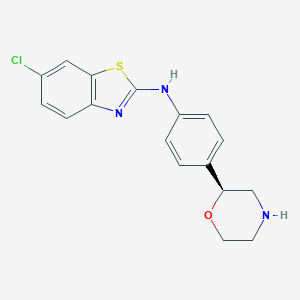 CHEMBL3701925 CHEMBL3701925 | C17H16ClN3OS | 345.845 | 5 / 2 | 3.9 | Yes |
| 20346 | 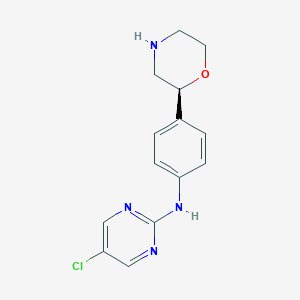 CHEMBL3702002 CHEMBL3702002 | C14H15ClN4O | 290.751 | 5 / 2 | 1.7 | Yes |
| 20347 | 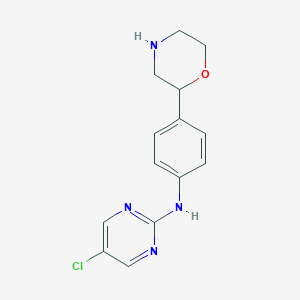 CHEMBL3701905 CHEMBL3701905 | C14H15ClN4O | 290.751 | 5 / 2 | 1.7 | Yes |
| 20350 | 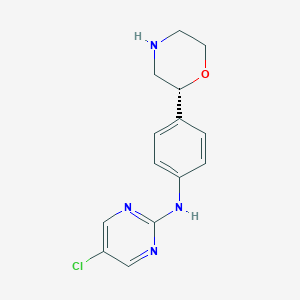 CHEMBL3702001 CHEMBL3702001 | C14H15ClN4O | 290.751 | 5 / 2 | 1.7 | Yes |
| 21465 | 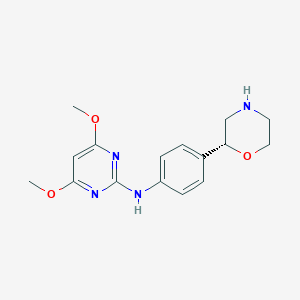 CHEMBL3641733 CHEMBL3641733 | C16H20N4O3 | 316.361 | 7 / 2 | 1.7 | Yes |
| 21772 |  CHEMBL3701903 CHEMBL3701903 | C21H24ClF3N2 | 396.882 | 5 / 2 | 6.1 | No |
| 26172 | 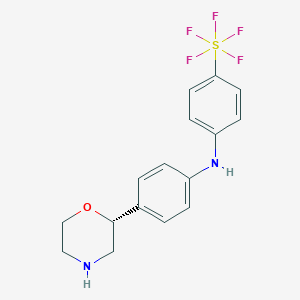 CHEMBL3641741 CHEMBL3641741 | C16H17F5N2OS | 380.377 | 8 / 2 | 5.9 | No |
| 26812 | 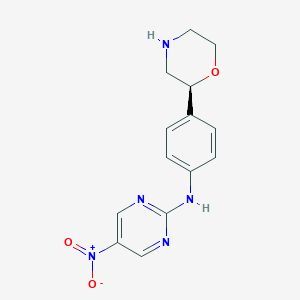 CHEMBL3701974 CHEMBL3701974 | C14H15N5O3 | 301.306 | 7 / 2 | 0.9 | Yes |
| 26830 | 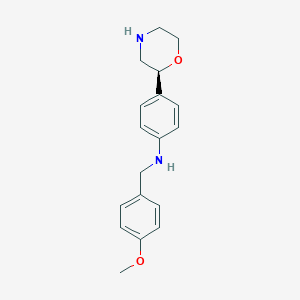 CHEMBL3701919 CHEMBL3701919 | C18H22N2O2 | 298.386 | 4 / 2 | 2.6 | Yes |
| 29384 | 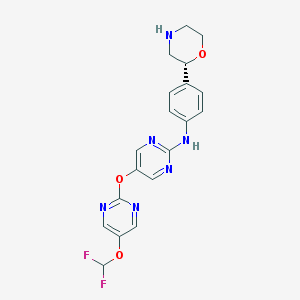 CHEMBL3641724 CHEMBL3641724 | C19H18F2N6O3 | 416.389 | 11 / 2 | 2.2 | No |
| 30178 | 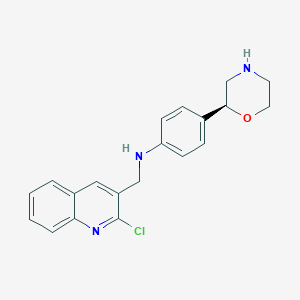 CHEMBL3701950 CHEMBL3701950 | C20H20ClN3O | 353.85 | 4 / 2 | 3.7 | Yes |
| 32164 | 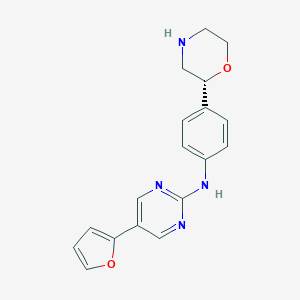 CHEMBL3641701 CHEMBL3641701 | C18H18N4O2 | 322.368 | 6 / 2 | 1.8 | Yes |
