

You can:
| Name | Metabotropic glutamate receptor 6 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm6 |
| Synonym | nob3 nob2 nerg1 mGluR6 mGlu6 receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 871 |
| Amino acid sequence | MGRLPVLLLWLAWWLSQAGIACGAGSVRLAGGLTLGGLFPVHARGAAGRACGALKKEQGVHRLEAMLYALDRVNADPELLPGVRLGARLLDTCSRDTYALEQALSFVQALIRGRGDGDEASVRCPGGVPPLRSAPPERVVAVVGASASSVSIMVANVLRLFAIPQISYASTAPELSDSTRYDFFSRVVPPDSYQAQAMVDIVRALGWNYVSTLASEGNYGESGVEAFVQISREAGGVCIAQSIKIPREPKPGEFHKVIRRLMETPNARGIIIFANEDDIRRVLEATRQANLTGHFLWVGSDSWGSKISPILNLEEEAVGAITILPKRASIDGFDQYFMTRSLENNRRNIWFAEFWEENFNCKLTSSGGQSDDSTRKCTGEERIGQDSAYEQEGKVQFVIDAVYAIAHALHSMHQALCPGHTGLCPAMEPTDGRTLLHYIRAVRFNGSAGTPVMFNENGDAPGRYDIFQYQATNGSASSGGYQAVGQWAEALRLDMEVLRWSGDPHEVPPSQCSLPCGPGERKKMVKGVPCCWHCEACDGYRFQVDEFTCEACPGDMRPTPNHTGCRPTPVVRLTWSSPWAALPLLLAVLGIMATTTIMATFMRHNDTPIVRASGRELSYVLLTGIFLIYAITFLMVAEPCAAICAARRLLLGLGTTLSYSALLTKTNRIYRIFEQGKRSVTPPPFISPTSQLVITFGLTSLQVVGVIAWLGAQPPHSVIDYEEQRTVDPEQARGVLKCDMSDLSLIGCLGYSLLLMVTCTVYAIKARGVPETFNEAKPIGFTMYTTCIIWLAFVPIFFGTAQSAEKIYIQTTTLTVSLSLSASVSLGMLYVPKTYVILFHPEQNVQKRKRSLKKTSTMAAPPQNENAEDAK |
| UniProt | P35349 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3842 |
| IUPHAR | 294 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 535989 | 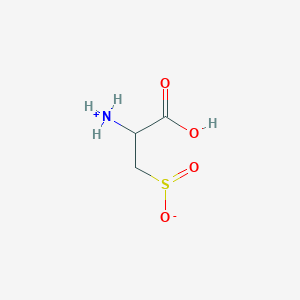 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 557553 | 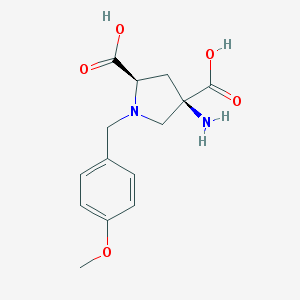 CHEMBL543097 CHEMBL543097 | C14H18N2O5 | 294.307 | 7 / 3 | -4.6 | Yes |
| 10399 | 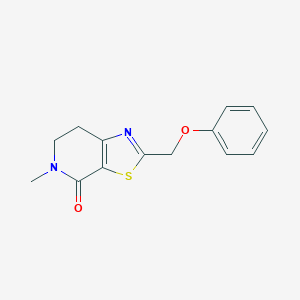 CHEMBL2426616 CHEMBL2426616 | C14H14N2O2S | 274.338 | 4 / 0 | 2.3 | Yes |
| 13328 | 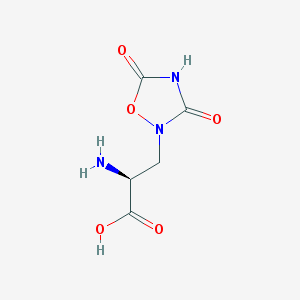 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 536572 | 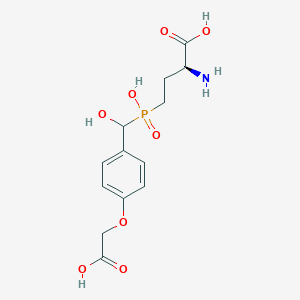 LSP4-2022 LSP4-2022 | C13H18NO8P | 347.26 | 9 / 5 | -3.8 | Yes |
