

You can:
| Name | Metabotropic glutamate receptor 3 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm3 |
| Synonym | mGluR3 mGlu3 receptor GPRC1C glutamate receptor |
| Disease | N/A for non-human GPCRs |
| Length | 879 |
| Amino acid sequence | MKMLTRLQILMLALFSKGFLLSLGDHNFMRREIKIEGDLVLGGLFPINEKGTGTEECGRINEDRGIQRLEAMLFAIDEINKDNYLLPGVKLGVHILDTCSRDTYALEQSLEFVRASLTKVDEAEYMCPDGSYAIQENIPLLIAGVIGGSYSSVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDYFARTVPPDFYQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFEQEARLRNICIATAEKVGRSNIRKSYDSVIRELLQKPNARVVVLFMRSDDSRELIAAANRVNASFTWVASDGWGAQESIVKGSEHVAYGAITLELASHPVRQFDRYFQSLNPYNNHRNPWFRDFWEQKFQCSLQNKRNHRQVCDKHLAIDSSNYEQESKIMFVVNAVYAMAHALHKMQRTLCPNTTKLCDAMKILDGKKLYKEYLLKINFTAPFNPNKGADSIVKFDTFGDGMGRYNVFNLQQTGGKYSYLKVGHWAETLSLDVDSIHWSRNSVPTSQCSDPCAPNEMKNMQPGDVCCWICIPCEPYEYLVDEFTCMDCGPGQWPTADLSGCYNLPEDYIKWEDAWAIGPVTIACLGFLCTCIVITVFIKHNNTPLVKASGRELCYILLFGVSLSYCMTFFFIAKPSPVICALRRLGLGTSFAICYSALLTKTNCIARIFDGVKNGAQRPKFISPSSQVFICLGLILVQIVMVSVWLILETPGTRRYTLPEKRETVILKCNVKDSSMLISLTYDVVLVILCTVYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVQTTTMCISVSLSGFVVLGCLFAPKVHIVLFQPQKNVVTHRLHLNRFSVSGTATTYSQSSASTYVPTVCNGREVLDSTTSSL |
| UniProt | P31422 |
| Protein Data Bank | 2e4u, 2e4v, 2e4w, 2e4x, 2e4y |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 2e4u. |
| BioLiP | BL0086116,BL0086117, BL0086120,BL0086121, BL0086118,BL0086119, BL0086114,BL0086115, BL0086112,BL0086113 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3067 |
| IUPHAR | 291 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 557307 | 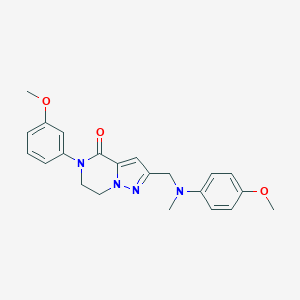 SCHEMBL17334385 SCHEMBL17334385 | C22H24N4O3 | 392.459 | 5 / 0 | 2.9 | Yes |
