

You can:
| Name | Glucagon-like peptide 1 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GLP1R |
| Synonym | glucagon-like peptide 1 receptor GLP-1R GLP-1-R GLP-1 receptor |
| Disease | Type 1/2 diabetes Type 1 diabetes Obesity Non-insulin dependent diabetes Non-alcoholic steatohepatitis [ Show all ] |
| Length | 463 |
| Amino acid sequence | MAGAPGPLRLALLLLGMVGRAGPRPQGATVSLWETVQKWREYRRQCQRSLTEDPPPATDLFCNRTFDEYACWPDGEPGSFVNVSCPWYLPWASSVPQGHVYRFCTAEGLWLQKDNSSLPWRDLSECEESKRGERSSPEEQLLFLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQQHQWDGLLSYQDSLSCRLVFLLMQYCVAANYYWLLVEGVYLYTLLAFSVLSEQWIFRLYVSIGWGVPLLFVVPWGIVKYLYEDEGCWTRNSNMNYWLIIRLPILFAIGVNFLIFVRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARGTLRFIKLFTELSFTSFQGLMVAILYCFVNNEVQLEFRKSWERWRLEHLHIQRDSSMKPLKCPTSSLSSGATAGSSMYTATCQASCS |
| UniProt | P43220 |
| Protein Data Bank | 5vex, 3c59, 3c5t, 5nx2, 3iol, 4zgm, 5otu, 5vew, 5otw, 5otx, 5otv |
| GPCR-HGmod model | P43220 |
| 3D structure model | This structure is from PDB ID 5vex. |
| BioLiP | BL0418498,BL0418499, BL0143794, BL0143795, BL0167479, BL0167480, BL0324354, BL0324355,BL0324356, BL0378791,BL0378792, BL0379513,BL0379514, BL0418500,BL0418501, BL0380967, BL0418494,BL0418495, BL0418496,BL0418497, BL0143732, BL0143731, BL0380966 |
| Therapeutic Target Database | T36075 |
| ChEMBL | CHEMBL1784 |
| IUPHAR | 249 |
| DrugBank | BE0000857 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 1 | 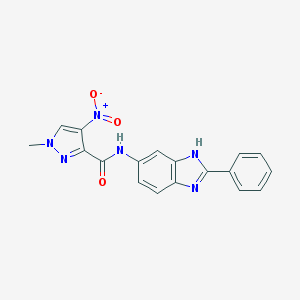 BAS 03421985 BAS 03421985 | C18H14N6O3 | 362.349 | 5 / 2 | 2.5 | Yes |
| 2 | 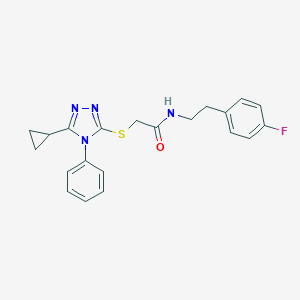 SMR000154917 SMR000154917 | C21H21FN4OS | 396.484 | 5 / 1 | 3.9 | Yes |
| 5 | 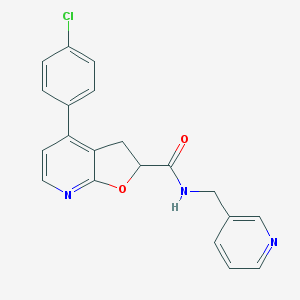 MLS003120225 MLS003120225 | C20H16ClN3O2 | 365.817 | 4 / 1 | 3.2 | Yes |
| 9 | 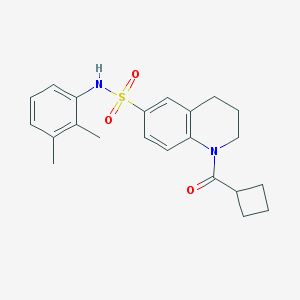 CHEMBL1354533 CHEMBL1354533 | C22H26N2O3S | 398.521 | 4 / 1 | 3.9 | Yes |
