

You can:
| Name | 5-hydroxytryptamine receptor 6 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HTR6 |
| Synonym | 5-HT6 receptor 5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled 5-HT-6 serotonin receptor 6 ST-B17 [ Show all ] |
| Disease | Schizophrenia Obesity Neurological disease Neurodegenerative disease Emesis [ Show all ] |
| Length | 440 |
| Amino acid sequence | MVPEPGPTANSTPAWGAGPPSAPGGSGWVAAALCVVIALTAAANSLLIALICTQPALRNTSNFFLVSLFTSDLMVGLVVMPPAMLNALYGRWVLARGLCLLWTAFDVMCCSASILNLCLISLDRYLLILSPLRYKLRMTPLRALALVLGAWSLAALASFLPLLLGWHELGHARPPVPGQCRLLASLPFVLVASGLTFFLPSGAICFTYCRILLAARKQAVQVASLTTGMASQASETLQVPRTPRPGVESADSRRLATKHSRKALKASLTLGILLGMFFVTWLPFFVANIVQAVCDCISPGLFDVLTWLGYCNSTMNPIIYPLFMRDFKRALGRFLPCPRCPRERQASLASPSLRTSHSGPRPGLSLQQVLPLPLPPDSDSDSDAGSGGSSGLRLTAQLLLPGEATQDPPLPTRAAAAVNFFNIDPAEPELRPHPLGIPTN |
| UniProt | P50406 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P50406 |
| 3D structure model | This predicted structure model is from GPCR-EXP P50406. |
| BioLiP | N/A |
| Therapeutic Target Database | T16691 |
| ChEMBL | CHEMBL3371 |
| IUPHAR | 11 |
| DrugBank | BE0000945 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 441668 | 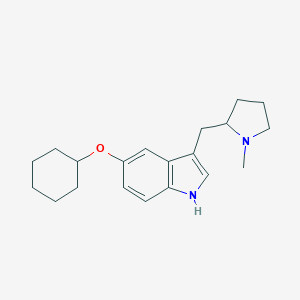 CHEMBL3329446 CHEMBL3329446 | C20H28N2O | 312.457 | 2 / 1 | 4.7 | Yes |
| 459234 | 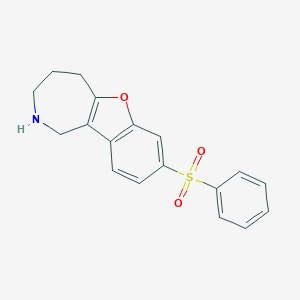 CHEMBL3692880 CHEMBL3692880 | C18H17NO3S | 327.398 | 4 / 1 | 2.9 | Yes |