| 32166 | 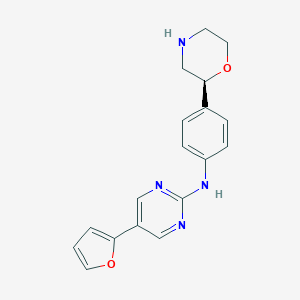 CHEMBL3641700 CHEMBL3641700 | C18H18N4O2 | 322.368 | 6 / 2 | 1.8 | Yes |
| 32369 | 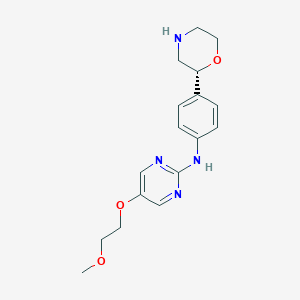 CHEMBL3641693 CHEMBL3641693 | C17H22N4O3 | 330.388 | 7 / 2 | 0.9 | Yes |
| 32371 | 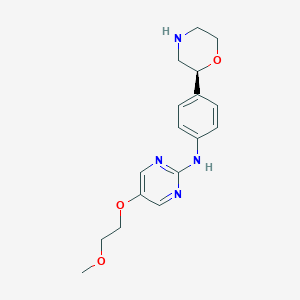 CHEMBL3641692 CHEMBL3641692 | C17H22N4O3 | 330.388 | 7 / 2 | 0.9 | Yes |
| 32667 | 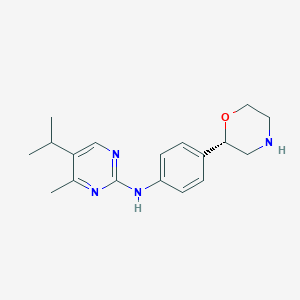 CHEMBL3701972 CHEMBL3701972 | C18H24N4O | 312.417 | 5 / 2 | 2.6 | Yes |
| 40721 | 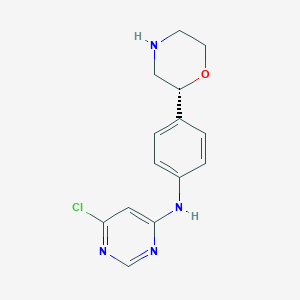 CHEMBL3641749 CHEMBL3641749 | C14H15ClN4O | 290.751 | 5 / 2 | 2.1 | Yes |
| 40778 | 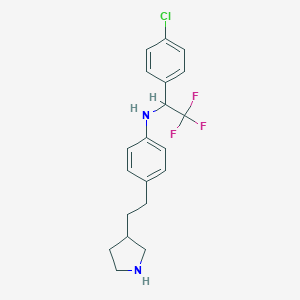 CHEMBL3701902 CHEMBL3701902 | C20H22ClF3N2 | 382.855 | 5 / 2 | 5.8 | No |
| 43595 | 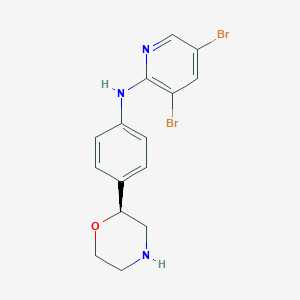 CHEMBL3701960 CHEMBL3701960 | C15H15Br2N3O | 413.113 | 4 / 2 | 3.1 | Yes |
| 45187 | 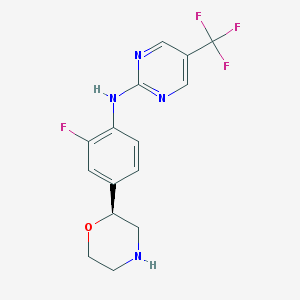 BDBM6698 BDBM6698 | C15H14F4N4O | 342.298 | 9 / 2 | 2.1 | Yes |
| 45190 | 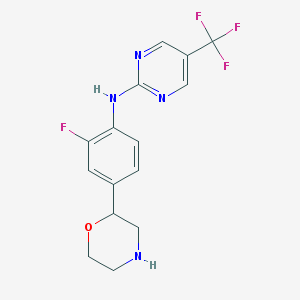 US8802673, 134 US8802673, 134 | C15H14F4N4O | 342.298 | 9 / 2 | 2.1 | Yes |
| 45191 | 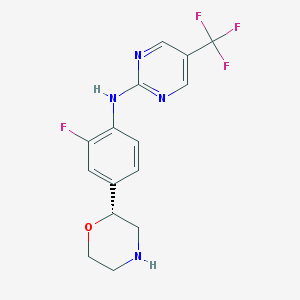 BDBM6566 BDBM6566 | C15H14F4N4O | 342.298 | 9 / 2 | 2.1 | Yes |
| 47188 | 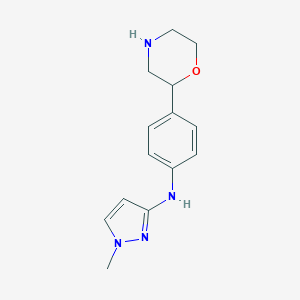 CHEMBL3641694 CHEMBL3641694 | C14H18N4O | 258.325 | 4 / 2 | 1.1 | Yes |