| 13 |  MLS001098390 MLS001098390 | C22H27BrN4O4 | 491.386 | 5 / 2 | 4.3 | Yes |
| 20 |  SMR000082495 SMR000082495 | C23H24N4O3 | 404.47 | 4 / 1 | 1.9 | Yes |
| 22 |  CHEMBL1734241 CHEMBL1734241 | C18H19F3N4O4S2 | 476.489 | 10 / 2 | 2.1 | Yes |
| 24 |  AC1NKC6O AC1NKC6O | C24H34N2O4 | 414.546 | 5 / 2 | 3.7 | Yes |
| 28 | 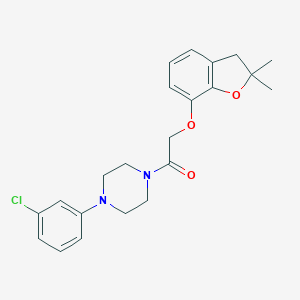 AC1P9W8Y AC1P9W8Y | C22H25ClN2O3 | 400.903 | 4 / 0 | 4.3 | Yes |
| 40 | 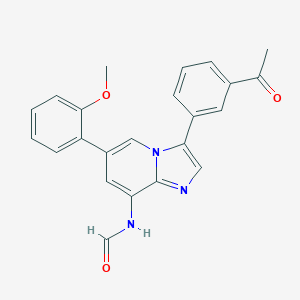 MLS003124119 MLS003124119 | C23H19N3O3 | 385.423 | 4 / 1 | 3.9 | Yes |
| 44 | 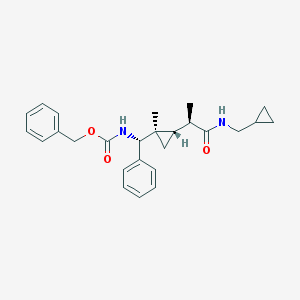 UPCMLD04ASTW002121 UPCMLD04ASTW002121 | C26H32N2O3 | 420.553 | 3 / 2 | 4.6 | Yes |
| 52 | 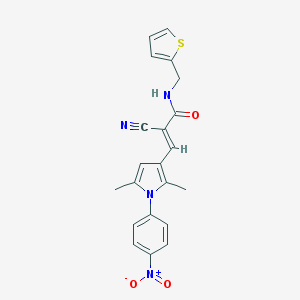 SMR000115985 SMR000115985 | C21H18N4O3S | 406.46 | 5 / 1 | 3.8 | Yes |
| 55 | 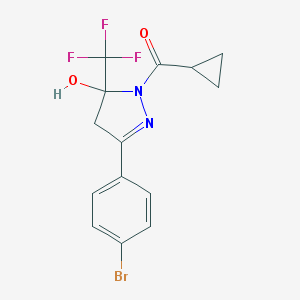 3-(4-bromophenyl)-1-(cyclopropylcarbonyl)-5-(trifluoromethyl)-4,5-dihydro-1H-pyrazol-5-ol 3-(4-bromophenyl)-1-(cyclopropylcarbonyl)-5-(trifluoromethyl)-4,5-dihydro-1H-pyrazol-5-ol | C14H12BrF3N2O2 | 377.161 | 6 / 1 | 3.3 | Yes |
| 58 | 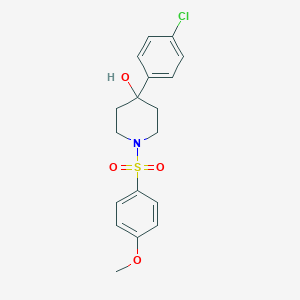 SMR000006315 SMR000006315 | C18H20ClNO4S | 381.871 | 5 / 1 | 2.8 | Yes |
| 61 | 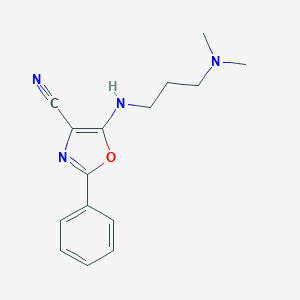 5-{[3-(dimethylamino)propyl]amino}-2-phenyl-1,3-oxazole-4-carbonitrile 5-{[3-(dimethylamino)propyl]amino}-2-phenyl-1,3-oxazole-4-carbonitrile | C15H18N4O | 270.336 | 5 / 1 | 3.3 | Yes |
| 65 | 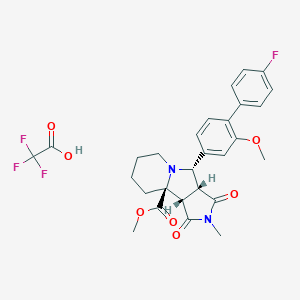 SMR001217115 SMR001217115 | C28H28F4N2O7 | 580.533 | 12 / 1 | N/A | No |
| 66 |  MLS000094053 MLS000094053 | C16H16FN5O2S | 361.395 | 8 / 0 | 3.5 | Yes |
| 100 |  2-(4-chlorophenyl)-N'-(3-pyridinylmethylene)-4-quinolinecarbohydrazide 2-(4-chlorophenyl)-N'-(3-pyridinylmethylene)-4-quinolinecarbohydrazide | C22H15ClN4O | 386.839 | 4 / 1 | 4.3 | Yes |
| 105 |  SMR000038563 SMR000038563 | C25H31N7O3 | 477.569 | 6 / 1 | 2.4 | Yes |