| 127 | 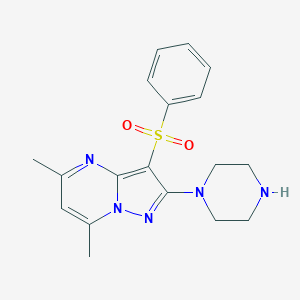 CHEMBL1762574 CHEMBL1762574 | C18H21N5O2S | 371.459 | 6 / 1 | 1.7 | Yes |
| 228 | 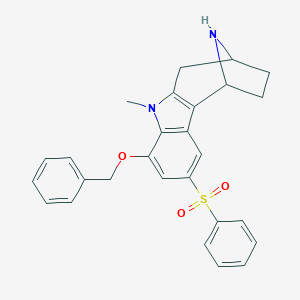 CHEMBL3664770 CHEMBL3664770 | C27H26N2O3S | 458.576 | 4 / 1 | 4.3 | Yes |
| 295 | 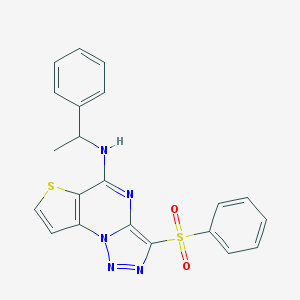 CHEMBL1170086 CHEMBL1170086 | C21H17N5O2S2 | 435.52 | 7 / 1 | 4.2 | Yes |
| 458 | 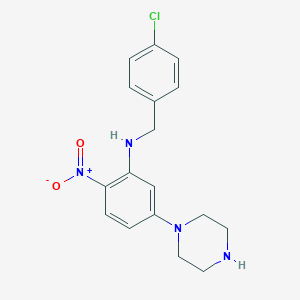 CHEMBL249034 CHEMBL249034 | C17H19ClN4O2 | 346.815 | 5 / 2 | 3.9 | Yes |
| 529 | 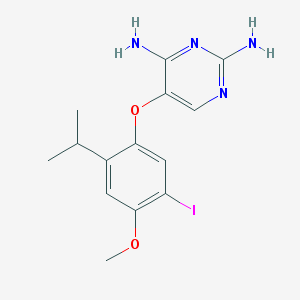 AF-353 AF-353 | C14H17IN4O2 | 400.22 | 6 / 2 | 2.8 | Yes |
| 584 | 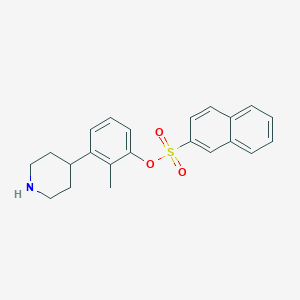 tolylpiperidine, 18n tolylpiperidine, 18n | C22H23NO3S | 381.49 | 4 / 1 | 4.7 | Yes |
| 683 | 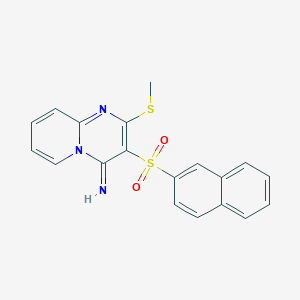 CHEMBL358948 CHEMBL358948 | C19H15N3O2S2 | 381.468 | 5 / 1 | 3.2 | Yes |
| 459241 | 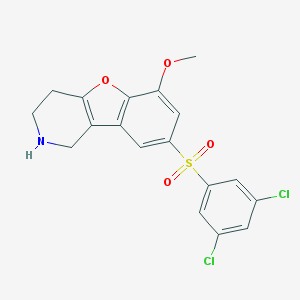 CHEMBL3692996 CHEMBL3692996 | C18H15Cl2NO4S | 412.281 | 5 / 1 | 3.8 | Yes |
| 960 | 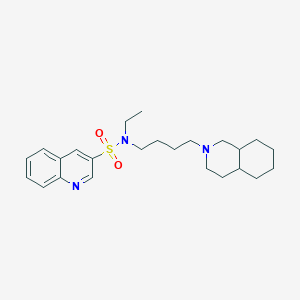 CHEMBL1917358 CHEMBL1917358 | C24H35N3O2S | 429.623 | 5 / 0 | 4.9 | Yes |
| 1007 | 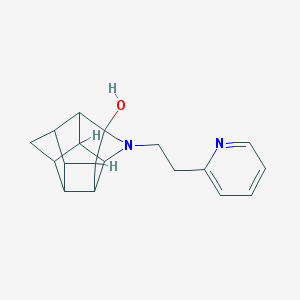 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1471 | 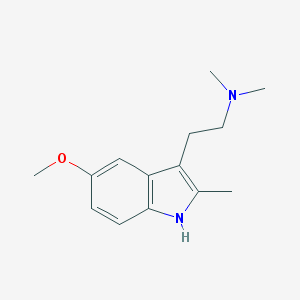 4365 CT 4365 CT | C14H20N2O | 232.327 | 2 / 1 | 2.7 | Yes |