| 48101 | 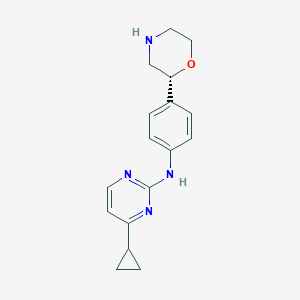 CHEMBL3641739 CHEMBL3641739 | C17H20N4O | 296.374 | 5 / 2 | 1.7 | Yes |
| 48675 | 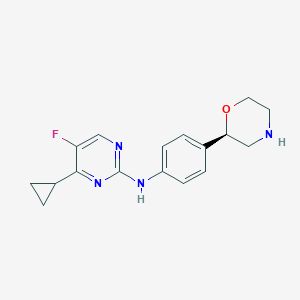 CHEMBL3641740 CHEMBL3641740 | C17H19FN4O | 314.364 | 6 / 2 | 1.8 | Yes |
| 49775 | 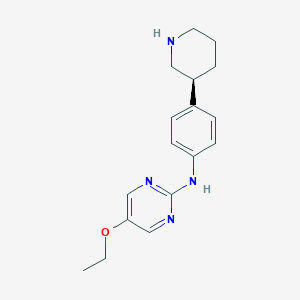 CHEMBL3702012 CHEMBL3702012 | C17H22N4O | 298.39 | 5 / 2 | 2.5 | Yes |
| 51493 | 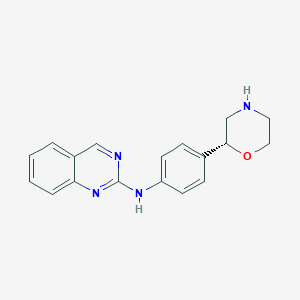 CHEMBL3641727 CHEMBL3641727 | C18H18N4O | 306.369 | 5 / 2 | 2.4 | Yes |
| 55665 | 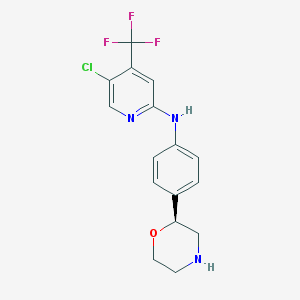 CHEMBL3640008 CHEMBL3640008 | C16H15ClF3N3O | 357.761 | 7 / 2 | 3.3 | Yes |
| 57772 | 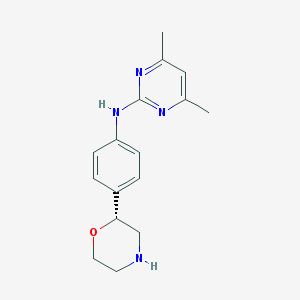 CHEMBL3641732 CHEMBL3641732 | C16H20N4O | 284.363 | 5 / 2 | 1.9 | Yes |
| 64552 | 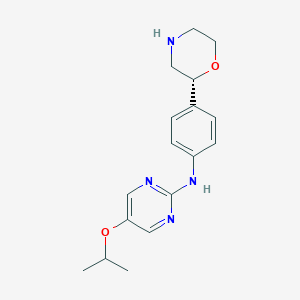 CHEMBL3702021 CHEMBL3702021 | C17H22N4O2 | 314.389 | 6 / 2 | 1.9 | Yes |
| 64554 | 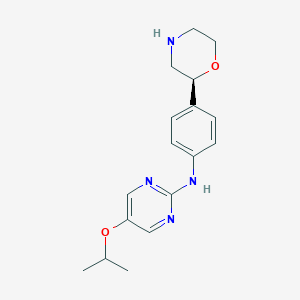 CHEMBL3702020 CHEMBL3702020 | C17H22N4O2 | 314.389 | 6 / 2 | 1.9 | Yes |
| 68872 | 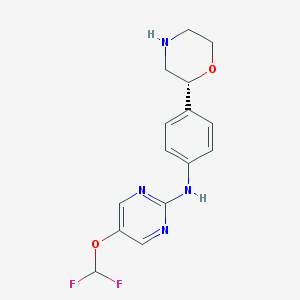 CHEMBL3641729 CHEMBL3641729 | C15H16F2N4O2 | 322.316 | 8 / 2 | 2.0 | Yes |
| 73935 | 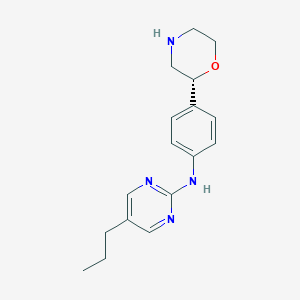 CHEMBL3701991 CHEMBL3701991 | C17H22N4O | 298.39 | 5 / 2 | 2.4 | Yes |
| 73936 | 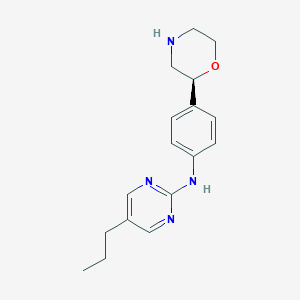 CHEMBL3701920 CHEMBL3701920 | C17H22N4O | 298.39 | 5 / 2 | 2.4 | Yes |
| 74477 | 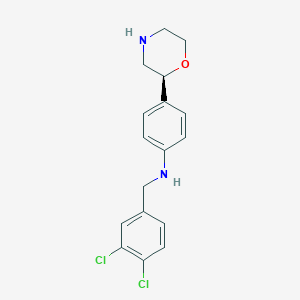 CHEMBL3701929 CHEMBL3701929 | C17H18Cl2N2O | 337.244 | 3 / 2 | 3.7 | Yes |