| 522261 | 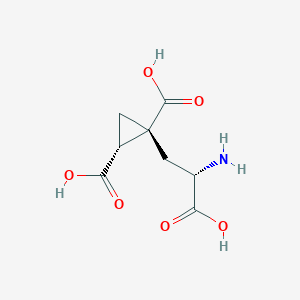 CHEMBL3786667 CHEMBL3786667 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522274 | 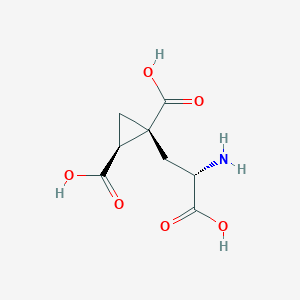 CHEMBL3787264 CHEMBL3787264 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522279 | 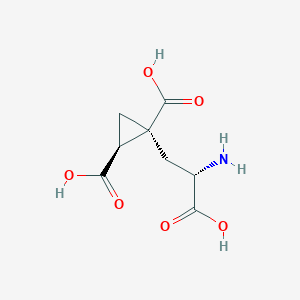 CHEMBL3786026 CHEMBL3786026 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522287 | 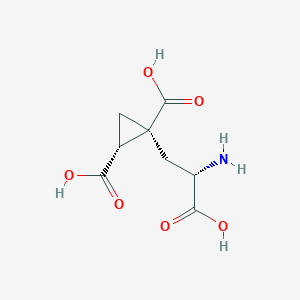 CHEMBL3786938 CHEMBL3786938 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 33520 | 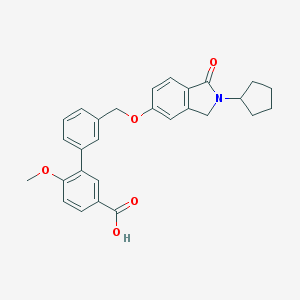 CHEMBL2179631 CHEMBL2179631 | C28H27NO5 | 457.526 | 5 / 1 | 4.8 | Yes |
| 36077 | 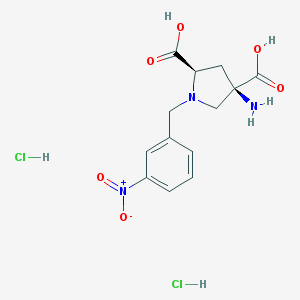 CHEMBL539760 CHEMBL539760 | C13H17Cl2N3O6 | 382.194 | 8 / 5 | N/A | N/A |
| 36686 | 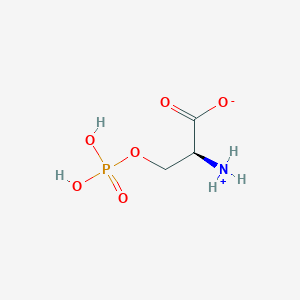 CID 57689797 CID 57689797 | C3H8NO6P | 185.072 | 6 / 3 | -4.5 | Yes |
| 42848 | 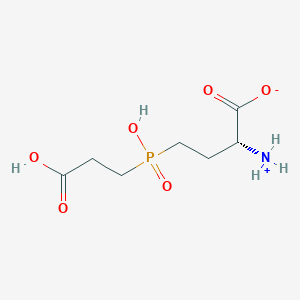 MolPort-000-743-361 MolPort-000-743-361 | C7H14NO6P | 239.164 | 6 / 3 | -4.2 | Yes |
| 42852 | 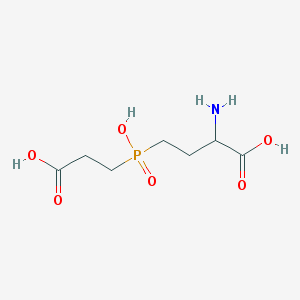 AC1MWWXC AC1MWWXC | C7H14NO6P | 239.164 | 7 / 4 | -4.8 | Yes |
| 42857 | 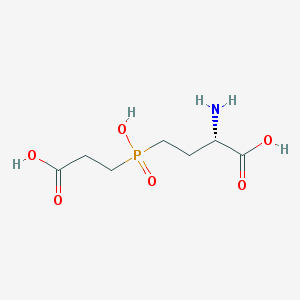 CHEMBL1089515 CHEMBL1089515 | C7H14NO6P | 239.164 | 7 / 4 | -4.8 | Yes |
| 522874 | 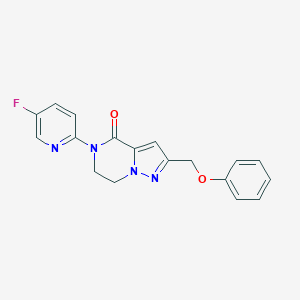 CHEMBL3746457 CHEMBL3746457 | C18H15FN4O2 | 338.342 | 5 / 0 | 2.2 | Yes |