| 1734 | 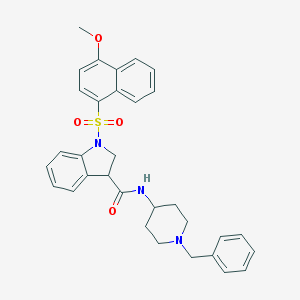 CHEMBL1085836 CHEMBL1085836 | C32H33N3O4S | 555.693 | 6 / 1 | 5.0 | No |
| 2164 | 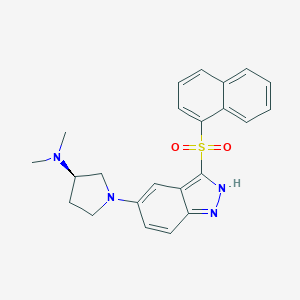 CHEMBL598444 CHEMBL598444 | C23H24N4O2S | 420.531 | 5 / 1 | 4.3 | Yes |
| 2166 | 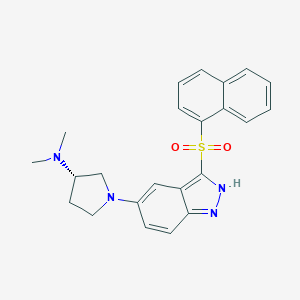 CHEMBL597009 CHEMBL597009 | C23H24N4O2S | 420.531 | 5 / 1 | 4.3 | Yes |
| 2183 |  CHEMBL458002 CHEMBL458002 | C29H28ClN5O2 | 514.026 | 5 / 2 | 4.3 | No |
| 2220 |  SCHEMBL6925318 SCHEMBL6925318 | C21H22N2O3S | 382.478 | 4 / 1 | 2.8 | Yes |
| 2270 |  1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2296 |  CHEMBL2165544 CHEMBL2165544 | C22H23F3N2O3S | 452.492 | 7 / 0 | 4.9 | Yes |
| 2920 |  CHEMBL368783 CHEMBL368783 | C20H22N2O | 306.409 | 2 / 1 | 4.1 | Yes |
| 3155 |  AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 547929 |  CHEMBL3941102 CHEMBL3941102 | C28H31F3N4O2 | 512.577 | 8 / 1 | 3.9 | No |
| 3401 |  CHEMBL1642882 CHEMBL1642882 | C18H19ClN4O2S | 390.886 | 5 / 3 | 3.4 | Yes |
| 3503 | 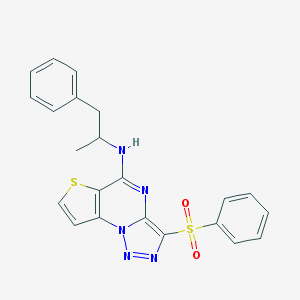 CHEMBL1173061 CHEMBL1173061 | C22H19N5O2S2 | 449.547 | 7 / 1 | 4.7 | Yes |
| 3627 | 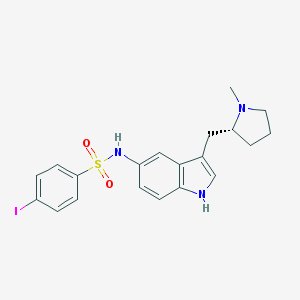 CHEMBL609994 CHEMBL609994 | C20H22IN3O2S | 495.379 | 4 / 2 | 4.1 | Yes |
| 3628 | 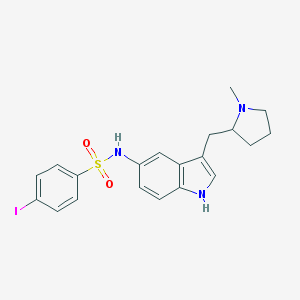 BDBM50159192 BDBM50159192 | C20H22IN3O2S | 495.379 | 4 / 2 | 4.1 | Yes |