| 76115 | 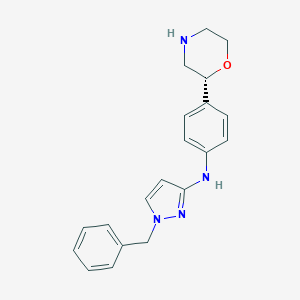 CHEMBL3641714 CHEMBL3641714 | C20H22N4O | 334.423 | 4 / 2 | 2.7 | Yes |
| 76117 | 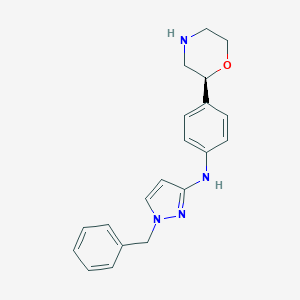 CHEMBL3641713 CHEMBL3641713 | C20H22N4O | 334.423 | 4 / 2 | 2.7 | Yes |
| 76601 | 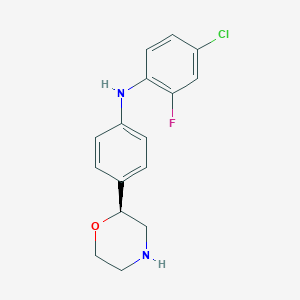 CHEMBL3701958 CHEMBL3701958 | C16H16ClFN2O | 306.765 | 4 / 2 | 3.2 | Yes |
| 84092 | 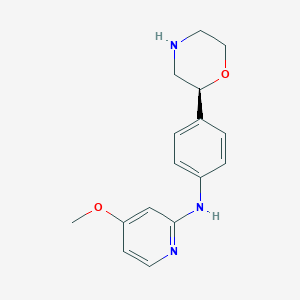 CHEMBL3701938 CHEMBL3701938 | C16H19N3O2 | 285.347 | 5 / 2 | 1.7 | Yes |
| 85140 | 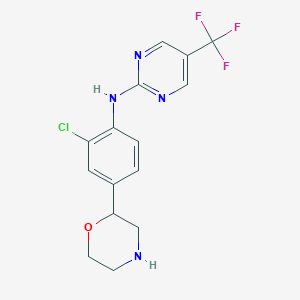 US8802673, 169 US8802673, 169 | C15H14ClF3N4O | 358.749 | 8 / 2 | 2.6 | Yes |
| 85828 | 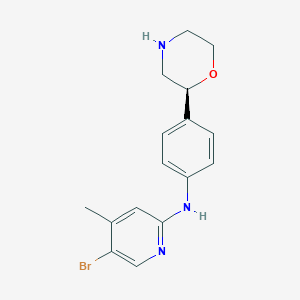 CHEMBL3701961 CHEMBL3701961 | C16H18BrN3O | 348.244 | 4 / 2 | 2.8 | Yes |
| 92929 | 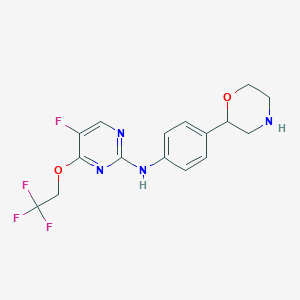 CHEMBL3641738 CHEMBL3641738 | C16H16F4N4O2 | 372.324 | 10 / 2 | 2.6 | Yes |
| 94014 | 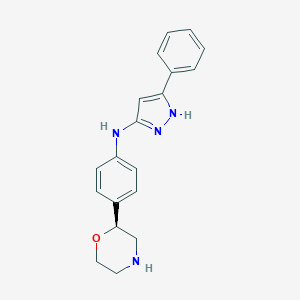 CHEMBL3702011 CHEMBL3702011 | C19H20N4O | 320.396 | 4 / 3 | 2.8 | Yes |
| 99060 | 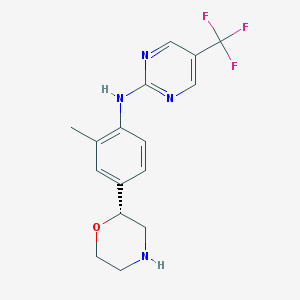 US8802673, 167 US8802673, 167 | C16H17F3N4O | 338.334 | 8 / 2 | 2.4 | Yes |
| 99062 | 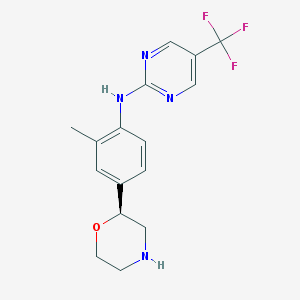 US8802673, 167 US8802673, 167 | C16H17F3N4O | 338.334 | 8 / 2 | 2.4 | Yes |
| 99138 | 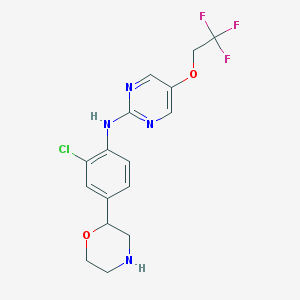 CHEMBL3641734 CHEMBL3641734 | C16H16ClF3N4O2 | 388.775 | 9 / 2 | 2.8 | Yes |