| 519957 | 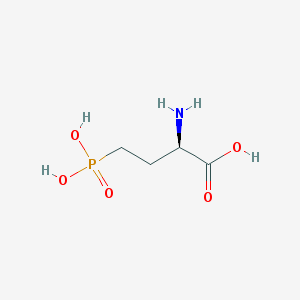 D-AP4 D-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 57277 | 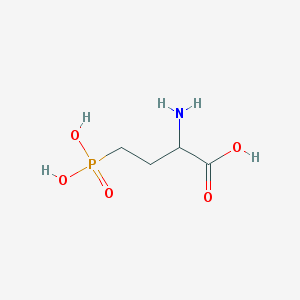 2-amino-4-phosphonobutanoic acid 2-amino-4-phosphonobutanoic acid | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 57291 | 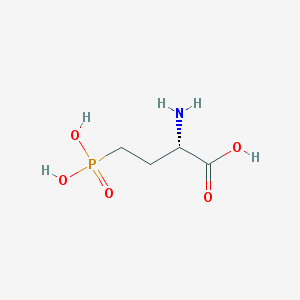 L-AP4 L-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 62854 | 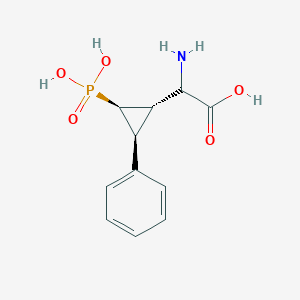 CHEMBL392419 CHEMBL392419 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 62867 | 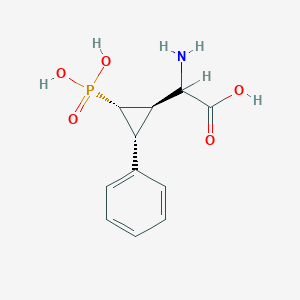 CHEMBL392420 CHEMBL392420 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 559361 | 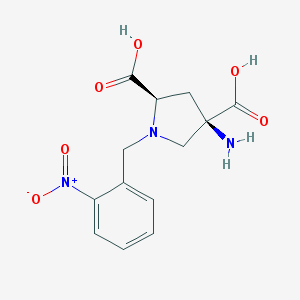 CHEMBL544508 CHEMBL544508 | C13H15N3O6 | 309.278 | 8 / 3 | -4.7 | Yes |
| 81848 | 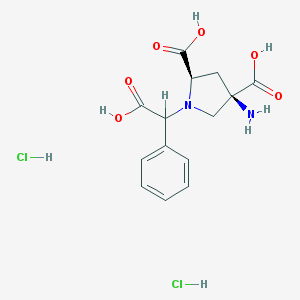 CHEMBL538489 CHEMBL538489 | C14H18Cl2N2O6 | 381.206 | 8 / 6 | N/A | No |
| 82269 | 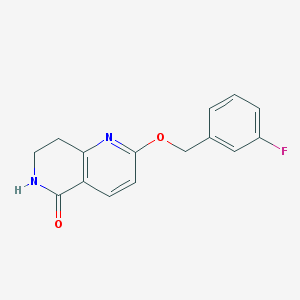 CHEMBL3298273 CHEMBL3298273 | C15H13FN2O2 | 272.279 | 4 / 1 | 2.1 | Yes |
| 553693 | 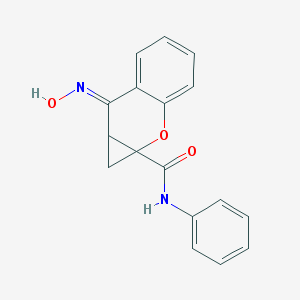 PHCCC PHCCC | C17H14N2O3 | 294.31 | 4 / 2 | 2.6 | Yes |
| 85070 | 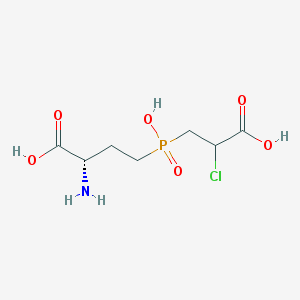 CHEMBL1076865 CHEMBL1076865 | C7H13ClNO6P | 273.606 | 7 / 4 | -4.1 | Yes |
| 560007 | 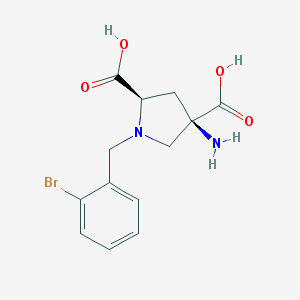 CHEMBL552984 CHEMBL552984 | C13H15BrN2O4 | 343.177 | 6 / 3 | -3.8 | Yes |
| 553764 | 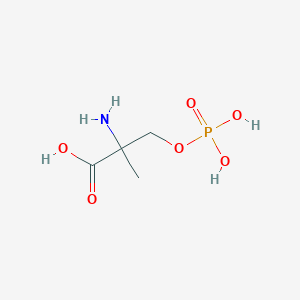 MSOP MSOP | C4H10NO6P | 199.099 | 7 / 4 | -5.0 | Yes |
| 109309 | 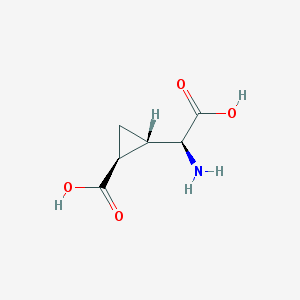 (2S,1'S,2'S)-2-(carboxycyclopropyl)glycine (2S,1'S,2'S)-2-(carboxycyclopropyl)glycine | C6H9NO4 | 159.141 | 5 / 3 | -3.4 | Yes |
| 538737 | 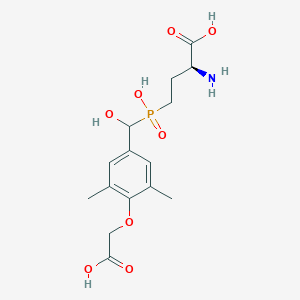 CHEMBL3912541 CHEMBL3912541 | C15H22NO8P | 375.314 | 9 / 5 | -3.1 | Yes |
| 560797 | 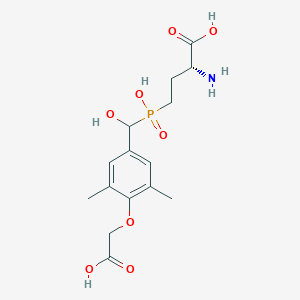 US9212196, Derivative 10 US9212196, Derivative 10 | C15H22NO8P | 375.314 | 9 / 5 | -3.1 | Yes |