| 3730 | 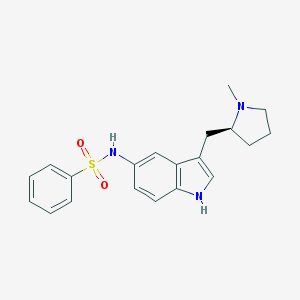 CHEMBL606706 CHEMBL606706 | C20H23N3O2S | 369.483 | 4 / 2 | 3.4 | Yes |
| 3731 | 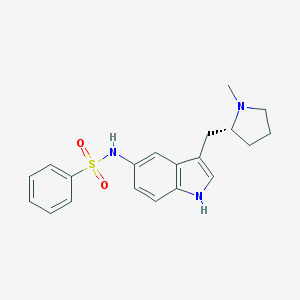 CHEMBL606546 CHEMBL606546 | C20H23N3O2S | 369.483 | 4 / 2 | 3.4 | Yes |
| 3732 | 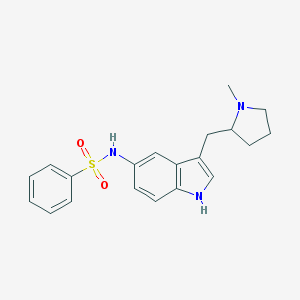 BDBM50159207 BDBM50159207 | C20H23N3O2S | 369.483 | 4 / 2 | 3.4 | Yes |
| 3817 | 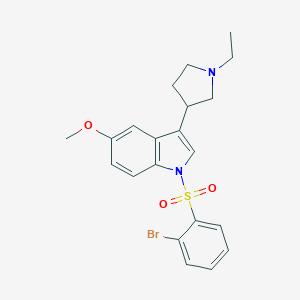 SCHEMBL6530223 SCHEMBL6530223 | C21H23BrN2O3S | 463.39 | 4 / 0 | 4.5 | Yes |
| 3832 | 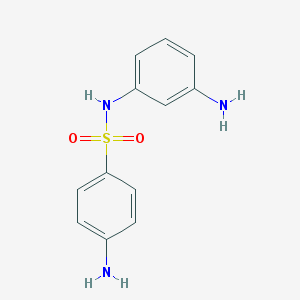 NSC37904 NSC37904 | C12H13N3O2S | 263.315 | 5 / 3 | 0.9 | Yes |
| 4027 | 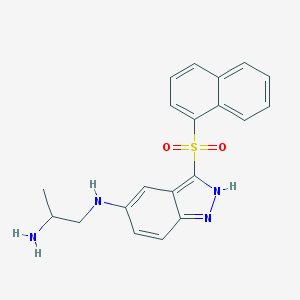 CHEMBL1642849 CHEMBL1642849 | C20H20N4O2S | 380.466 | 5 / 3 | 3.4 | Yes |
| 4093 | 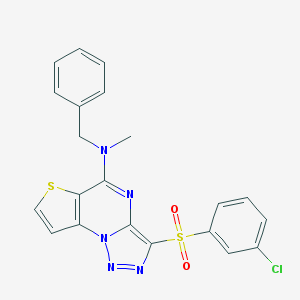 CHEMBL1173578 CHEMBL1173578 | C21H16ClN5O2S2 | 469.962 | 7 / 0 | 4.6 | Yes |
| 4307 | 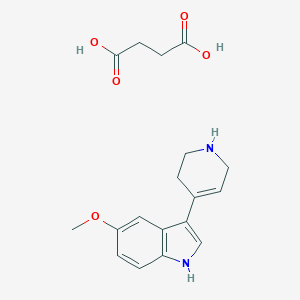 Butanedioic acid, compd. with 5-methoxy-3-(1,2,3,6-tetrahydro-4-pyridinyl)-1H-indole (1:1) Butanedioic acid, compd. with 5-methoxy-3-(1,2,3,6-tetrahydro-4-pyridinyl)-1H-indole (1:1) | C18H22N2O5 | 346.383 | 6 / 4 | N/A | N/A |
| 4468 | 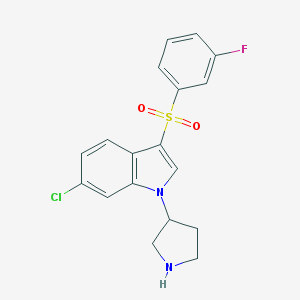 CHEMBL1080694 CHEMBL1080694 | C18H16ClFN2O2S | 378.846 | 4 / 1 | 3.2 | Yes |
| 4524 | 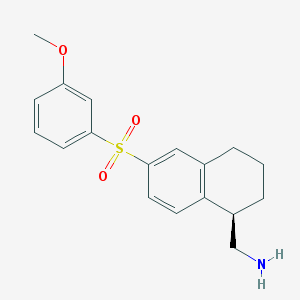 CHEMBL1086325 CHEMBL1086325 | C18H21NO3S | 331.43 | 4 / 1 | 2.7 | Yes |