| 231 | 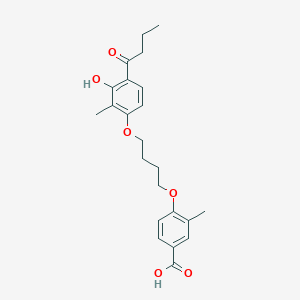 CHEMBL3287722 CHEMBL3287722 | C23H28O6 | 400.471 | 6 / 2 | 5.2 | No |
| 939 | 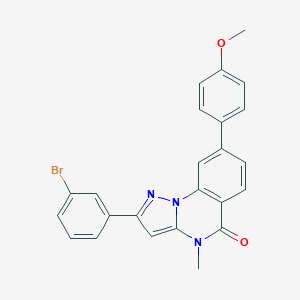 CHEMBL3288658 CHEMBL3288658 | C24H18BrN3O2 | 460.331 | 3 / 0 | 5.3 | No |
| 1837 | 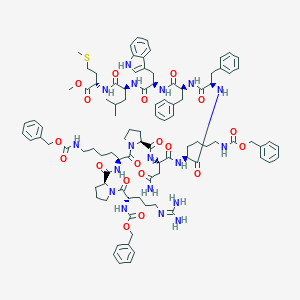 CHEMBL2304114 CHEMBL2304114 | C97H126N18O19S | 1880.24 | 21 / 15 | 7.8 | No |
| 557396 | 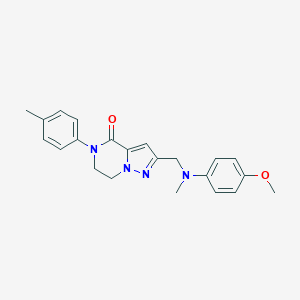 SCHEMBL17334442 SCHEMBL17334442 | C22H24N4O2 | 376.46 | 4 / 0 | 3.3 | Yes |
| 557411 | 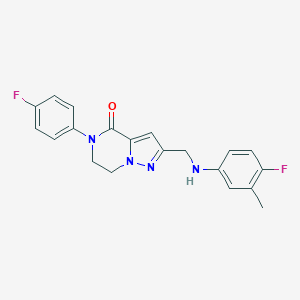 SCHEMBL17334231 SCHEMBL17334231 | C20H18F2N4O | 368.388 | 5 / 1 | 3.4 | Yes |
| 5896 | 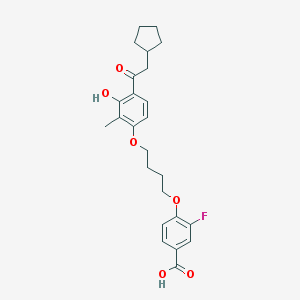 CHEMBL3287694 CHEMBL3287694 | C25H29FO6 | 444.499 | 7 / 2 | 6.3 | No |
| 557499 | 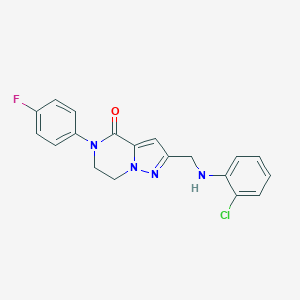 SCHEMBL17334341 SCHEMBL17334341 | C19H16ClFN4O | 370.812 | 4 / 1 | 3.5 | Yes |
| 9566 | 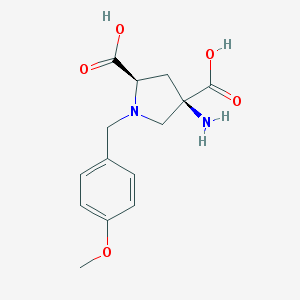 CHEMBL543097 CHEMBL543097 | C14H18N2O5 | 294.307 | 7 / 3 | -4.6 | Yes |
| 10717 | 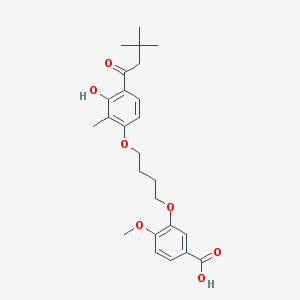 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 521768 | 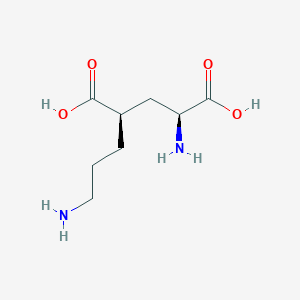 CHEMBL2312685 CHEMBL2312685 | C8H16N2O4 | 204.226 | 6 / 4 | -6.8 | Yes |
| 11664 | 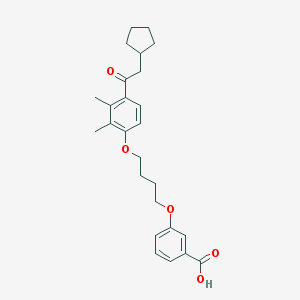 CHEMBL3287674 CHEMBL3287674 | C26H32O5 | 424.537 | 5 / 1 | 6.3 | No |
| 13326 | 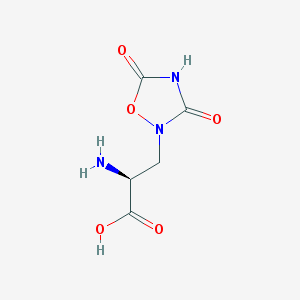 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 464488 | 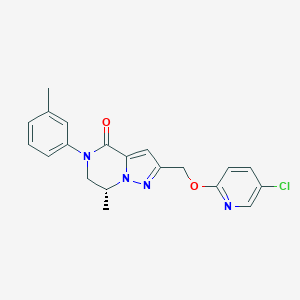 CHEMBL3617636 CHEMBL3617636 | C20H19ClN4O2 | 382.848 | 4 / 0 | 3.5 | Yes |