| 107 |  MLS001361981 MLS001361981 | C28H30F4N2O6 | 566.55 | 11 / 2 | N/A | No |
| 113 | 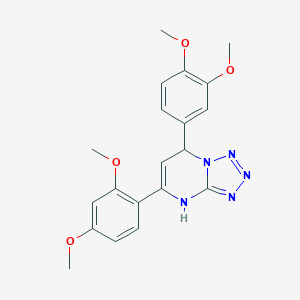 5-(2,4-dimethoxyphenyl)-7-(3,4-dimethoxyphenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine 5-(2,4-dimethoxyphenyl)-7-(3,4-dimethoxyphenyl)-4,7-dihydrotetrazolo[1,5-a]pyrimidine | C20H21N5O4 | 395.419 | 8 / 1 | 3.2 | Yes |
| 120 | 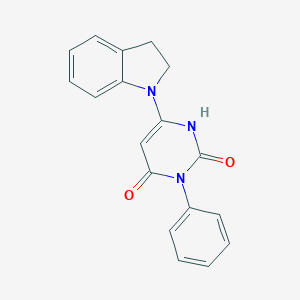 SMR000016661 SMR000016661 | C18H15N3O2 | 305.337 | 3 / 1 | 2.7 | Yes |
| 137 | 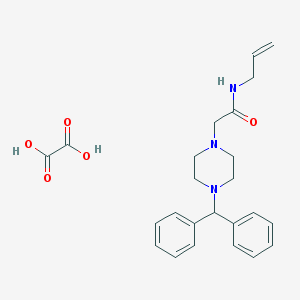 N-Allyl-2-(4-benzhydryl-piperazin-1-yl)-acetamide N-Allyl-2-(4-benzhydryl-piperazin-1-yl)-acetamide | C24H29N3O5 | 439.512 | 7 / 3 | N/A | N/A |
| 145 | 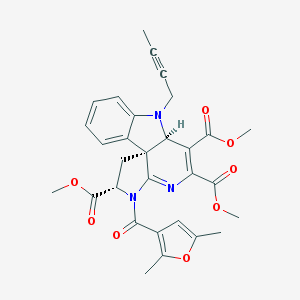 MLS000829697 MLS000829697 | C30H29N3O8 | 559.575 | 10 / 0 | 2.8 | No |
| 146 | 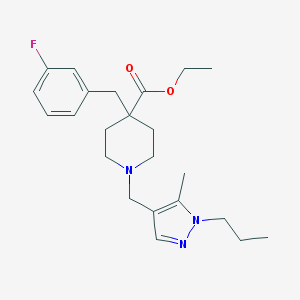 MLS000735125 MLS000735125 | C23H32FN3O2 | 401.526 | 5 / 0 | 3.9 | Yes |
| 154 | 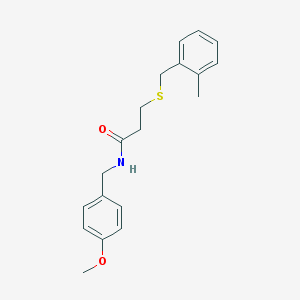 IFLab1_004408 IFLab1_004408 | C19H23NO2S | 329.458 | 3 / 1 | 3.4 | Yes |
| 155 | 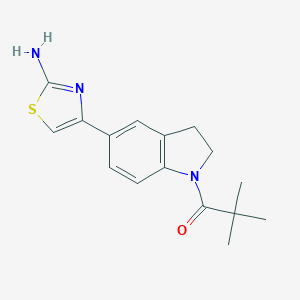 MLS000527948 MLS000527948 | C16H19N3OS | 301.408 | 4 / 1 | 3.1 | Yes |
| 158 | 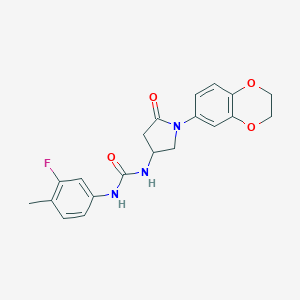 MLS001235152 MLS001235152 | C20H20FN3O4 | 385.395 | 5 / 2 | 1.9 | Yes |
| 162 | 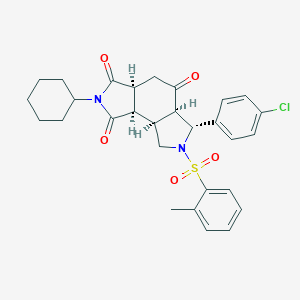 MLS003178907 MLS003178907 | C29H31ClN2O5S | 555.086 | 6 / 0 | 3.9 | No |
| 163 | 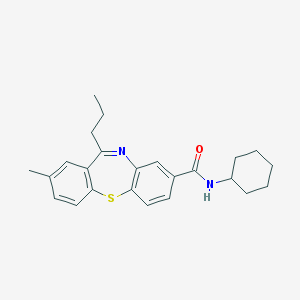 CHEMBL1396550 CHEMBL1396550 | C24H28N2OS | 392.561 | 3 / 1 | 5.8 | No |
| 166 | 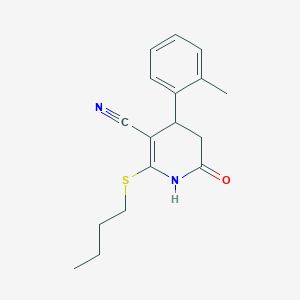 MLS000103088 MLS000103088 | C17H20N2OS | 300.42 | 3 / 1 | 3.2 | Yes |
| 179 | 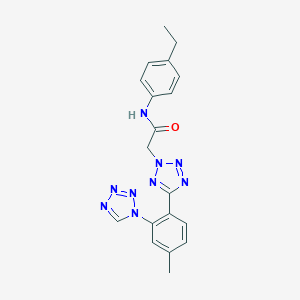 AC1LRY9S AC1LRY9S | C19H19N9O | 389.423 | 7 / 1 | 3.2 | Yes |
| 180 | 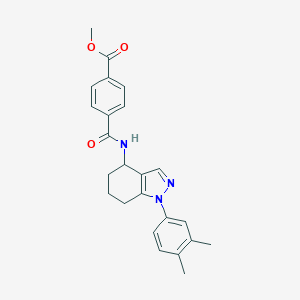 MLS001072616 MLS001072616 | C24H25N3O3 | 403.482 | 4 / 1 | 4.1 | Yes |
| 184 | 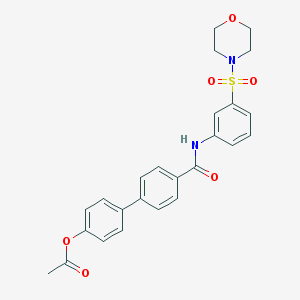 AC1MOBES AC1MOBES | C25H24N2O6S | 480.535 | 7 / 1 | 2.9 | Yes |