| 547944 | 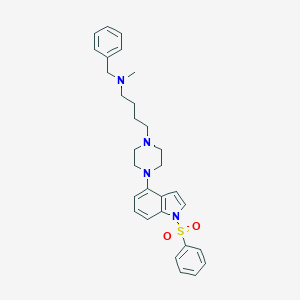 CHEMBL3883921 CHEMBL3883921 | C30H36N4O2S | 516.704 | 5 / 0 | 5.4 | No |
| 4611 | 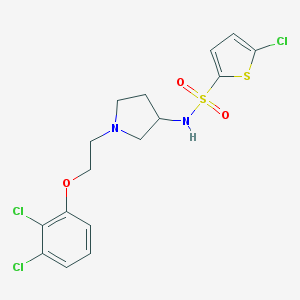 CHEMBL2159474 CHEMBL2159474 | C16H17Cl3N2O3S2 | 455.793 | 6 / 1 | 5.0 | Yes |
| 4660 | 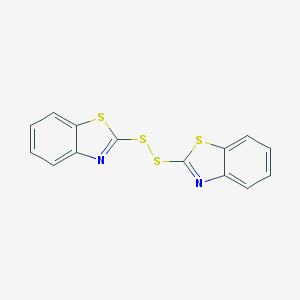 120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 4765 |  CHEMBL3664787 CHEMBL3664787 | C24H25N3O2S | 419.543 | 3 / 1 | 3.2 | Yes |
| 521602 |  CHEMBL3785626 CHEMBL3785626 | C14H21N5O2S | 323.415 | 5 / 2 | 0.9 | Yes |
| 459263 |  CHEMBL3692841 CHEMBL3692841 | C17H15NO3S | 313.371 | 4 / 1 | 2.6 | Yes |
| 4953 |  5HT6-ligand-1 5HT6-ligand-1 | C20H22BrN3O2S | 448.379 | 4 / 0 | 3.6 | Yes |
| 5096 | 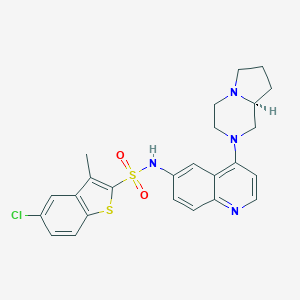 CHEMBL94988 CHEMBL94988 | C25H25ClN4O2S2 | 513.071 | 7 / 1 | 5.5 | No |
| 5226 | 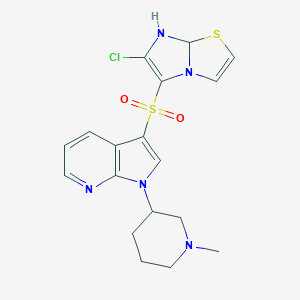 CID 44433197 CID 44433197 | C18H20ClN5O2S2 | 437.961 | 7 / 1 | 2.7 | Yes |
| 5253 | 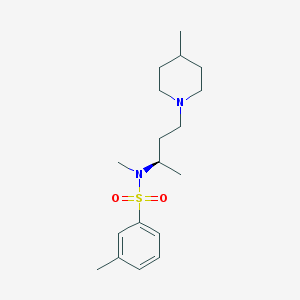 SB-258719 SB-258719 | C18H30N2O2S | 338.51 | 4 / 0 | 3.5 | Yes |
| 5294 | 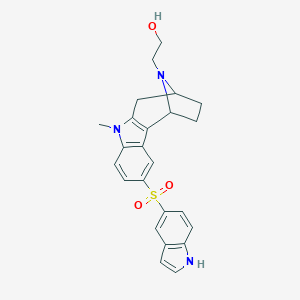 SCHEMBL6924367 SCHEMBL6924367 | C24H25N3O3S | 435.542 | 4 / 2 | 2.7 | Yes |
| 5299 | 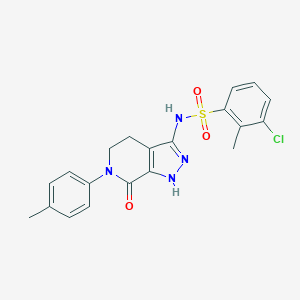 CHEMBL2407545 CHEMBL2407545 | C20H19ClN4O3S | 430.907 | 5 / 2 | 3.8 | Yes |
| 5470 | 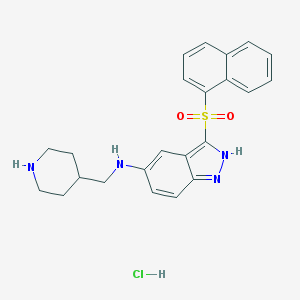 CHEMBL1642853 CHEMBL1642853 | C23H25ClN4O2S | 456.989 | 5 / 4 | N/A | N/A |