| 464528 | 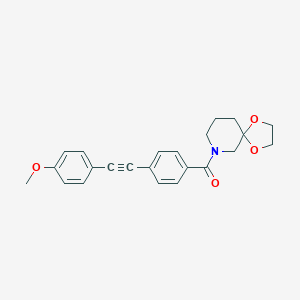 CHEMBL3182249 CHEMBL3182249 | C23H23NO4 | 377.44 | 4 / 0 | 3.3 | Yes |
| 464659 | 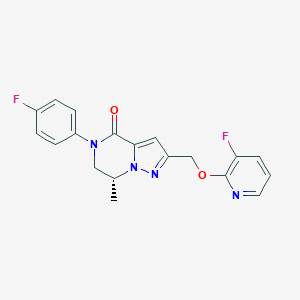 CHEMBL3617514 CHEMBL3617514 | C19H16F2N4O2 | 370.36 | 6 / 0 | 2.7 | Yes |
| 16563 |  CHEMBL3287725 CHEMBL3287725 | C25H32O6 | 428.525 | 6 / 2 | 6.1 | No |
| 18050 |  CHEMBL2304032 CHEMBL2304032 | C80H112N16O17S2 | 1633.99 | 20 / 13 | 4.3 | No |
| 18056 |  CHEMBL2079568 CHEMBL2079568 | C80H112N16O17S2 | 1633.99 | 20 / 14 | 4.7 | No |
| 18629 |  CHEMBL3287682 CHEMBL3287682 | C26H31ClO5 | 458.979 | 5 / 1 | 7.0 | No |
| 557854 |  SCHEMBL17334239 SCHEMBL17334239 | C19H15F3N4O | 372.351 | 6 / 1 | 3.1 | Yes |
| 19408 |  CHEMBL185210 CHEMBL185210 | C19H18FNO5 | 359.353 | 7 / 3 | -0.7 | Yes |
| 557867 |  SCHEMBL17334388 SCHEMBL17334388 | C21H21FN4O2 | 380.423 | 5 / 1 | 3.3 | Yes |
| 557869 |  SCHEMBL17334300 SCHEMBL17334300 | C21H21FN4O2 | 380.423 | 5 / 1 | 3.3 | Yes |
| 20313 | 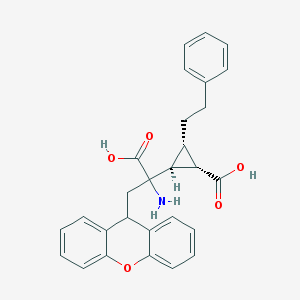 CHEMBL319279 CHEMBL319279 | C28H27NO5 | 457.526 | 6 / 3 | 2.1 | Yes |
| 21929 | 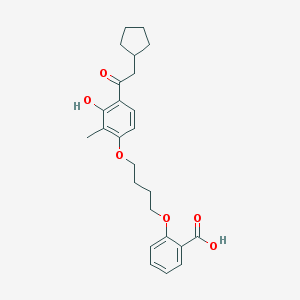 CHEMBL3287680 CHEMBL3287680 | C25H30O6 | 426.509 | 6 / 2 | 6.2 | No |
| 22398 | 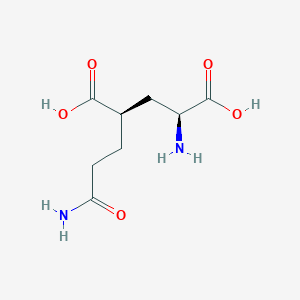 CHEMBL479804 CHEMBL479804 | C8H14N2O5 | 218.209 | 6 / 4 | -3.3 | Yes |
| 557969 | 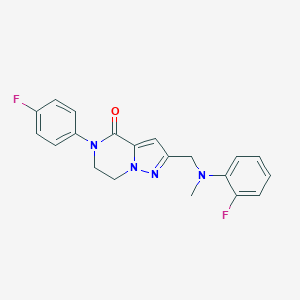 SCHEMBL17334434 SCHEMBL17334434 | C20H18F2N4O | 368.388 | 5 / 0 | 3.1 | Yes |
| 465786 | 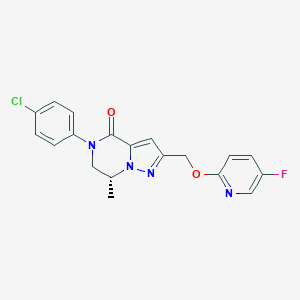 CHEMBL3617616 CHEMBL3617616 | C19H16ClFN4O2 | 386.811 | 5 / 0 | 3.2 | Yes |
| 23637 | 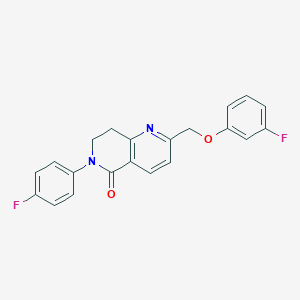 CHEMBL3298463 CHEMBL3298463 | C21H16F2N2O2 | 366.368 | 5 / 0 | 3.6 | Yes |