| 560881 | 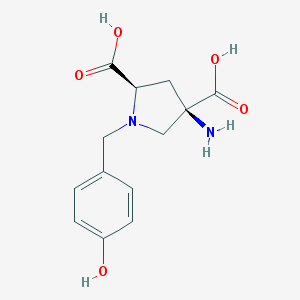 CHEMBL543568 CHEMBL543568 | C13H16N2O5 | 280.28 | 7 / 4 | -4.9 | Yes |
| 116403 | 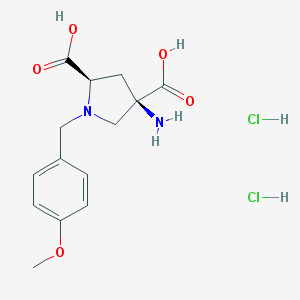 CHEMBL543097 CHEMBL543097 | C14H20Cl2N2O5 | 367.223 | 7 / 5 | N/A | N/A |
| 553879 | 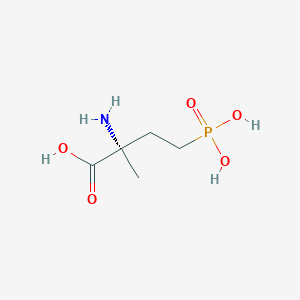 MAP4 MAP4 | C5H12NO5P | 197.127 | 6 / 4 | -4.6 | Yes |
| 122379 | 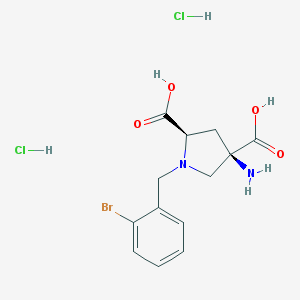 CHEMBL552984 CHEMBL552984 | C13H17BrCl2N2O4 | 416.093 | 6 / 5 | N/A | N/A |
| 128448 | 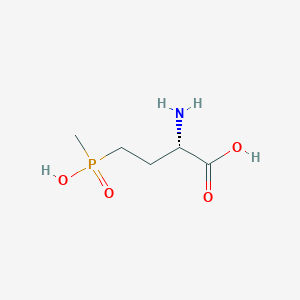 L-PHOSPHINOTHRICIN L-PHOSPHINOTHRICIN | C5H12NO4P | 181.128 | 5 / 3 | -5.0 | Yes |
| 130324 | 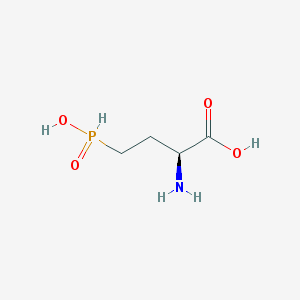 demethylphosphinothricin demethylphosphinothricin | C4H10NO4P | 167.101 | 5 / 3 | -5.1 | Yes |
| 561396 | 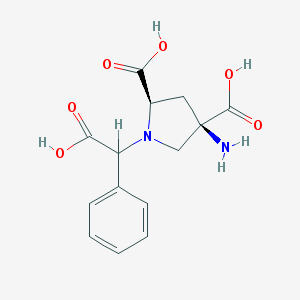 CHEMBL538489 CHEMBL538489 | C14H16N2O6 | 308.29 | 8 / 4 | -4.7 | Yes |
| 135013 | 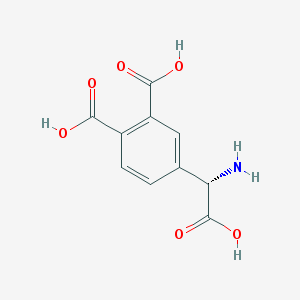 (S)-3,4-DCPG (S)-3,4-DCPG | C10H9NO6 | 239.183 | 7 / 4 | -2.7 | Yes |
| 561521 | 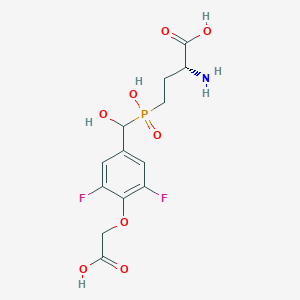 US9212196, Derivative 28 US9212196, Derivative 28 | C13H16F2NO8P | 383.241 | 11 / 5 | -3.6 | No |
| 539424 | 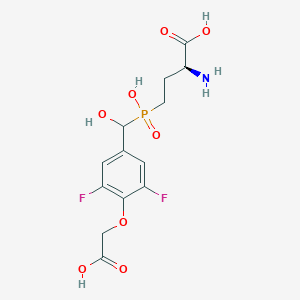 CHEMBL3896442 CHEMBL3896442 | C13H16F2NO8P | 383.241 | 11 / 5 | -3.6 | No |
| 140037 | 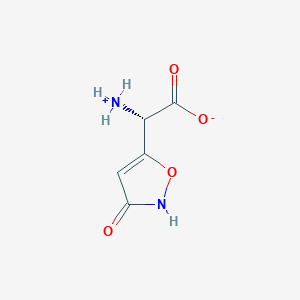 AC1ODZEZ AC1ODZEZ | C5H6N2O4 | 158.113 | 4 / 2 | -3.2 | Yes |
| 539802 | 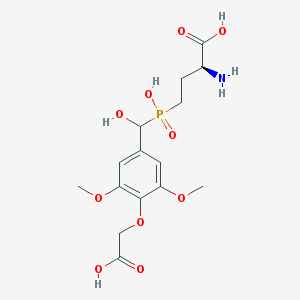 CHEMBL3982807 CHEMBL3982807 | C15H22NO10P | 407.312 | 11 / 5 | -3.9 | No |
| 561929 | 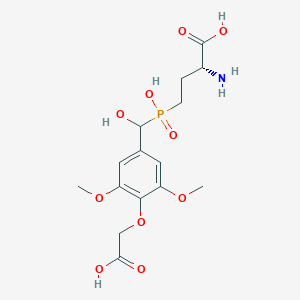 US9212196, Derivative 4 US9212196, Derivative 4 | C15H22NO10P | 407.312 | 11 / 5 | -3.9 | No |
| 562102 | 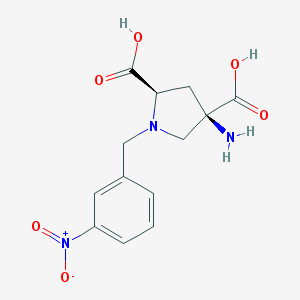 CHEMBL539760 CHEMBL539760 | C13H15N3O6 | 309.278 | 8 / 3 | -4.7 | Yes |