| 5665 | 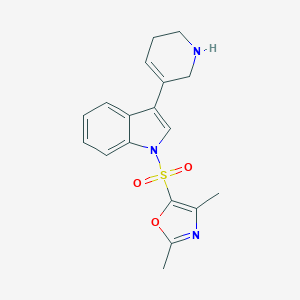 CHEMBL380777 CHEMBL380777 | C18H19N3O3S | 357.428 | 5 / 1 | 2.5 | Yes |
| 5678 | 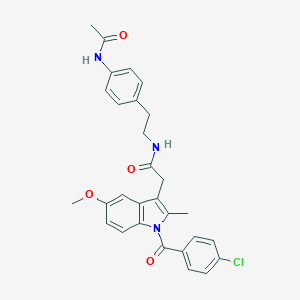 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 5780 |  CHEMBL568673 CHEMBL568673 | C17H17Cl2N3O2S | 398.302 | 4 / 0 | 3.3 | Yes |
| 5846 |  CHEMBL246500 CHEMBL246500 | C20H22N4O3S | 398.481 | 7 / 1 | 1.9 | Yes |
| 463600 |  CHEMBL3696970 CHEMBL3696970 | C20H21NO5S | 387.45 | 6 / 2 | 2.3 | Yes |
| 6217 |  CHEMBL2178747 CHEMBL2178747 | C22H29N3O3S | 415.552 | 6 / 1 | 3.6 | Yes |
| 6230 | 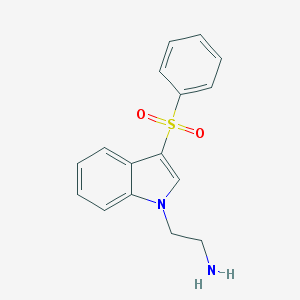 CHEMBL188182 CHEMBL188182 | C16H16N2O2S | 300.376 | 3 / 1 | 1.8 | Yes |
| 6269 | 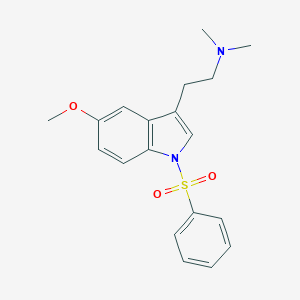 CHEMBL76237 CHEMBL76237 | C19H22N2O3S | 358.456 | 4 / 0 | 3.5 | Yes |
| 6313 | 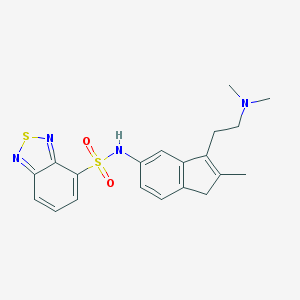 CHEMBL514383 CHEMBL514383 | C20H22N4O2S2 | 414.542 | 7 / 1 | 2.9 | Yes |
| 441954 | 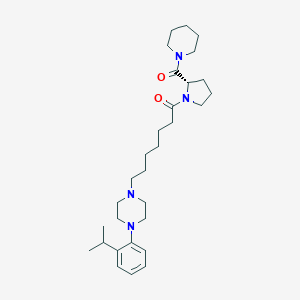 CHEMBL3409037 CHEMBL3409037 | C30H48N4O2 | 496.74 | 4 / 0 | 4.9 | Yes |
| 6687 | 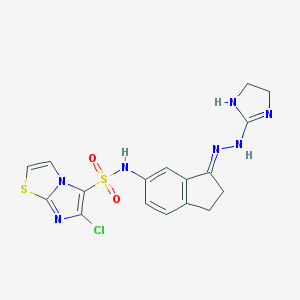 CHEMBL574403 CHEMBL574403 | C17H16ClN7O2S2 | 449.932 | 7 / 3 | 2.7 | Yes |
| 6844 | 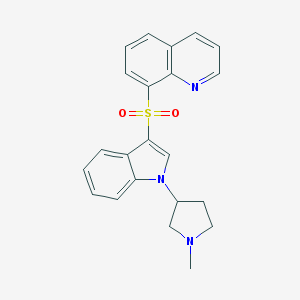 CHEMBL1080899 CHEMBL1080899 | C22H21N3O2S | 391.489 | 4 / 0 | 3.2 | Yes |
| 6847 | 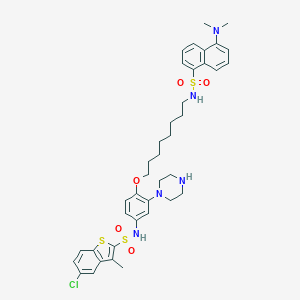 CHEMBL1256255 CHEMBL1256255 | C39H48ClN5O5S3 | 798.473 | 11 / 3 | 8.8 | No |