| 23808 | 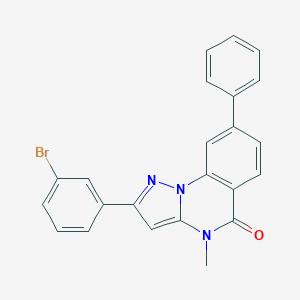 CHEMBL3288657 CHEMBL3288657 | C23H16BrN3O | 430.305 | 2 / 0 | 5.3 | No |
| 522263 | 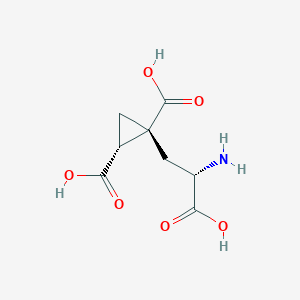 CHEMBL3786667 CHEMBL3786667 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522268 | 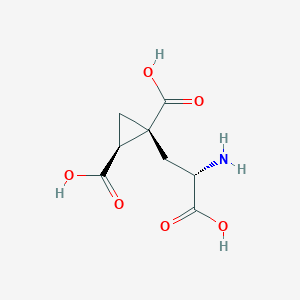 CHEMBL3787264 CHEMBL3787264 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522281 | 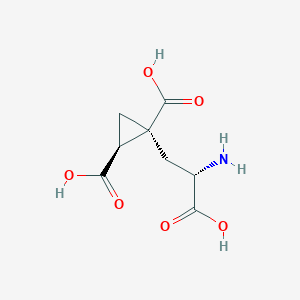 CHEMBL3786026 CHEMBL3786026 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522286 | 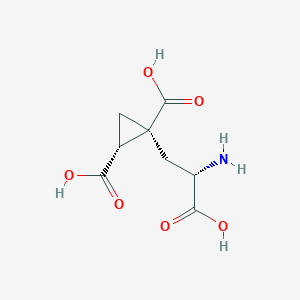 CHEMBL3786938 CHEMBL3786938 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 466190 | 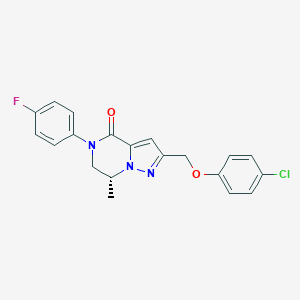 CHEMBL3617505 CHEMBL3617505 | C20H17ClFN3O2 | 385.823 | 4 / 0 | 3.9 | Yes |
| 29402 | 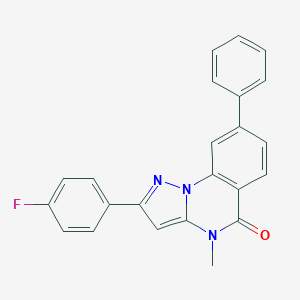 CHEMBL3286420 CHEMBL3286420 | C23H16FN3O | 369.399 | 3 / 0 | 4.7 | Yes |
| 29461 | 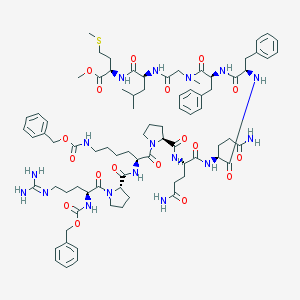 CHEMBL2304129 CHEMBL2304129 | C81H113N17O18S | 1644.96 | 20 / 13 | 3.2 | No |
| 466555 | 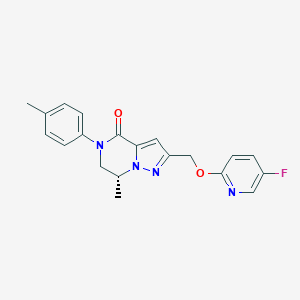 CHEMBL3617619 CHEMBL3617619 | C20H19FN4O2 | 366.396 | 5 / 0 | 2.9 | Yes |
| 32183 | 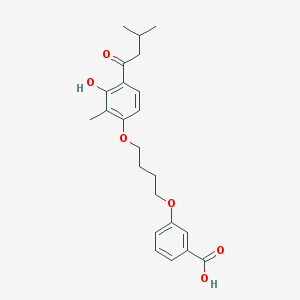 CHEMBL3287675 CHEMBL3287675 | C23H28O6 | 400.471 | 6 / 2 | 5.3 | No |
| 32246 | 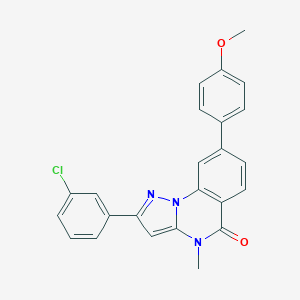 CHEMBL3288655 CHEMBL3288655 | C24H18ClN3O2 | 415.877 | 3 / 0 | 5.2 | No |
| 466975 | 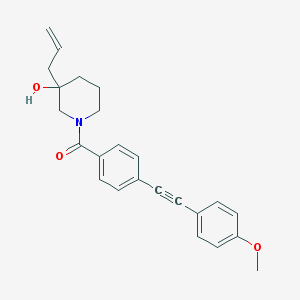 CHEMBL3182491 CHEMBL3182491 | C24H25NO3 | 375.468 | 3 / 1 | 4.2 | Yes |
| 36076 | 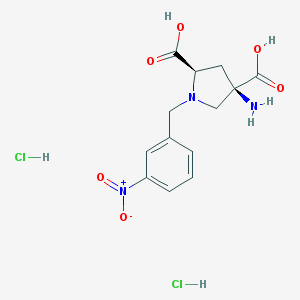 CHEMBL539760 CHEMBL539760 | C13H17Cl2N3O6 | 382.194 | 8 / 5 | N/A | N/A |