| 158297 | 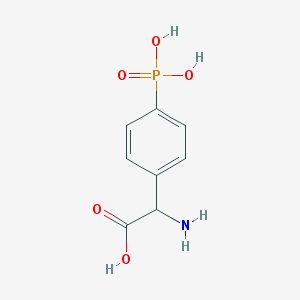 (RS)-PPG (RS)-PPG | C8H10NO5P | 231.144 | 6 / 4 | -3.7 | Yes |
| 161798 | 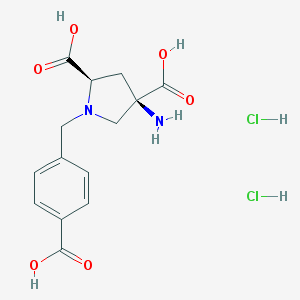 CHEMBL539761 CHEMBL539761 | C14H18Cl2N2O6 | 381.206 | 8 / 6 | N/A | No |
| 562416 | 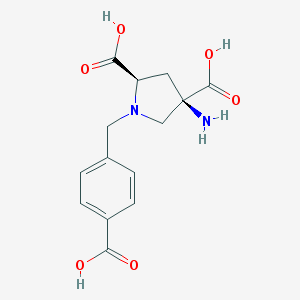 CHEMBL539761 CHEMBL539761 | C14H16N2O6 | 308.29 | 8 / 4 | -5.0 | Yes |
| 181958 | 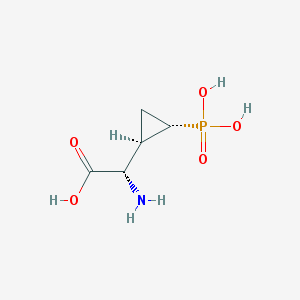 CHEMBL197976 CHEMBL197976 | C5H10NO5P | 195.111 | 6 / 4 | -4.7 | Yes |
| 181962 | 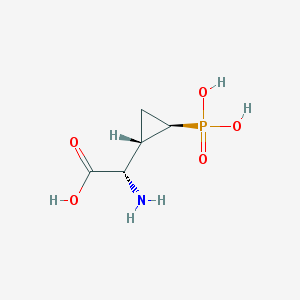 CHEMBL199626 CHEMBL199626 | C5H10NO5P | 195.111 | 6 / 4 | -4.7 | Yes |
| 181973 | 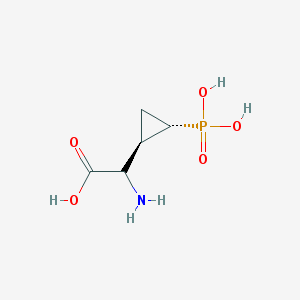 CHEMBL442076 CHEMBL442076 | C5H10NO5P | 195.111 | 6 / 4 | -4.7 | Yes |
| 182957 | 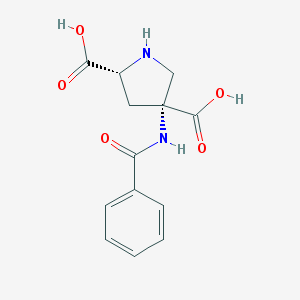 CHEMBL345381 CHEMBL345381 | C13H14N2O5 | 278.264 | 6 / 4 | -2.3 | Yes |
| 185812 | 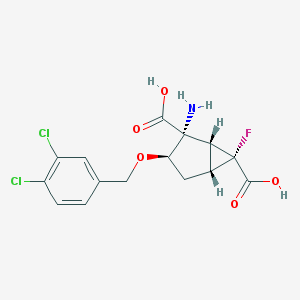 MGS-0039 MGS-0039 | C15H14Cl2FNO5 | 378.177 | 7 / 3 | -0.7 | Yes |
| 187019 | 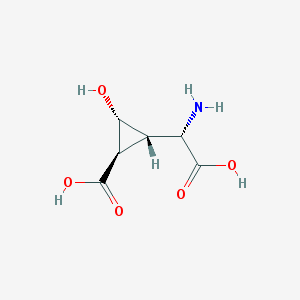 CHEMBL140197 CHEMBL140197 | C6H9NO5 | 175.14 | 6 / 4 | -3.9 | Yes |
| 188380 | 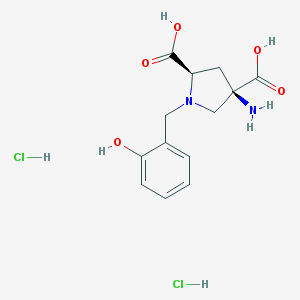 CHEMBL540015 CHEMBL540015 | C13H18Cl2N2O5 | 353.196 | 7 / 6 | N/A | No |
| 189223 | 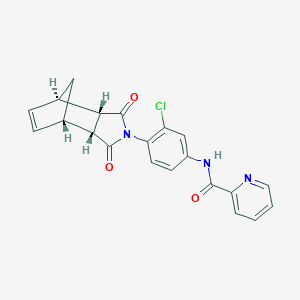 MLS003171619 MLS003171619 | C21H16ClN3O3 | 393.827 | 4 / 1 | 2.6 | Yes |
| 541002 | 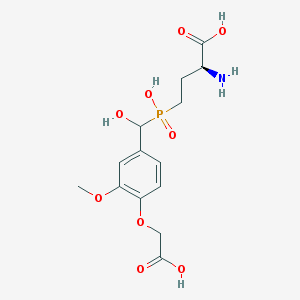 CHEMBL3935255 CHEMBL3935255 | C14H20NO9P | 377.286 | 10 / 5 | -3.9 | Yes |
| 563329 | 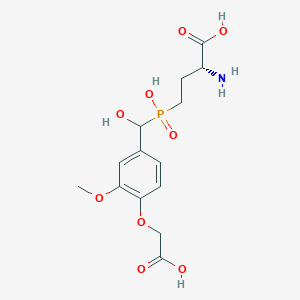 US9212196, Derivative 3 US9212196, Derivative 3 | C14H20NO9P | 377.286 | 10 / 5 | -3.9 | Yes |
| 192703 | 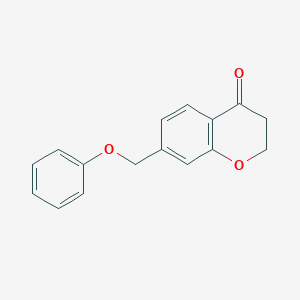 CHEMBL1682820 CHEMBL1682820 | C16H14O3 | 254.285 | 3 / 0 | 2.8 | Yes |