| 103763 | 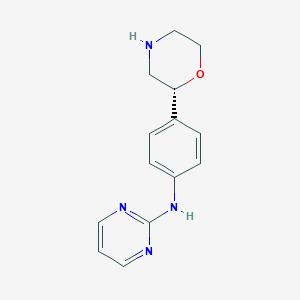 CHEMBL3641726 CHEMBL3641726 | C14H16N4O | 256.309 | 5 / 2 | 1.3 | Yes |
| 110238 | 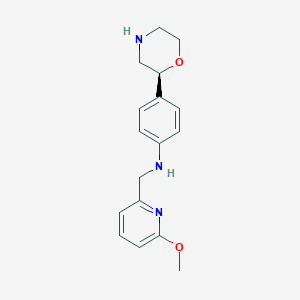 CHEMBL3701927 CHEMBL3701927 | C17H21N3O2 | 299.374 | 5 / 2 | 1.7 | Yes |
| 113680 | 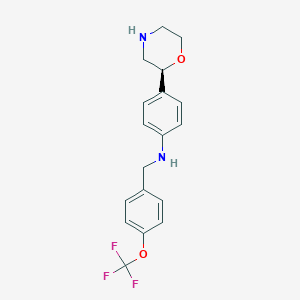 CHEMBL3701924 CHEMBL3701924 | C18H19F3N2O2 | 352.357 | 7 / 2 | 3.6 | Yes |
| 113780 | 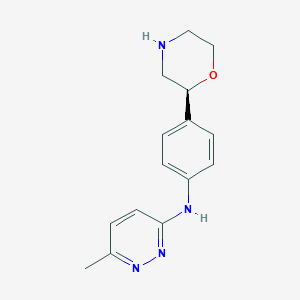 CHEMBL3641712 CHEMBL3641712 | C15H18N4O | 270.336 | 5 / 2 | 1.1 | Yes |
| 116280 | 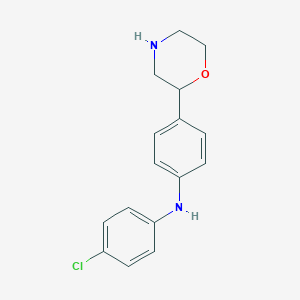 CHEMBL3701907 CHEMBL3701907 | C16H17ClN2O | 288.775 | 3 / 2 | 3.3 | Yes |
| 125497 | 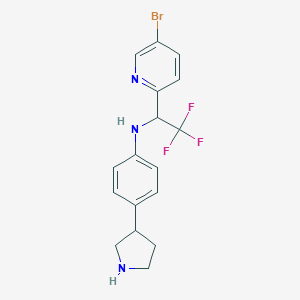 CHEMBL3701901 CHEMBL3701901 | C17H17BrF3N3 | 400.243 | 6 / 2 | 4.0 | Yes |
| 128716 | 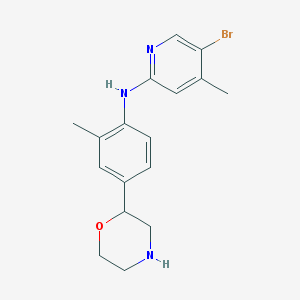 CHEMBL3701983 CHEMBL3701983 | C17H20BrN3O | 362.271 | 4 / 2 | 3.2 | Yes |
| 129669 | 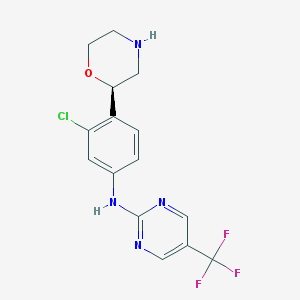 CHEMBL3641754 CHEMBL3641754 | C15H14ClF3N4O | 358.749 | 8 / 2 | 2.6 | Yes |
| 129829 | 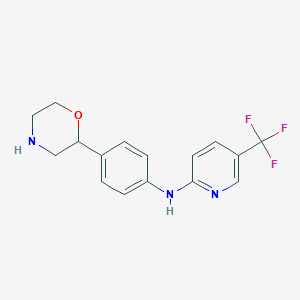 CHEMBL3701910 CHEMBL3701910 | C16H16F3N3O | 323.319 | 7 / 2 | 2.6 | Yes |
| 138742 | 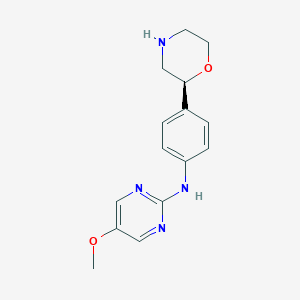 CHEMBL3701911 CHEMBL3701911 | C15H18N4O2 | 286.335 | 6 / 2 | 1.1 | Yes |
| 140425 | 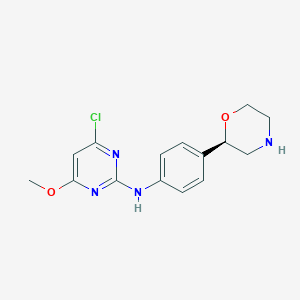 CHEMBL3641730 CHEMBL3641730 | C15H17ClN4O2 | 320.777 | 6 / 2 | 2.4 | Yes |