| 189 | 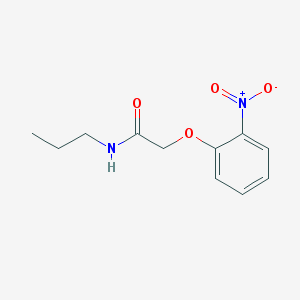 2-(2-nitrophenoxy)-N-propylacetamide 2-(2-nitrophenoxy)-N-propylacetamide | C11H14N2O4 | 238.243 | 4 / 1 | 1.6 | Yes |
| 191 | 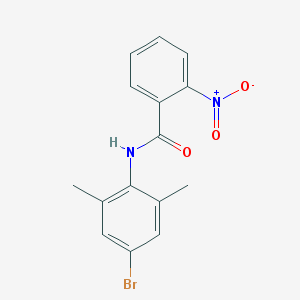 N-(4-bromo-2,6-dimethylphenyl)-2-nitrobenzamide N-(4-bromo-2,6-dimethylphenyl)-2-nitrobenzamide | C15H13BrN2O3 | 349.184 | 3 / 1 | 3.2 | Yes |
| 192 | 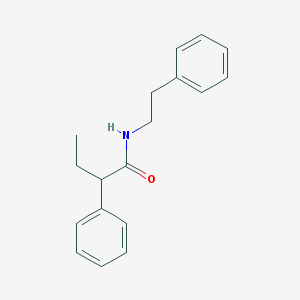 2-phenyl-N-(2-phenylethyl)butanamide 2-phenyl-N-(2-phenylethyl)butanamide | C18H21NO | 267.372 | 1 / 1 | 4.0 | Yes |
| 193 | 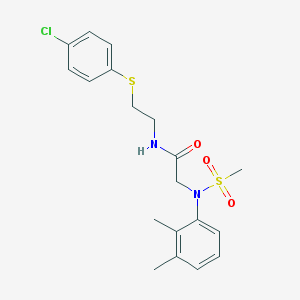 N-{2-[(4-chlorophenyl)sulfanyl]ethyl}-N~2~-(2,3-dimethylphenyl)-N~2~-(methylsulfonyl)glycinamide N-{2-[(4-chlorophenyl)sulfanyl]ethyl}-N~2~-(2,3-dimethylphenyl)-N~2~-(methylsulfonyl)glycinamide | C19H23ClN2O3S2 | 426.974 | 5 / 1 | 4.0 | Yes |
| 195 | 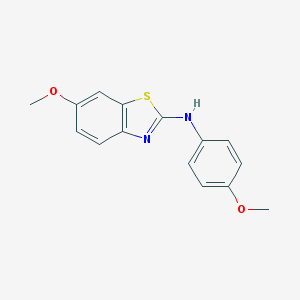 MLS000684722 MLS000684722 | C15H14N2O2S | 286.349 | 5 / 1 | 4.2 | Yes |
| 199 | 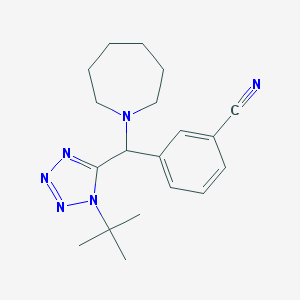 3-[Azepan-1-yl-(1-tert-butyl-1H-tetrazol-5-yl)-methyl]-benzonitrile 3-[Azepan-1-yl-(1-tert-butyl-1H-tetrazol-5-yl)-methyl]-benzonitrile | C19H26N6 | 338.459 | 5 / 0 | 2.9 | Yes |
| 204 | 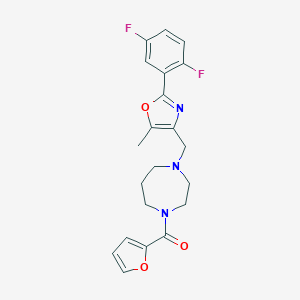 MLS000733515 MLS000733515 | C21H21F2N3O3 | 401.414 | 7 / 0 | 3.2 | Yes |
| 205 | 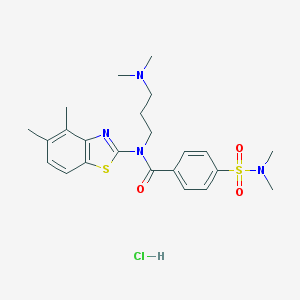 MLS000684946 MLS000684946 | C23H31ClN4O3S2 | 511.096 | 7 / 1 | N/A | No |
| 210 | 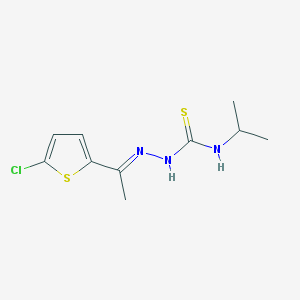 MLS000392662 MLS000392662 | C10H14ClN3S2 | 275.813 | 3 / 2 | 3.6 | Yes |
| 217 | 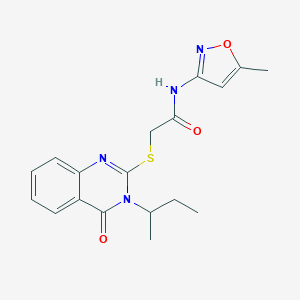 MLS000569662 MLS000569662 | C18H20N4O3S | 372.443 | 6 / 1 | 3.2 | Yes |
| 220 | 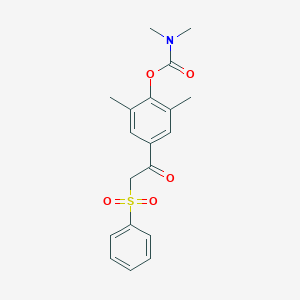 2,6-dimethyl-4-[2-(phenylsulfonyl)acetyl]phenyl N,N-dimethylcarbamate 2,6-dimethyl-4-[2-(phenylsulfonyl)acetyl]phenyl N,N-dimethylcarbamate | C19H21NO5S | 375.439 | 5 / 0 | 3.1 | Yes |