| 37241 | 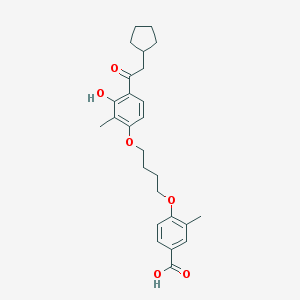 CHEMBL3287692 CHEMBL3287692 | C26H32O6 | 440.536 | 6 / 2 | 6.5 | No |
| 558412 | 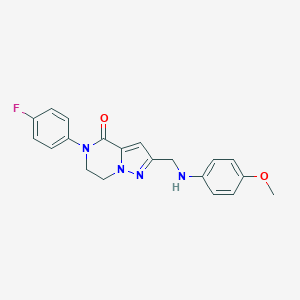 SCHEMBL17334380 SCHEMBL17334380 | C20H19FN4O2 | 366.396 | 5 / 1 | 2.9 | Yes |
| 40237 | 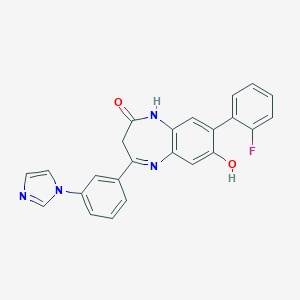 CHEMBL405895 CHEMBL405895 | C24H17FN4O2 | 412.424 | 5 / 2 | 3.5 | Yes |
| 41508 | 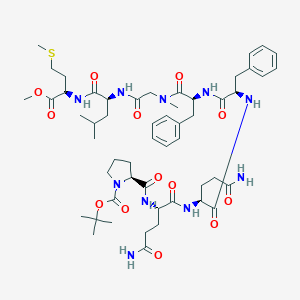 CHEMBL2304128 CHEMBL2304128 | C53H78N10O13S | 1095.32 | 14 / 8 | 2.4 | No |
| 43740 | 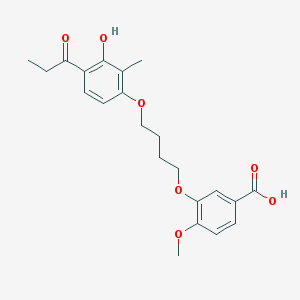 CHEMBL3287709 CHEMBL3287709 | C22H26O7 | 402.443 | 7 / 2 | 4.5 | Yes |
| 45467 |  CHEMBL1629864 CHEMBL1629864 | C21H15Cl2N3O | 396.271 | 3 / 1 | 4.6 | Yes |
| 48601 |  CHEMBL2304098 CHEMBL2304098 | C76H106N20O14S | 1555.87 | 18 / 18 | -1.6 | No |
| 558788 |  SCHEMBL17334298 SCHEMBL17334298 | C21H20ClFN4O2 | 414.865 | 5 / 0 | 3.6 | Yes |
| 50636 |  CHEMBL3288644 CHEMBL3288644 | C22H15FN4O | 370.387 | 4 / 0 | 3.6 | Yes |
| 52431 | 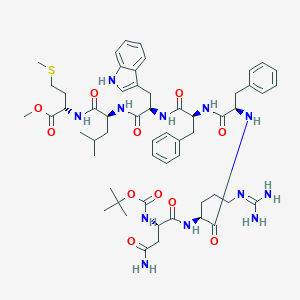 CHEMBL406889 CHEMBL406889 | C56H78N12O11S | 1127.37 | 13 / 11 | 3.9 | No |
| 52615 | 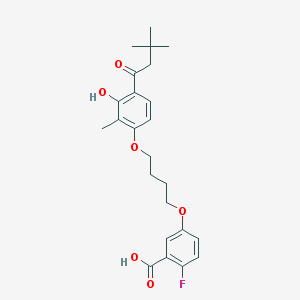 CHEMBL3287726 CHEMBL3287726 | C24H29FO6 | 432.488 | 7 / 2 | 5.8 | No |
| 469504 | 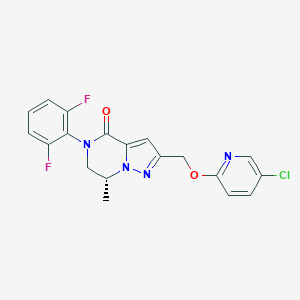 CHEMBL3617646 CHEMBL3617646 | C19H15ClF2N4O2 | 404.802 | 6 / 0 | 3.3 | Yes |
| 53756 | 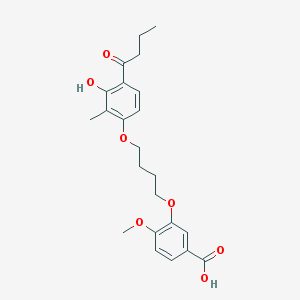 CHEMBL3287719 CHEMBL3287719 | C23H28O7 | 416.47 | 7 / 2 | 4.8 | Yes |
| 558956 | 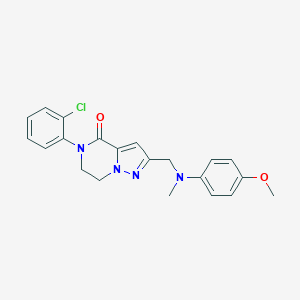 SCHEMBL17334418 SCHEMBL17334418 | C21H21ClN4O2 | 396.875 | 4 / 0 | 3.5 | Yes |
| 55585 | 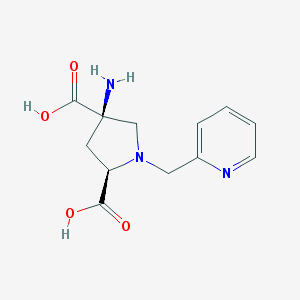 CHEMBL543811 CHEMBL543811 | C12H15N3O4 | 265.269 | 7 / 3 | -5.6 | Yes |