| 145152 | 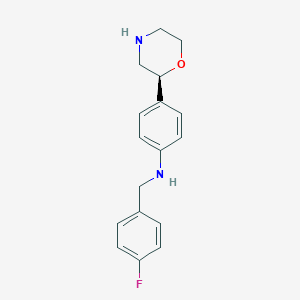 CHEMBL3701930 CHEMBL3701930 | C17H19FN2O | 286.35 | 4 / 2 | 2.7 | Yes |
| 145605 | 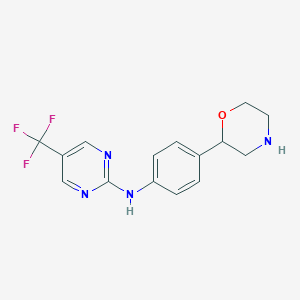 CHEMBL3701918 CHEMBL3701918 | C15H15F3N4O | 324.307 | 8 / 2 | 2.0 | Yes |
| 145607 | 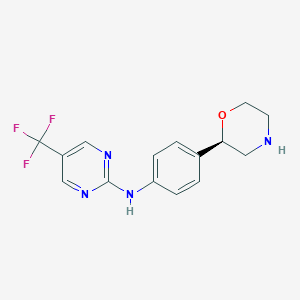 CHEMBL3701993 CHEMBL3701993 | C15H15F3N4O | 324.307 | 8 / 2 | 2.0 | Yes |
| 148856 | 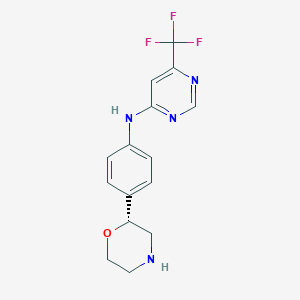 CHEMBL3641748 CHEMBL3641748 | C15H15F3N4O | 324.307 | 8 / 2 | 2.0 | Yes |
| 148943 | 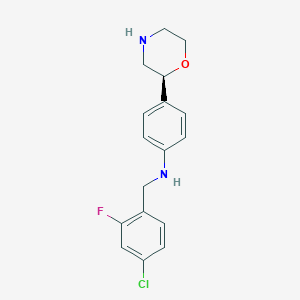 CHEMBL3701934 CHEMBL3701934 | C17H18ClFN2O | 320.792 | 4 / 2 | 3.2 | Yes |
| 149438 | 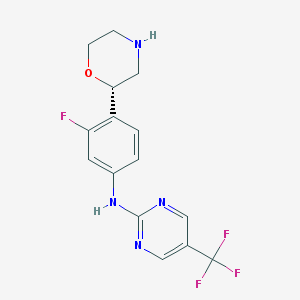 CHEMBL3641747 CHEMBL3641747 | C15H14F4N4O | 342.298 | 9 / 2 | 2.1 | Yes |
| 149441 | 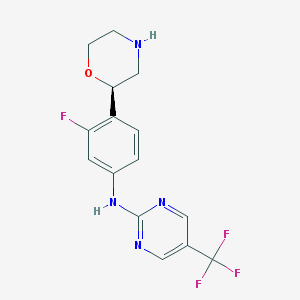 CHEMBL3641746 CHEMBL3641746 | C15H14F4N4O | 342.298 | 9 / 2 | 2.1 | Yes |
| 149624 | 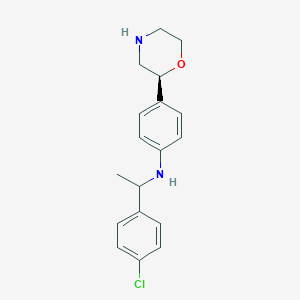 CHEMBL3701969 CHEMBL3701969 | C18H21ClN2O | 316.829 | 3 / 2 | 3.6 | Yes |
| 150885 | 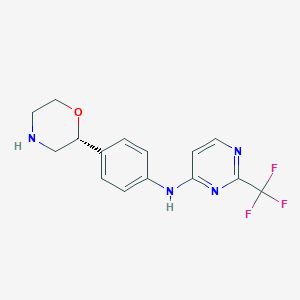 CHEMBL3641750 CHEMBL3641750 | C15H15F3N4O | 324.307 | 8 / 2 | 2.0 | Yes |
| 151877 | 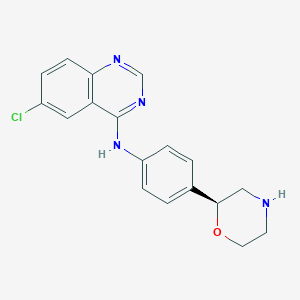 CHEMBL3701949 CHEMBL3701949 | C18H17ClN4O | 340.811 | 5 / 2 | 3.1 | Yes |
| 152160 | 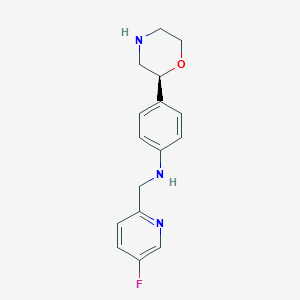 CHEMBL3701936 CHEMBL3701936 | C16H18FN3O | 287.338 | 5 / 2 | 1.5 | Yes |
| 156143 | 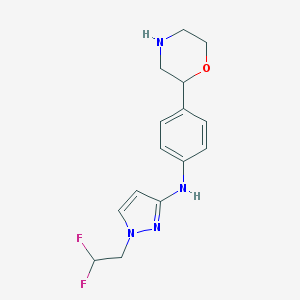 CHEMBL3641703 CHEMBL3641703 | C15H18F2N4O | 308.333 | 6 / 2 | 1.9 | Yes |