| 222 | 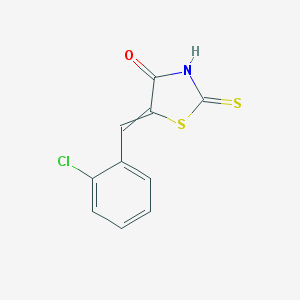 MLS002639376 MLS002639376 | C10H6ClNOS2 | 255.734 | 3 / 1 | 3.4 | Yes |
| 557312 | 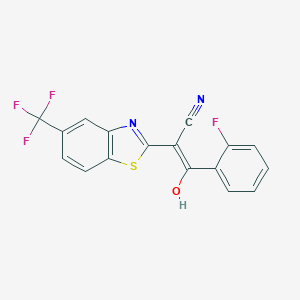 AC1NWZI7 AC1NWZI7 | C17H8F4N2OS | 364.318 | 8 / 1 | 4.6 | Yes |
| 557314 | 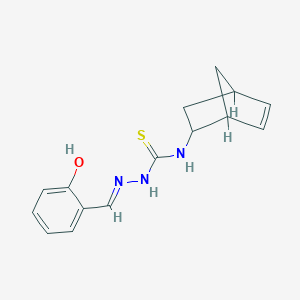 MLS000849868 MLS000849868 | C15H17N3OS | 287.381 | 3 / 3 | 2.5 | Yes |
| 557315 | 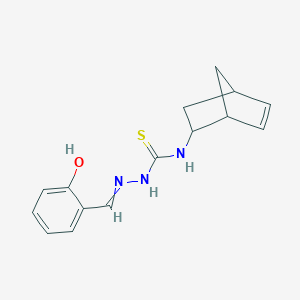 MLS003115380 MLS003115380 | C15H17N3OS | 287.381 | 3 / 3 | 2.5 | Yes |
| 232 | 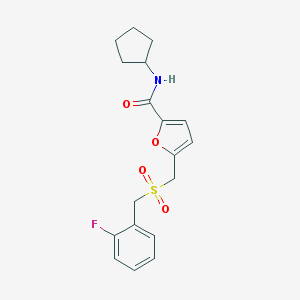 AC1NNPKR AC1NNPKR | C18H20FNO4S | 365.419 | 5 / 1 | 2.7 | Yes |
| 247 | 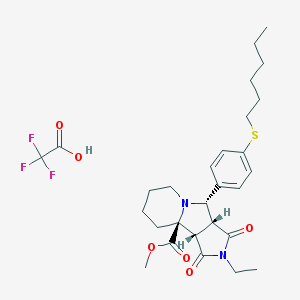 MLS001362111 MLS001362111 | C28H37F3N2O6S | 586.667 | 11 / 1 | N/A | No |
| 248 | 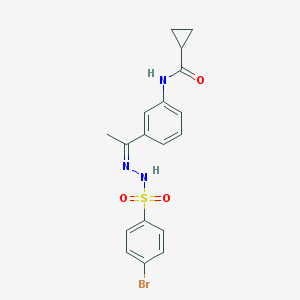 AC1NUSAF AC1NUSAF | C18H18BrN3O3S | 436.324 | 5 / 2 | 3.1 | Yes |
| 252 |  MLS000912240 MLS000912240 | C19H14ClN3O3 | 367.789 | 4 / 2 | 3.7 | Yes |
| 253 |  4-ethyl-N-[3-(trifluoromethyl)phenyl]piperazine-1-carbothioamide 4-ethyl-N-[3-(trifluoromethyl)phenyl]piperazine-1-carbothioamide | C14H18F3N3S | 317.374 | 5 / 1 | 2.8 | Yes |
| 254 |  CHEMBL1320515 CHEMBL1320515 | C18H17ClN2O2S | 360.856 | 3 / 1 | 2.9 | Yes |
| 255 |  BAS 01923086 BAS 01923086 | C20H23FN2O3S | 390.473 | 5 / 1 | 3.1 | Yes |
| 256 | 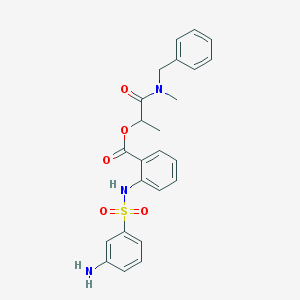 SMR000243071 SMR000243071 | C24H25N3O5S | 467.54 | 7 / 2 | 3.6 | Yes |
| 257 | 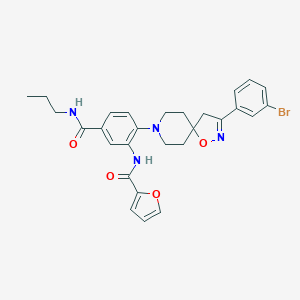 MLS001074397 MLS001074397 | C28H29BrN4O4 | 565.468 | 6 / 2 | 5.1 | No |
| 263 | 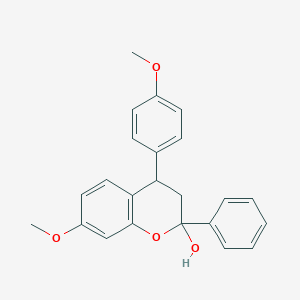 MLS000882561 MLS000882561 | C23H22O4 | 362.425 | 4 / 1 | 4.4 | Yes |
| 264 | 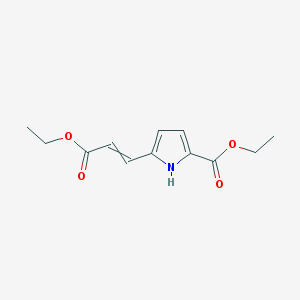 MLS003106901 MLS003106901 | C12H15NO4 | 237.255 | 4 / 1 | 1.9 | Yes |
| 268 | 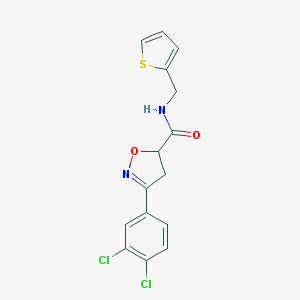 BAS 09528871 BAS 09528871 | C15H12Cl2N2O2S | 355.233 | 4 / 1 | 3.6 | Yes |