| 57286 | 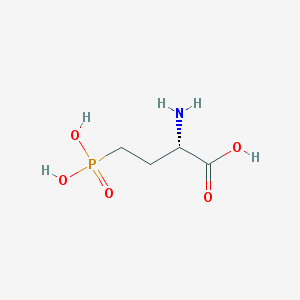 L-AP4 L-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 470031 | 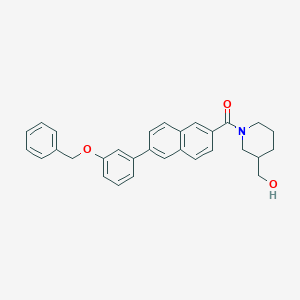 CHEMBL3184264 CHEMBL3184264 | C30H29NO3 | 451.566 | 3 / 1 | 5.8 | No |
| 470117 |  CHEMBL3617645 CHEMBL3617645 | C19H15ClF2N4O2 | 404.802 | 6 / 0 | 3.3 | Yes |
| 58962 |  CHEMBL1629865 CHEMBL1629865 | C22H16F3N3O | 395.385 | 6 / 1 | 4.3 | Yes |
| 59248 |  CHEMBL3288666 CHEMBL3288666 | C24H18ClN3O2 | 415.877 | 3 / 0 | 5.2 | No |
| 59331 |  CHEMBL3288637 CHEMBL3288637 | C23H17N3O | 351.409 | 2 / 0 | 4.6 | Yes |
| 61160 | 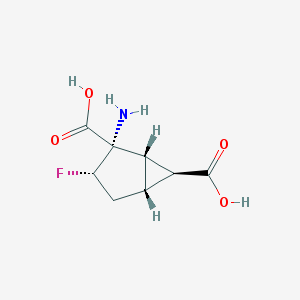 CHEMBL334014 CHEMBL334014 | C8H10FNO4 | 203.169 | 6 / 3 | -3.2 | Yes |
| 62813 | 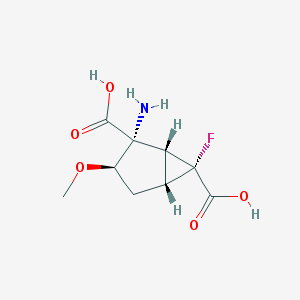 CHEMBL183956 CHEMBL183956 | C9H12FNO5 | 233.195 | 7 / 3 | -3.4 | Yes |
| 63097 | 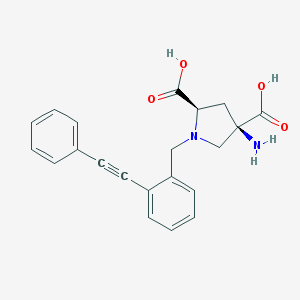 CHEMBL539516 CHEMBL539516 | C21H20N2O4 | 364.401 | 6 / 3 | -2.5 | Yes |
| 470691 | 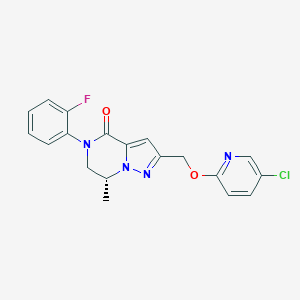 CHEMBL3617630 CHEMBL3617630 | C19H16ClFN4O2 | 386.811 | 5 / 0 | 3.2 | Yes |
| 559253 | 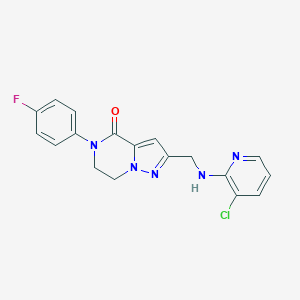 SCHEMBL17334340 SCHEMBL17334340 | C18H15ClFN5O | 371.8 | 5 / 1 | 2.8 | Yes |
| 64229 | 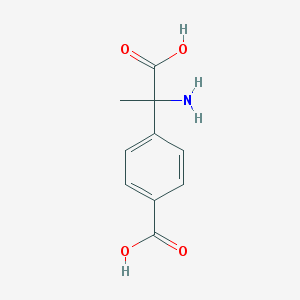 (RS)-MCPG (RS)-MCPG | C10H11NO4 | 209.201 | 5 / 3 | -2.0 | Yes |
| 559286 | 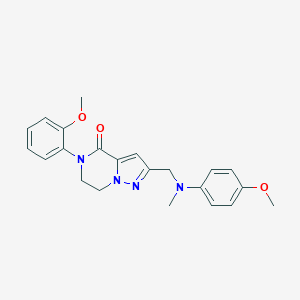 SCHEMBL17334338 SCHEMBL17334338 | C22H24N4O3 | 392.459 | 5 / 0 | 2.9 | Yes |
| 559296 | 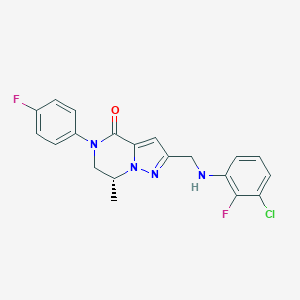 BDBM196629 BDBM196629 | C20H17ClF2N4O | 402.83 | 5 / 1 | 4.0 | Yes |
| 559297 | 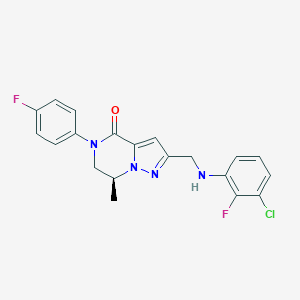 BDBM196630 BDBM196630 | C20H17ClF2N4O | 402.83 | 5 / 1 | 4.0 | Yes |