| 6927 | 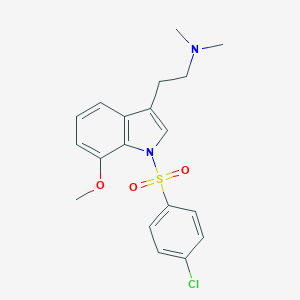 CHEMBL75029 CHEMBL75029 | C19H21ClN2O3S | 392.898 | 4 / 0 | 4.1 | Yes |
| 6944 | 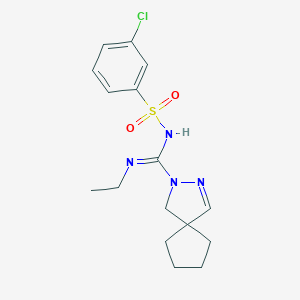 CHEMBL1834347 CHEMBL1834347 | C16H21ClN4O2S | 368.88 | 4 / 1 | 2.7 | Yes |
| 463710 | 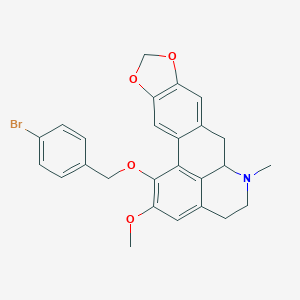 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 536143 | 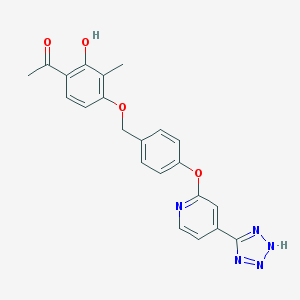 CHEMBL3899832 CHEMBL3899832 | C22H19N5O4 | 417.425 | 8 / 2 | 3.6 | Yes |
| 7044 | 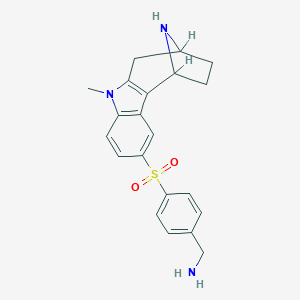 SCHEMBL6925846 SCHEMBL6925846 | C21H23N3O2S | 381.494 | 4 / 2 | 1.7 | Yes |
| 7058 | 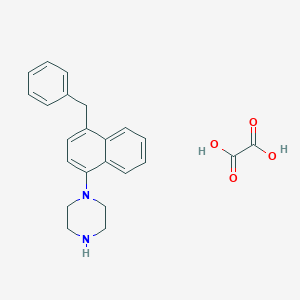 CHEMBL180945 CHEMBL180945 | C23H24N2O4 | 392.455 | 6 / 3 | N/A | N/A |
| 7137 | 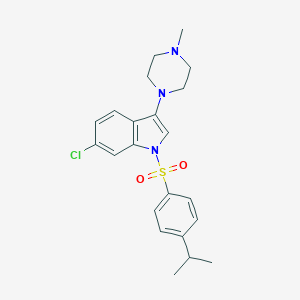 CHEMBL1642428 CHEMBL1642428 | C22H26ClN3O2S | 431.979 | 4 / 0 | 4.9 | Yes |
| 7161 | 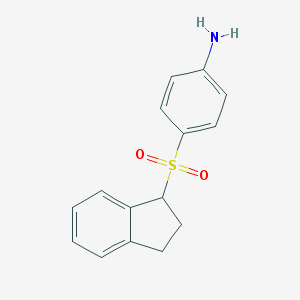 CHEMBL191972 CHEMBL191972 | C15H15NO2S | 273.35 | 3 / 1 | 2.4 | Yes |
| 7176 | 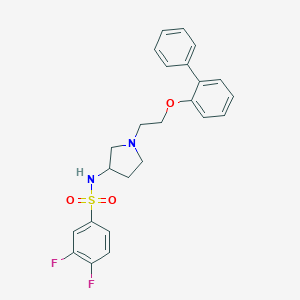 CHEMBL2159481 CHEMBL2159481 | C24H24F2N2O3S | 458.524 | 7 / 1 | 4.6 | Yes |
| 459284 | 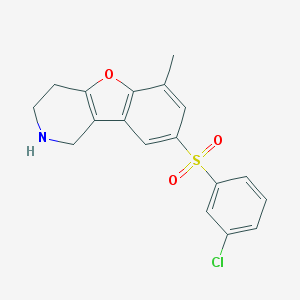 CHEMBL3693000 CHEMBL3693000 | C18H16ClNO3S | 361.84 | 4 / 1 | 3.6 | Yes |
| 7413 | 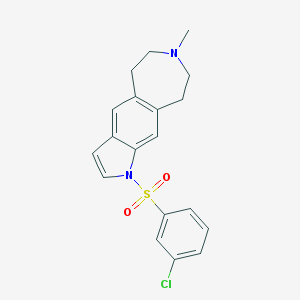 CHEMBL518591 CHEMBL518591 | C19H19ClN2O2S | 374.883 | 3 / 0 | 4.2 | Yes |