| 281 | 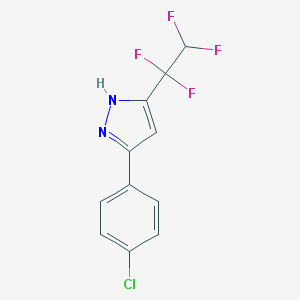 3-(4-chlorophenyl)-5-(1,1,2,2-tetrafluoroethyl)-1H-pyrazole 3-(4-chlorophenyl)-5-(1,1,2,2-tetrafluoroethyl)-1H-pyrazole | C11H7ClF4N2 | 278.635 | 5 / 1 | 3.8 | Yes |
| 283 | 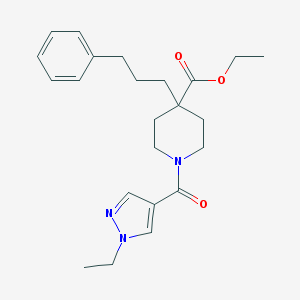 MLS000734874 MLS000734874 | C23H31N3O3 | 397.519 | 4 / 0 | 3.4 | Yes |
| 284 | 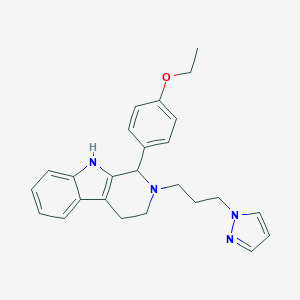 1-(4-ethoxyphenyl)-2-[3-(1H-pyrazol-1-yl)propyl]-2,3,4,9-tetrahydro-1H-beta-carboline 1-(4-ethoxyphenyl)-2-[3-(1H-pyrazol-1-yl)propyl]-2,3,4,9-tetrahydro-1H-beta-carboline | C25H28N4O | 400.526 | 3 / 1 | 4.3 | Yes |
| 285 | 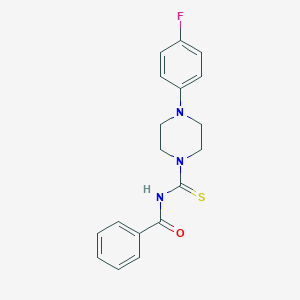 AC1LE41X AC1LE41X | C18H18FN3OS | 343.42 | 4 / 1 | 3.3 | Yes |
| 286 | 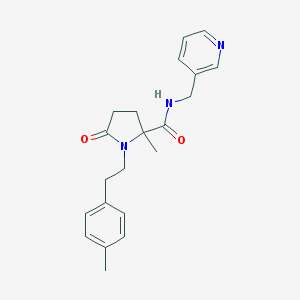 SMR000131481 SMR000131481 | C21H25N3O2 | 351.45 | 3 / 1 | 2.1 | Yes |
| 296 | 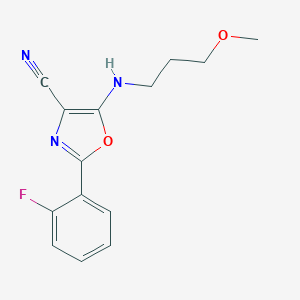 SMR000091688 SMR000091688 | C14H14FN3O2 | 275.283 | 6 / 1 | 3.2 | Yes |
| 306 | 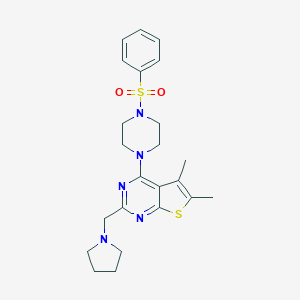 MLS002157455 MLS002157455 | C23H29N5O2S2 | 471.638 | 8 / 0 | 3.8 | Yes |
| 307 | 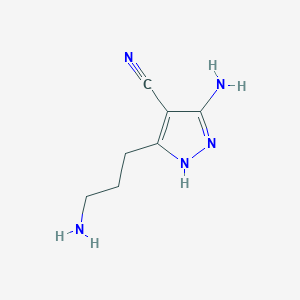 5-amino-3-(3-aminopropyl)-1H-pyrazole-4-carbonitrile 5-amino-3-(3-aminopropyl)-1H-pyrazole-4-carbonitrile | C7H11N5 | 165.2 | 4 / 3 | -0.1 | Yes |
| 309 | 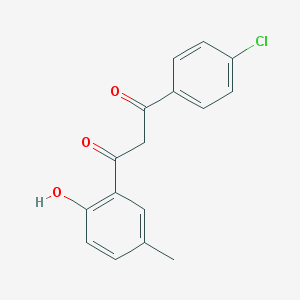 1-(4-chlorophenyl)-3-(2-hydroxy-5-methylphenyl)propane-1,3-dione 1-(4-chlorophenyl)-3-(2-hydroxy-5-methylphenyl)propane-1,3-dione | C16H13ClO3 | 288.727 | 3 / 1 | 4.3 | Yes |
| 310 | 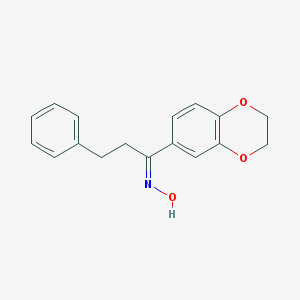 AC1NT5A7 AC1NT5A7 | C17H17NO3 | 283.327 | 4 / 1 | 3.5 | Yes |
| 311 | 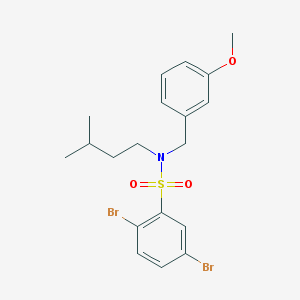 MLS002768222 MLS002768222 | C19H23Br2NO3S | 505.265 | 4 / 0 | 5.6 | No |
| 313 | 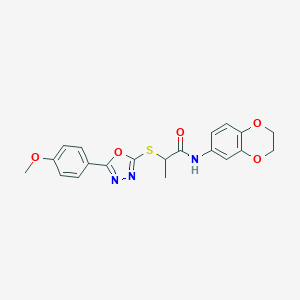 SMR000065140 SMR000065140 | C20H19N3O5S | 413.448 | 8 / 1 | 3.2 | Yes |
| 314 | 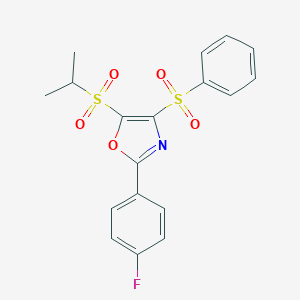 CHEMBL1559360 CHEMBL1559360 | C18H16FNO5S2 | 409.446 | 7 / 0 | 3.6 | Yes |