| 65348 | 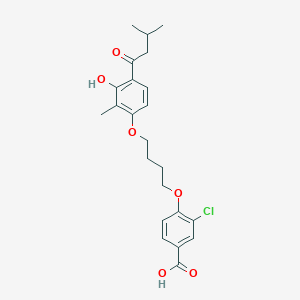 CHEMBL3287697 CHEMBL3287697 | C23H27ClO6 | 434.913 | 6 / 2 | 5.9 | No |
| 66454 | 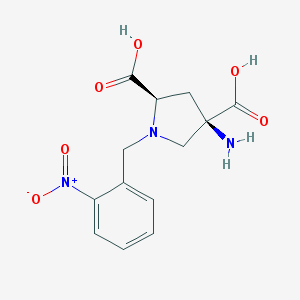 CHEMBL544508 CHEMBL544508 | C13H15N3O6 | 309.278 | 8 / 3 | -4.7 | Yes |
| 66541 | 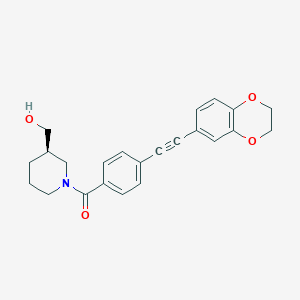 CHEMBL2385873 CHEMBL2385873 | C23H23NO4 | 377.44 | 4 / 1 | 3.2 | Yes |
| 471249 |  CHEMBL3617524 CHEMBL3617524 | C20H19FN4O3 | 382.395 | 6 / 0 | 2.9 | Yes |
| 68414 |  CHEMBL3287721 CHEMBL3287721 | C22H25FO6 | 404.434 | 7 / 2 | 5.0 | Yes |
| 69277 |  CHEMBL3288680 CHEMBL3288680 | C26H18N4O | 402.457 | 3 / 0 | 4.8 | Yes |
| 559472 |  SCHEMBL17334431 SCHEMBL17334431 | C19H15ClF3N5O2 | 437.807 | 8 / 1 | 3.7 | Yes |
| 70063 | 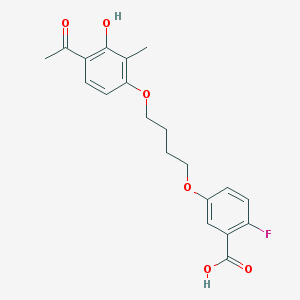 CHEMBL3287706 CHEMBL3287706 | C20H21FO6 | 376.38 | 7 / 2 | 4.1 | Yes |
| 70573 | 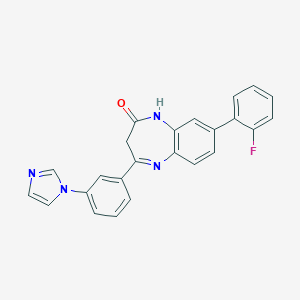 CHEMBL260636 CHEMBL260636 | C24H17FN4O | 396.425 | 4 / 1 | 3.9 | Yes |
| 559535 | 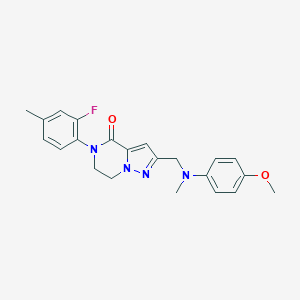 SCHEMBL17334417 SCHEMBL17334417 | C22H23FN4O2 | 394.45 | 5 / 0 | 3.4 | Yes |
| 75159 | 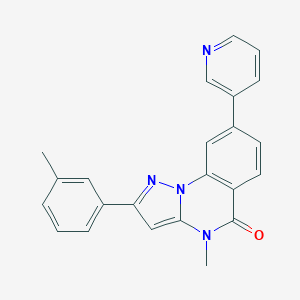 CHEMBL3288653 CHEMBL3288653 | C23H18N4O | 366.424 | 3 / 0 | 3.9 | Yes |
| 472241 | 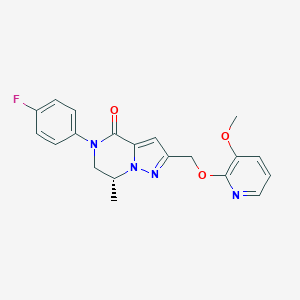 CHEMBL3617516 CHEMBL3617516 | C20H19FN4O3 | 382.395 | 6 / 0 | 2.5 | Yes |
| 76660 | 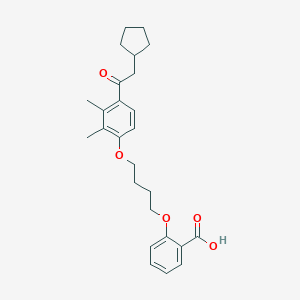 CHEMBL3287678 CHEMBL3287678 | C26H32O5 | 424.537 | 5 / 1 | 6.3 | No |
| 76744 | 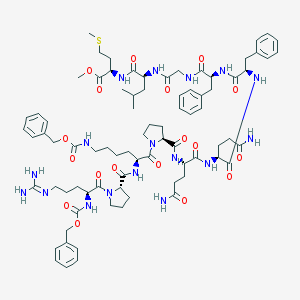 CHEMBL2304111 CHEMBL2304111 | C80H111N17O18S | 1630.93 | 20 / 14 | 3.0 | No |