| 193425 | 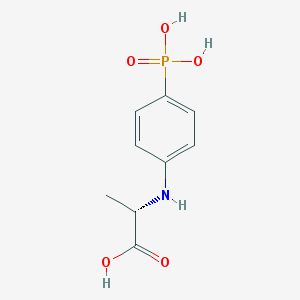 CHEMBL18439 CHEMBL18439 | C9H12NO5P | 245.171 | 6 / 4 | -0.1 | Yes |
| 199221 | 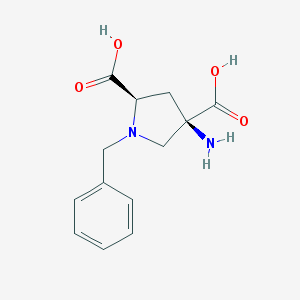 1-Benzyl-APDC 1-Benzyl-APDC | C13H16N2O4 | 264.281 | 6 / 3 | -4.5 | Yes |
| 201014 | 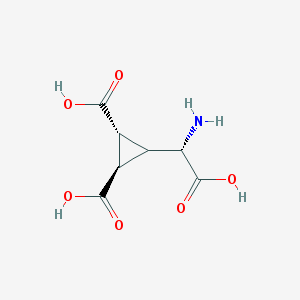 Dcg-IV Dcg-IV | C7H9NO6 | 203.15 | 7 / 4 | -4.3 | Yes |
| 208555 | 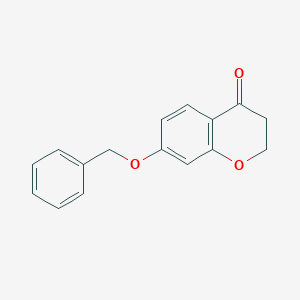 7-(benzyloxy)-2,3-dihydro-4H-chromen-4-one 7-(benzyloxy)-2,3-dihydro-4H-chromen-4-one | C16H14O3 | 254.285 | 3 / 0 | 2.8 | Yes |
| 218734 | 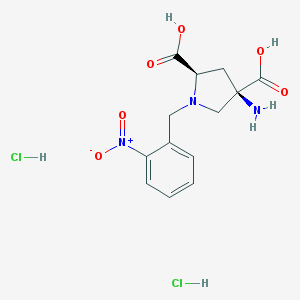 CHEMBL544508 CHEMBL544508 | C13H17Cl2N3O6 | 382.194 | 8 / 5 | N/A | N/A |
| 235588 | 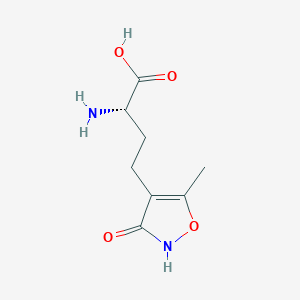 CHEMBL315268 CHEMBL315268 | C8H12N2O4 | 200.194 | 5 / 3 | -2.9 | Yes |
| 235590 | 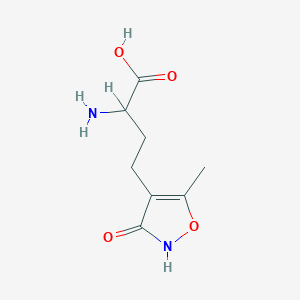 homo-AMPA homo-AMPA | C8H12N2O4 | 200.194 | 5 / 3 | -2.9 | Yes |
| 235599 | 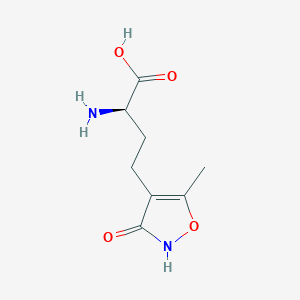 CHEMBL262416 CHEMBL262416 | C8H12N2O4 | 200.194 | 5 / 3 | -2.9 | Yes |
| 542297 | 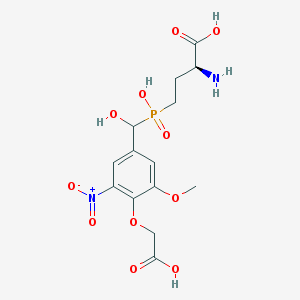 CHEMBL3966924 CHEMBL3966924 | C14H19N2O11P | 422.283 | 12 / 5 | -4.0 | No |
| 564754 | 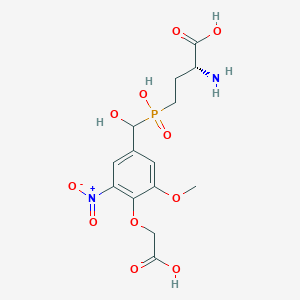 US9212196, Derivative 5 US9212196, Derivative 5 | C14H19N2O11P | 422.283 | 12 / 5 | -4.0 | No |
| 564839 | 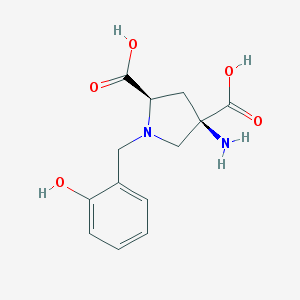 CHEMBL540015 CHEMBL540015 | C13H16N2O5 | 280.28 | 7 / 4 | -4.9 | Yes |
| 250530 | 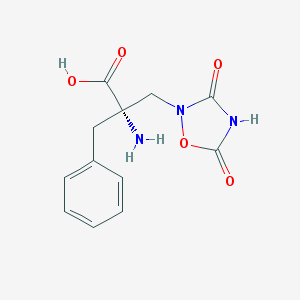 CHEMBL140862 CHEMBL140862 | C12H13N3O5 | 279.252 | 6 / 3 | -2.1 | Yes |
| 253257 | 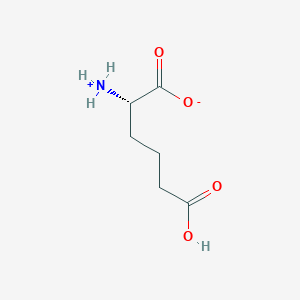 CID 59066632 CID 59066632 | C6H11NO4 | 161.157 | 4 / 2 | -2.5 | Yes |