| 156223 | 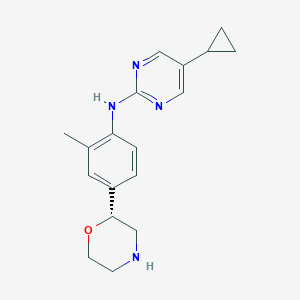 CHEMBL3702008 CHEMBL3702008 | C18H22N4O | 310.401 | 5 / 2 | 2.4 | Yes |
| 156225 | 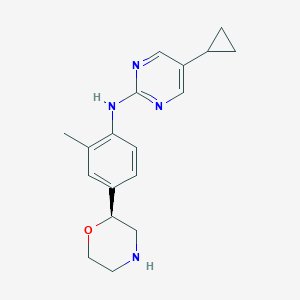 CHEMBL3702009 CHEMBL3702009 | C18H22N4O | 310.401 | 5 / 2 | 2.4 | Yes |
| 156227 | 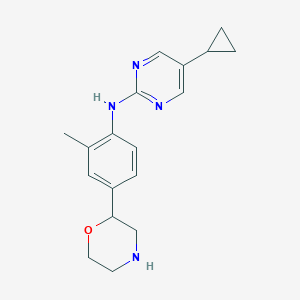 CHEMBL3701985 CHEMBL3701985 | C18H22N4O | 310.401 | 5 / 2 | 2.4 | Yes |
| 171436 | 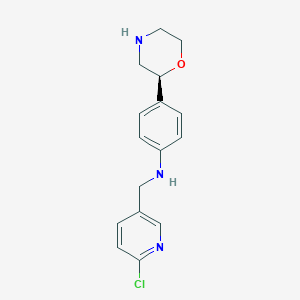 CHEMBL3701931 CHEMBL3701931 | C16H18ClN3O | 303.79 | 4 / 2 | 2.3 | Yes |
| 172516 | 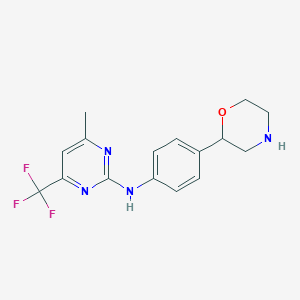 CHEMBL3641728 CHEMBL3641728 | C16H17F3N4O | 338.334 | 8 / 2 | 2.4 | Yes |
| 172865 | 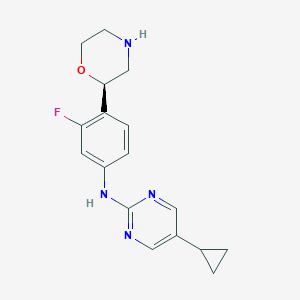 CHEMBL3641742 CHEMBL3641742 | C17H19FN4O | 314.364 | 6 / 2 | 2.1 | Yes |
| 172867 | 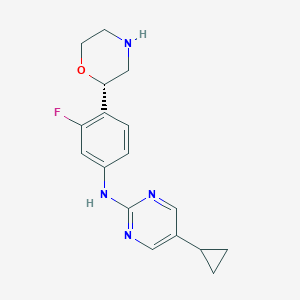 CHEMBL3641745 CHEMBL3641745 | C17H19FN4O | 314.364 | 6 / 2 | 2.1 | Yes |
| 175323 | 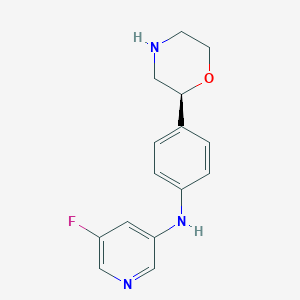 CHEMBL3701942 CHEMBL3701942 | C15H16FN3O | 273.311 | 5 / 2 | 1.5 | Yes |
| 183705 | 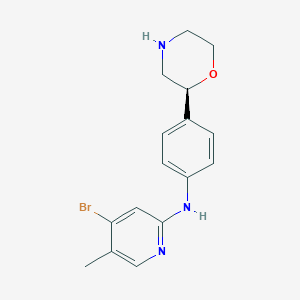 CHEMBL3701963 CHEMBL3701963 | C16H18BrN3O | 348.244 | 4 / 2 | 2.8 | Yes |
| 186387 | 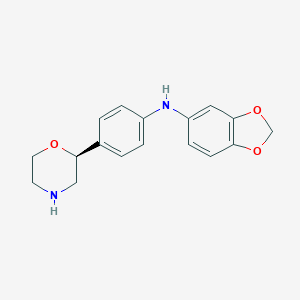 CHEMBL3701964 CHEMBL3701964 | C17H18N2O3 | 298.342 | 5 / 2 | 2.3 | Yes |
| 187517 | 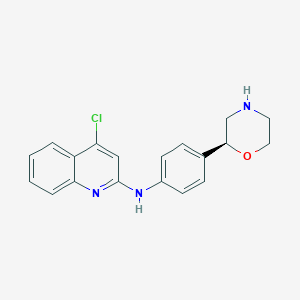 CHEMBL3701953 CHEMBL3701953 | C19H18ClN3O | 339.823 | 4 / 2 | 3.7 | Yes |