| 559690 | 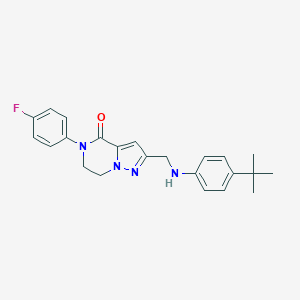 SCHEMBL17334264 SCHEMBL17334264 | C23H25FN4O | 392.478 | 4 / 1 | 4.6 | Yes |
| 559699 | 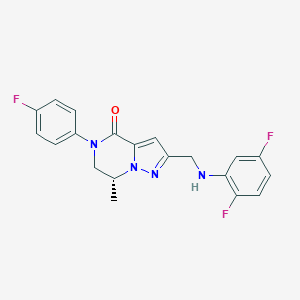 SCHEMBL17334351 SCHEMBL17334351 | C20H17F3N4O | 386.378 | 6 / 1 | 3.5 | Yes |
| 559701 | 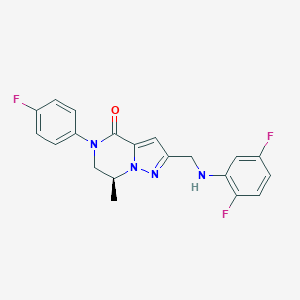 SCHEMBL17334372 SCHEMBL17334372 | C20H17F3N4O | 386.378 | 6 / 1 | 3.5 | Yes |
| 472520 | 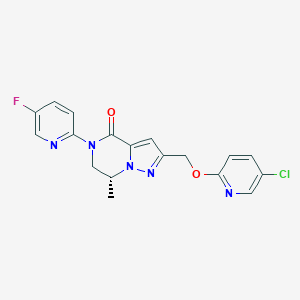 CHEMBL3617654 CHEMBL3617654 | C18H15ClFN5O2 | 387.799 | 6 / 0 | 2.4 | Yes |
| 559742 | 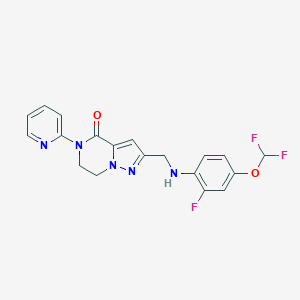 SCHEMBL17334408 SCHEMBL17334408 | C19H16F3N5O2 | 403.365 | 8 / 1 | 3.1 | Yes |
| 78702 | 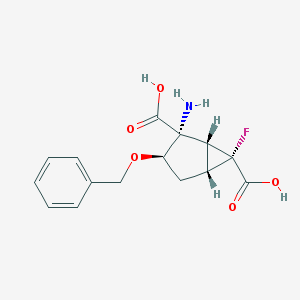 CHEMBL186214 CHEMBL186214 | C15H16FNO5 | 309.293 | 7 / 3 | -1.9 | Yes |
| 559784 | 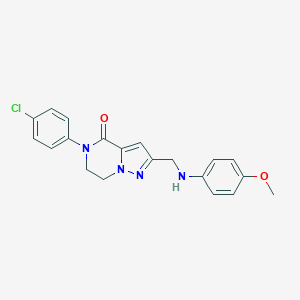 SCHEMBL17334282 SCHEMBL17334282 | C20H19ClN4O2 | 382.848 | 4 / 1 | 3.4 | Yes |
| 80290 | 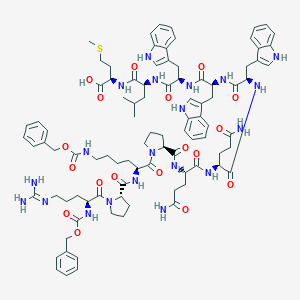 CHEMBL2304089 CHEMBL2304089 | C92H118N20O18S | 1824.14 | 20 / 18 | 5.0 | No |
| 472834 | 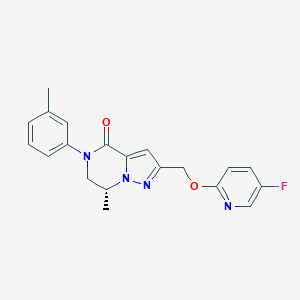 CHEMBL3617618 CHEMBL3617618 | C20H19FN4O2 | 366.396 | 5 / 0 | 2.9 | Yes |
| 472858 | 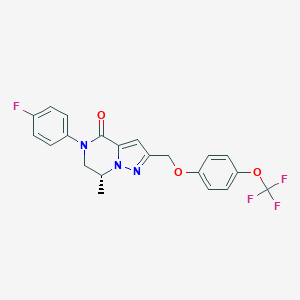 CHEMBL3617513 CHEMBL3617513 | C21H17F4N3O3 | 435.379 | 8 / 0 | 4.5 | Yes |
| 559798 | 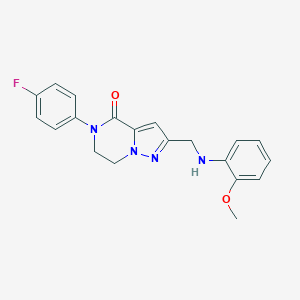 SCHEMBL17334378 SCHEMBL17334378 | C20H19FN4O2 | 366.396 | 5 / 1 | 2.9 | Yes |
| 559812 | 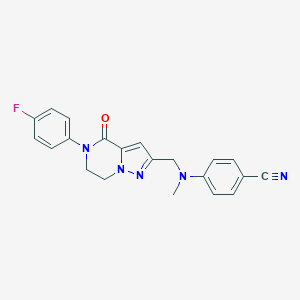 SCHEMBL17334406 SCHEMBL17334406 | C21H18FN5O | 375.407 | 5 / 0 | 2.8 | Yes |
| 81846 | 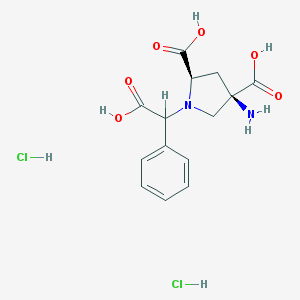 CHEMBL538489 CHEMBL538489 | C14H18Cl2N2O6 | 381.206 | 8 / 6 | N/A | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417