| 315 | 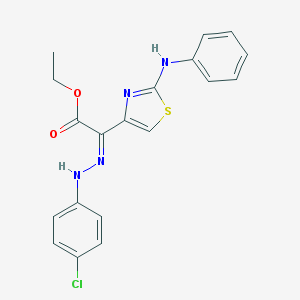 SMR000155256 SMR000155256 | C19H17ClN4O2S | 400.881 | 7 / 2 | 6.6 | No |
| 317 | 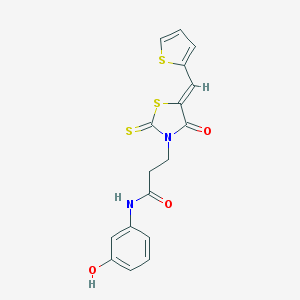 CHEMBL179161 CHEMBL179161 | C17H14N2O3S3 | 390.49 | 6 / 2 | 3.1 | Yes |
| 328 |  N-[2-(benzyloxy)-5-bromobenzyl]-3-[(1-methyl-1H-tetrazol-5-yl)sulfanyl]propan-1-amine N-[2-(benzyloxy)-5-bromobenzyl]-3-[(1-methyl-1H-tetrazol-5-yl)sulfanyl]propan-1-amine | C19H22BrN5OS | 448.383 | 6 / 1 | 4.0 | Yes |
| 329 |  MLS000757188 MLS000757188 | C19H25NO2 | 299.414 | 2 / 1 | 6.1 | No |
| 330 |  AC1LLI6B AC1LLI6B | C21H26BrNO2 | 404.348 | 3 / 0 | 5.1 | No |
| 334 |  MLS000336683 MLS000336683 | C21H20N4O3S | 408.476 | 7 / 2 | 3.6 | Yes |
| 339 | 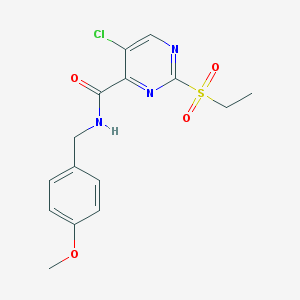 5-chloro-2-(ethylsulfonyl)-N-(4-methoxybenzyl)pyrimidine-4-carboxamide 5-chloro-2-(ethylsulfonyl)-N-(4-methoxybenzyl)pyrimidine-4-carboxamide | C15H16ClN3O4S | 369.82 | 6 / 1 | 1.8 | Yes |
| 345 | 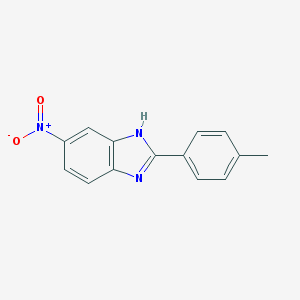 5-Nitro-2-p-tolyl-1H-benzoimidazole 5-Nitro-2-p-tolyl-1H-benzoimidazole | C14H11N3O2 | 253.261 | 3 / 1 | 3.8 | Yes |
| 346 | 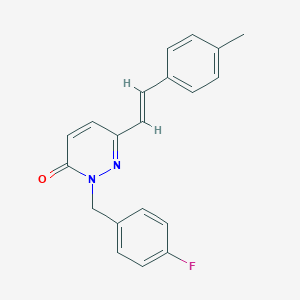 2-(4-fluorobenzyl)-6-(4-methylstyryl)-3(2H)-pyridazinone 2-(4-fluorobenzyl)-6-(4-methylstyryl)-3(2H)-pyridazinone | C20H17FN2O | 320.367 | 3 / 0 | 3.8 | Yes |
| 350 | 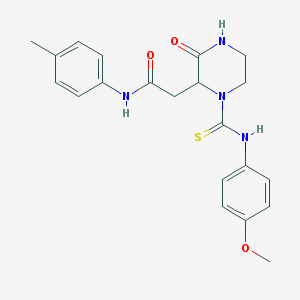 2-{1-[(4-methoxyphenyl)carbamothioyl]-3-oxopiperazin-2-yl}-N-(4-methylphenyl)acetamide 2-{1-[(4-methoxyphenyl)carbamothioyl]-3-oxopiperazin-2-yl}-N-(4-methylphenyl)acetamide | C21H24N4O3S | 412.508 | 4 / 3 | 2.0 | Yes |
| 352 | 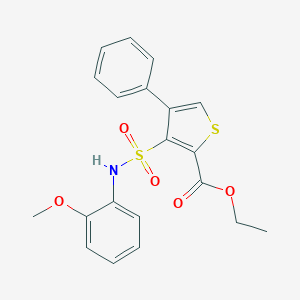 CHEMBL1435065 CHEMBL1435065 | C20H19NO5S2 | 417.494 | 7 / 1 | 4.3 | Yes |
| 362 | 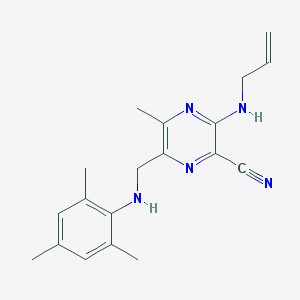 AC1OJ1IV AC1OJ1IV | C19H23N5 | 321.428 | 5 / 2 | 4.4 | Yes |
| 367 | 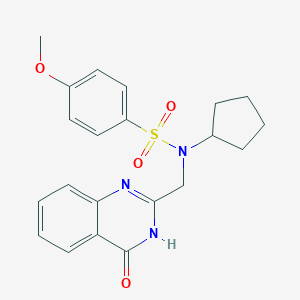 SMR000005572 SMR000005572 | C21H23N3O4S | 413.492 | 6 / 1 | 2.8 | Yes |
