

You can:
| Name | Metabotropic glutamate receptor 3 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm3 |
| Synonym | mGluR3 mGlu3 receptor GPRC1C glutamate receptor |
| Disease | N/A for non-human GPCRs |
| Length | 879 |
| Amino acid sequence | MKMLTRLQILMLALFSKGFLLSLGDHNFMRREIKIEGDLVLGGLFPINEKGTGTEECGRINEDRGIQRLEAMLFAIDEINKDNYLLPGVKLGVHILDTCSRDTYALEQSLEFVRASLTKVDEAEYMCPDGSYAIQENIPLLIAGVIGGSYSSVSIQVANLLRLFQIPQISYASTSAKLSDKSRYDYFARTVPPDFYQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFEQEARLRNICIATAEKVGRSNIRKSYDSVIRELLQKPNARVVVLFMRSDDSRELIAAANRVNASFTWVASDGWGAQESIVKGSEHVAYGAITLELASHPVRQFDRYFQSLNPYNNHRNPWFRDFWEQKFQCSLQNKRNHRQVCDKHLAIDSSNYEQESKIMFVVNAVYAMAHALHKMQRTLCPNTTKLCDAMKILDGKKLYKEYLLKINFTAPFNPNKGADSIVKFDTFGDGMGRYNVFNLQQTGGKYSYLKVGHWAETLSLDVDSIHWSRNSVPTSQCSDPCAPNEMKNMQPGDVCCWICIPCEPYEYLVDEFTCMDCGPGQWPTADLSGCYNLPEDYIKWEDAWAIGPVTIACLGFLCTCIVITVFIKHNNTPLVKASGRELCYILLFGVSLSYCMTFFFIAKPSPVICALRRLGLGTSFAICYSALLTKTNCIARIFDGVKNGAQRPKFISPSSQVFICLGLILVQIVMVSVWLILETPGTRRYTLPEKRETVILKCNVKDSSMLISLTYDVVLVILCTVYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVQTTTMCISVSLSGFVVLGCLFAPKVHIVLFQPQKNVVTHRLHLNRFSVSGTATTYSQSSASTYVPTVCNGREVLDSTTSSL |
| UniProt | P31422 |
| Protein Data Bank | 2e4u, 2e4v, 2e4w, 2e4x, 2e4y |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 2e4u. |
| BioLiP | BL0086116,BL0086117, BL0086120,BL0086121, BL0086118,BL0086119, BL0086114,BL0086115, BL0086112,BL0086113 |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3067 |
| IUPHAR | 291 |
| DrugBank | N/A |
| Name | CHEMBL334014 |
|---|---|
| Molecular formula | C8H10FNO4 |
| IUPAC name | (1S,2S,3S,5R,6S)-2-amino-3-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid |
| Molecular weight | 203.169 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 3 |
| XlogP | -3.2 |
| Synonyms | 234085-20-2 DTXSID50431168 Bicyclo[3.1.0]hexane-2,6-dicarboxylicacid, 2-amino-3-fluoro-, (1S,2S,3S,5R,6S)- (1s,2s,3s,5r,6s)-2-amino-3-fluorobicyclo[3.1.0]hexane-2,6-dicarboxylic acid CTK1A0318 [ Show all ] |
| Inchi Key | DIWVNJFXRKZAGI-FJNNMDBYSA-N |
| Inchi ID | InChI=1S/C8H10FNO4/c9-3-1-2-4(6(11)12)5(2)8(3,10)7(13)14/h2-5H,1,10H2,(H,11,12)(H,13,14)/t2-,3-,4-,5-,8-/m0/s1 |
| PubChem CID | 9815616 |
| ChEMBL | CHEMBL334014 |
| IUPHAR | N/A |
| BindingDB | 50094841 |
| DrugBank | N/A |
Structure | 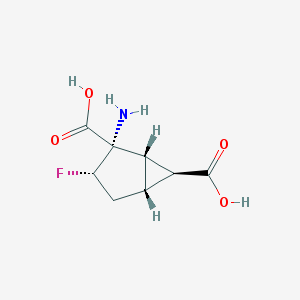 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC50 | <100000.0 nM | PMID11123999 | BindingDB,ChEMBL |
| EC50 | 45.4 nM | PMID11123999 | BindingDB,ChEMBL |
| Ki | 65.9 nM | PMID11123999 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417