| 253264 | 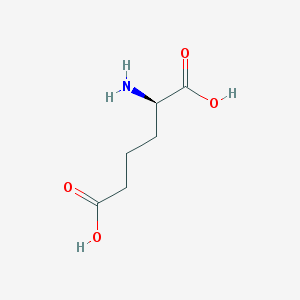 D-2-aminoadipic acid D-2-aminoadipic acid | C6H11NO4 | 161.157 | 5 / 3 | -3.1 | Yes |
| 542782 | 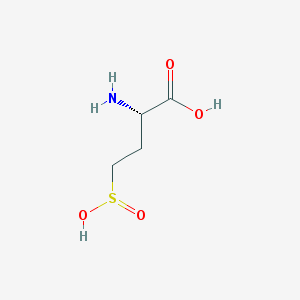 (2s)-2-amino-4-sulfinobutanoic acid (2s)-2-amino-4-sulfinobutanoic acid | C4H9NO4S | 167.179 | 6 / 3 | -3.8 | Yes |
| 554555 | 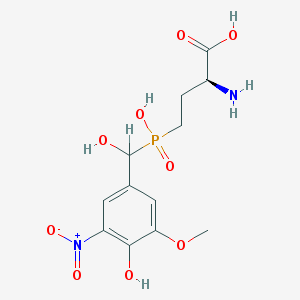 LSP1-2111 LSP1-2111 | C12H17N2O9P | 364.247 | 10 / 5 | -3.4 | Yes |
| 259127 | 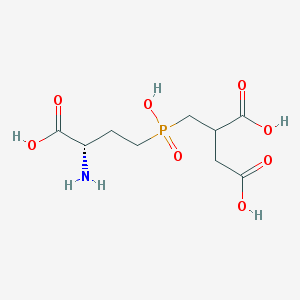 CHEMBL1092315 CHEMBL1092315 | C9H16NO8P | 297.2 | 9 / 5 | -5.3 | Yes |
| 265335 | 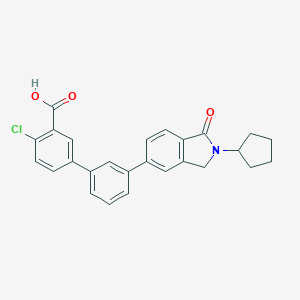 CHEMBL2179646 CHEMBL2179646 | C26H22ClNO3 | 431.916 | 3 / 1 | 5.6 | No |
| 529236 | 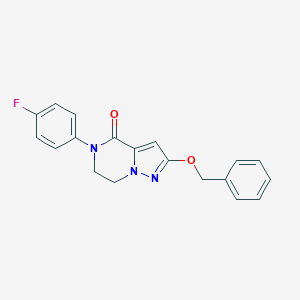 CHEMBL3747116 CHEMBL3747116 | C19H16FN3O2 | 337.354 | 4 / 0 | 3.2 | Yes |
| 543560 | 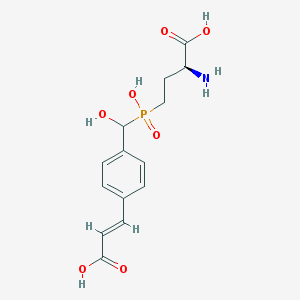 CHEMBL3982457 CHEMBL3982457 | C14H18NO7P | 343.272 | 8 / 5 | -3.5 | Yes |
| 565980 | 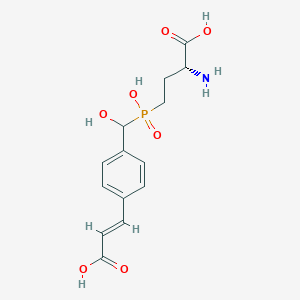 US9212196, Derivative 11 US9212196, Derivative 11 | C14H18NO7P | 343.272 | 8 / 5 | -3.5 | Yes |
| 282812 | 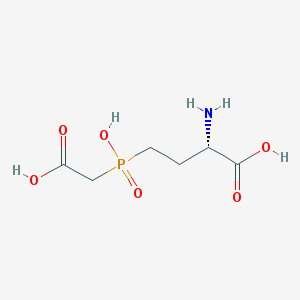 CHEMBL1089852 CHEMBL1089852 | C6H12NO6P | 225.137 | 7 / 4 | -4.7 | Yes |
| 566053 | 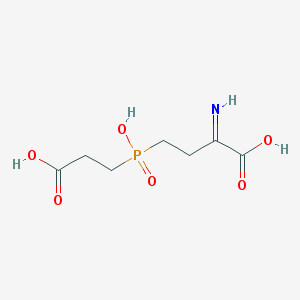 BDBM50314796 BDBM50314796 | C7H12NO6P | 237.148 | 7 / 4 | -2.1 | Yes |
| 292331 | 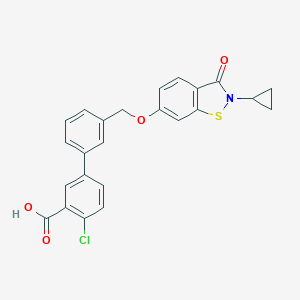 CHEMBL2179629 CHEMBL2179629 | C24H18ClNO4S | 451.921 | 5 / 1 | 5.3 | No |
| 310564 | 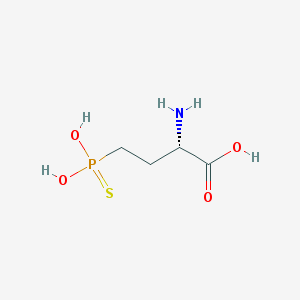 CHEMBL243925 CHEMBL243925 | C4H10NO4PS | 199.161 | 6 / 4 | -3.8 | Yes |
| 326890 | 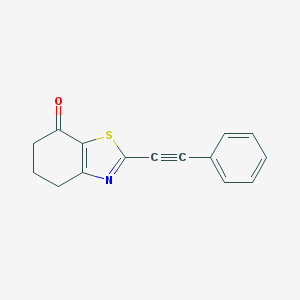 CHEMBL2426621 CHEMBL2426621 | C15H11NOS | 253.319 | 3 / 0 | 3.5 | Yes |
| 544827 | 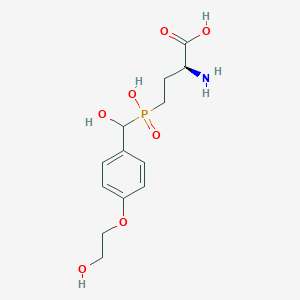 CHEMBL3896964 CHEMBL3896964 | C13H20NO7P | 333.277 | 8 / 5 | -4.1 | Yes |