| 372 | 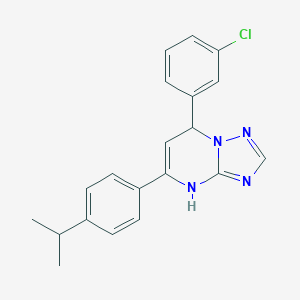 AC1O08MQ AC1O08MQ | C20H19ClN4 | 350.85 | 3 / 1 | 5.5 | No |
| 373 | 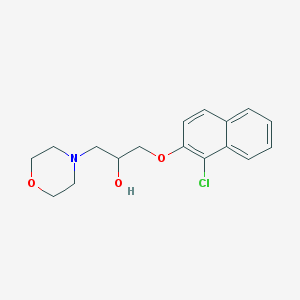 F1243-0137 F1243-0137 | C17H20ClNO3 | 321.801 | 4 / 1 | 2.8 | Yes |
| 381 | 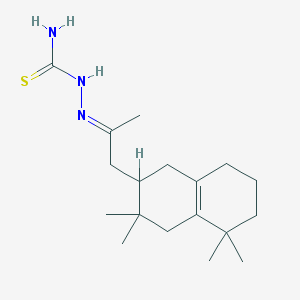 MLS000526436 MLS000526436 | C18H31N3S | 321.527 | 2 / 2 | 3.7 | Yes |
| 387 | 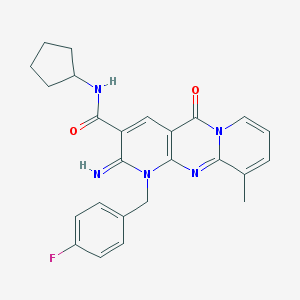 AC1MR1M1 AC1MR1M1 | C25H24FN5O2 | 445.498 | 5 / 2 | 2.7 | Yes |
| 391 | 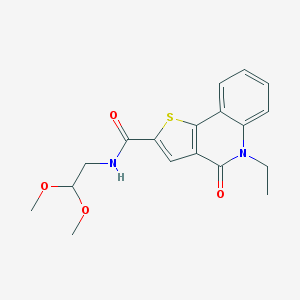 ChemDiv3_010013 ChemDiv3_010013 | C18H20N2O4S | 360.428 | 5 / 1 | 2.2 | Yes |
| 395 | 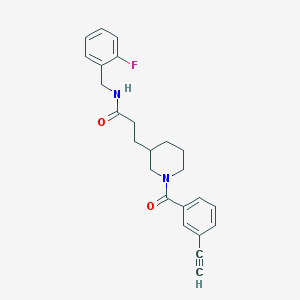 MLS001100937 MLS001100937 | C24H25FN2O2 | 392.474 | 3 / 1 | 3.6 | Yes |
| 399 | 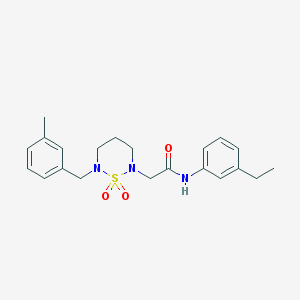 CHEMBL1417984 CHEMBL1417984 | C21H27N3O3S | 401.525 | 5 / 1 | 3.1 | Yes |
| 400 | 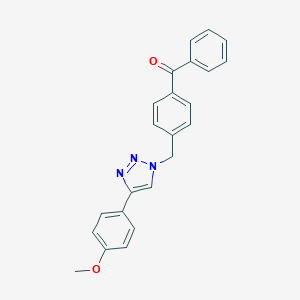 MLS000882705 MLS000882705 | C23H19N3O2 | 369.424 | 4 / 0 | 4.3 | Yes |
| 401 | 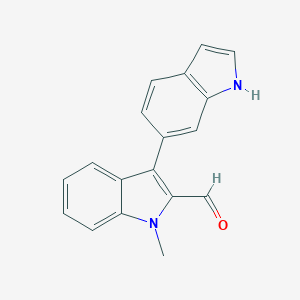 MLS002402777 MLS002402777 | C18H14N2O | 274.323 | 1 / 1 | 3.5 | Yes |
| 407 | 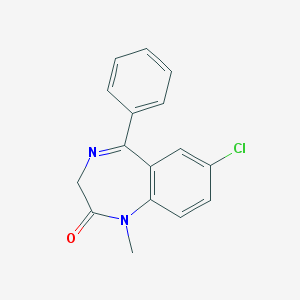 diazepam diazepam | C16H13ClN2O | 284.743 | 2 / 0 | 3.0 | Yes |
| 410 | 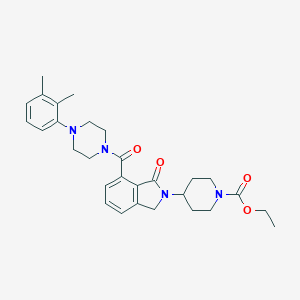 ethyl 4-(7-{[4-(2,3-dimethylphenyl)piperazin-1-yl]carbonyl}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-1-carboxylate ethyl 4-(7-{[4-(2,3-dimethylphenyl)piperazin-1-yl]carbonyl}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-1-carboxylate | C29H36N4O4 | 504.631 | 5 / 0 | 3.6 | No |
| 416 | 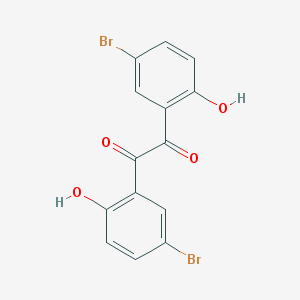 Dibromosalicil Dibromosalicil | C14H8Br2O4 | 400.022 | 4 / 2 | 4.7 | Yes |
| 417 | 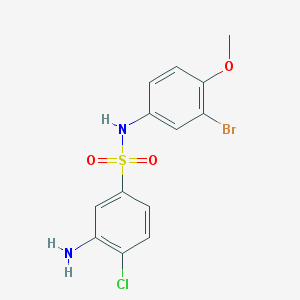 3-amino-N-(3-bromo-4-methoxyphenyl)-4-chlorobenzene-1-sulfonamide 3-amino-N-(3-bromo-4-methoxyphenyl)-4-chlorobenzene-1-sulfonamide | C13H12BrClN2O3S | 391.664 | 5 / 2 | 3.1 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417