| 191243 | 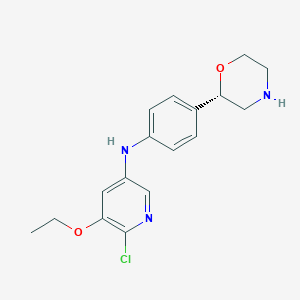 CHEMBL3701970 CHEMBL3701970 | C17H20ClN3O2 | 333.816 | 5 / 2 | 2.7 | Yes |
| 193924 | 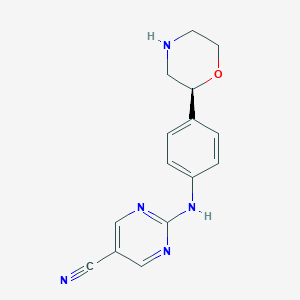 CHEMBL3701915 CHEMBL3701915 | C15H15N5O | 281.319 | 6 / 2 | 0.8 | Yes |
| 193961 | 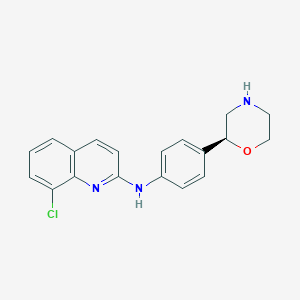 CHEMBL3701951 CHEMBL3701951 | C19H18ClN3O | 339.823 | 4 / 2 | 3.7 | Yes |
| 195800 | 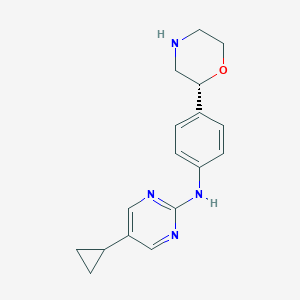 CHEMBL3701984 CHEMBL3701984 | C17H20N4O | 296.374 | 5 / 2 | 2.0 | Yes |
| 195803 | 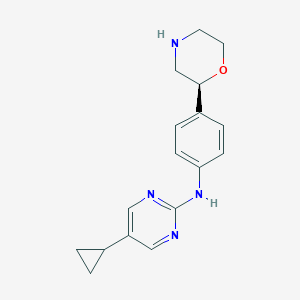 CHEMBL3701916 CHEMBL3701916 | C17H20N4O | 296.374 | 5 / 2 | 2.0 | Yes |
| 199703 | 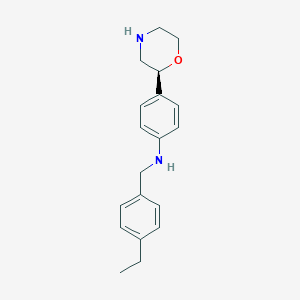 CHEMBL3701935 CHEMBL3701935 | C19H24N2O | 296.414 | 3 / 2 | 3.4 | Yes |
| 200170 | 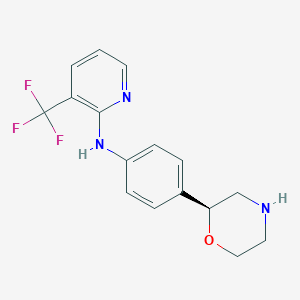 CHEMBL3701940 CHEMBL3701940 | C16H16F3N3O | 323.319 | 7 / 2 | 2.6 | Yes |
| 201466 | 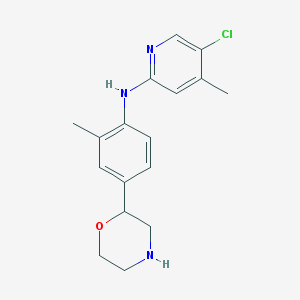 CHEMBL3701982 CHEMBL3701982 | C17H20ClN3O | 317.817 | 4 / 2 | 3.1 | Yes |
| 201668 | 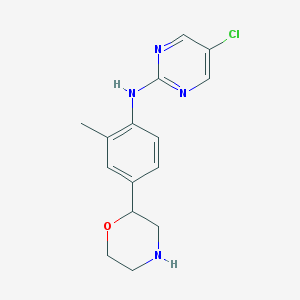 CHEMBL3701980 CHEMBL3701980 | C15H17ClN4O | 304.778 | 5 / 2 | 2.1 | Yes |
| 202064 | 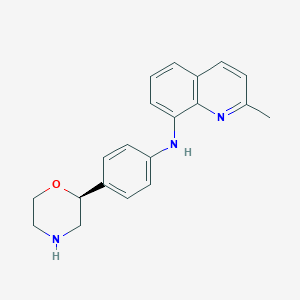 CHEMBL3701947 CHEMBL3701947 | C20H21N3O | 319.408 | 4 / 2 | 3.2 | Yes |
| 204992 | 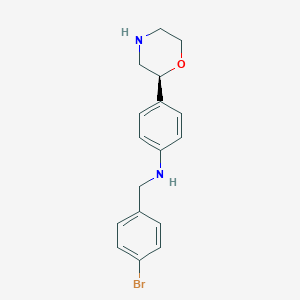 CHEMBL3701944 CHEMBL3701944 | C17H19BrN2O | 347.256 | 3 / 2 | 3.3 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417