| 567542 | 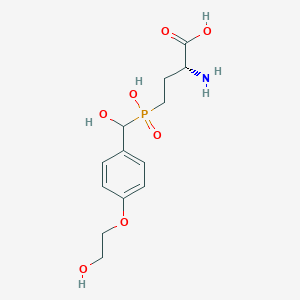 US9212196, Derivative 13 US9212196, Derivative 13 | C13H20NO7P | 333.277 | 8 / 5 | -4.1 | Yes |
| 337440 | 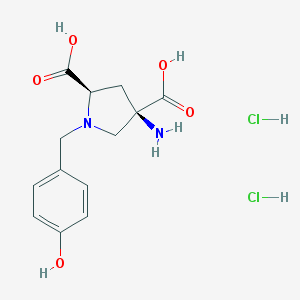 CHEMBL543568 CHEMBL543568 | C13H18Cl2N2O5 | 353.196 | 7 / 6 | N/A | No |
| 338064 |  CHEMBL3287724 CHEMBL3287724 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 347625 |  CHEMBL1093009 CHEMBL1093009 | C8H16NO6P | 253.191 | 7 / 4 | -4.5 | Yes |
| 348642 |  MLS003171622 MLS003171622 | C22H16ClN3O3 | 405.838 | 4 / 1 | 3.1 | Yes |
| 545459 |  L-Homocysteic acid L-Homocysteic acid | C4H9NO5S | 183.178 | 6 / 3 | -3.6 | Yes |
| 455829 | 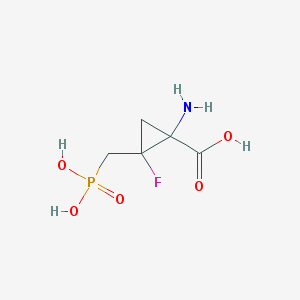 CHEMBL3427499 CHEMBL3427499 | C5H9FNO5P | 213.101 | 7 / 4 | -5.0 | Yes |
| 358356 | 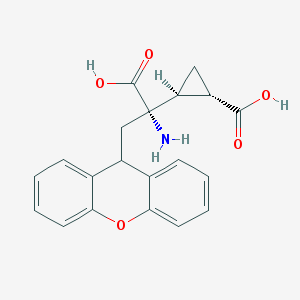 201943-63-7 201943-63-7 | C20H19NO5 | 353.374 | 6 / 3 | -0.2 | Yes |
| 369895 | 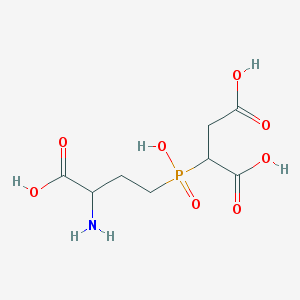 CHEMBL1092251 CHEMBL1092251 | C8H14NO8P | 283.173 | 9 / 5 | -5.2 | Yes |
| 373661 | 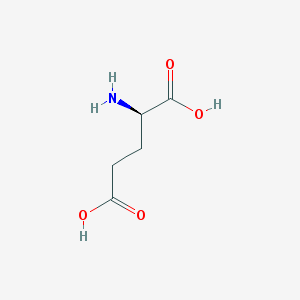 D-glutamic acid D-glutamic acid | C5H9NO4 | 147.13 | 5 / 3 | -3.7 | Yes |
| 373673 | 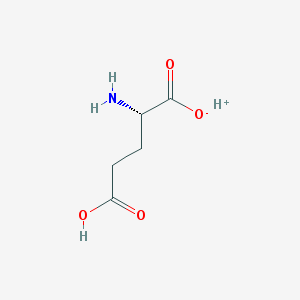 SCHEMBL11235794 SCHEMBL11235794 | C5H9NO4 | 147.13 | 5 / 3 | N/A | N/A |
| 569472 | 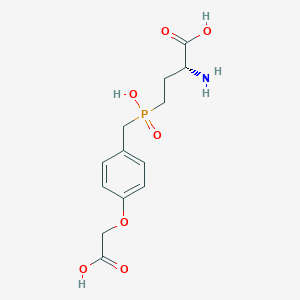 US9212196, Derivative 30 US9212196, Derivative 30 | C13H18NO7P | 331.261 | 8 / 4 | -3.3 | Yes |
| 546522 | 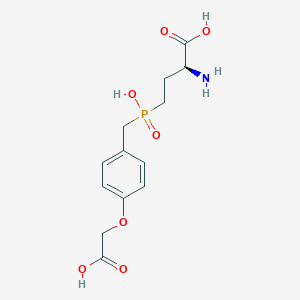 CHEMBL3919887 CHEMBL3919887 | C13H18NO7P | 331.261 | 8 / 4 | -3.3 | Yes |
| 395927 | 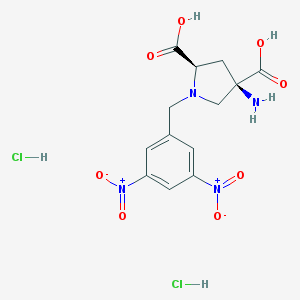 CHEMBL544272 CHEMBL544272 | C13H16Cl2N4O8 | 427.191 | 10 / 5 | N/A | N/A |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417