| 7542 | 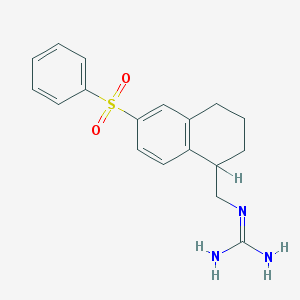 CHEMBL1086079 CHEMBL1086079 | C18H21N3O2S | 343.445 | 3 / 2 | 2.2 | Yes |
| 7668 | 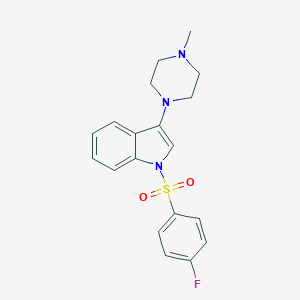 CHEMBL1642413 CHEMBL1642413 | C19H20FN3O2S | 373.446 | 5 / 0 | 3.3 | Yes |
| 7823 |  CHEMBL469668 CHEMBL469668 | C20H29F3N2O2 | 386.459 | 7 / 1 | 3.9 | Yes |
| 547979 |  CHEMBL3983496 CHEMBL3983496 | C32H41N3O4 | 531.697 | 6 / 1 | 5.2 | No |
| 521669 |  CHEMBL3786432 CHEMBL3786432 | C17H24N4O3S | 364.464 | 5 / 1 | 2.0 | Yes |
| 8013 |  CHEMBL1668564 CHEMBL1668564 | C16H18N4O3S | 346.405 | 6 / 1 | 2.1 | Yes |
| 8149 | 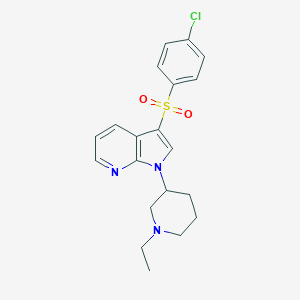 CHEMBL237179 CHEMBL237179 | C20H22ClN3O2S | 403.925 | 4 / 0 | 3.5 | Yes |
| 8202 | 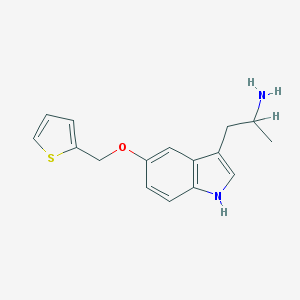 BW-723C86 BW-723C86 | C16H18N2OS | 286.393 | 3 / 2 | 3.0 | Yes |
| 8387 | 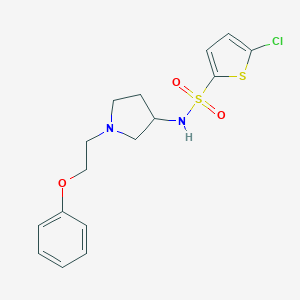 CHEMBL2159476 CHEMBL2159476 | C16H19ClN2O3S2 | 386.909 | 6 / 1 | 3.8 | Yes |
| 8470 | 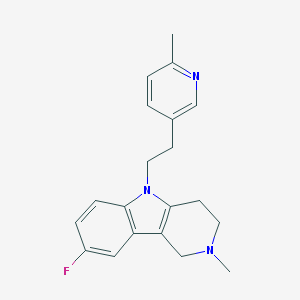 CHEMBL1783963 CHEMBL1783963 | C20H22FN3 | 323.415 | 3 / 0 | 3.2 | Yes |
| 8538 |  CHEMBL1642423 CHEMBL1642423 | C19H19Br2N3O2S | 513.248 | 4 / 0 | 4.5 | No |
| 521692 |  CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8626 |  CHEMBL2165557 CHEMBL2165557 | C22H26N2O3S | 398.521 | 4 / 0 | 4.4 | Yes |
| 8707 |  CHEMBL1922625 CHEMBL1922625 | C15H17N5O2S | 331.394 | 6 / 2 | 1.3 | Yes |
| 8818 | 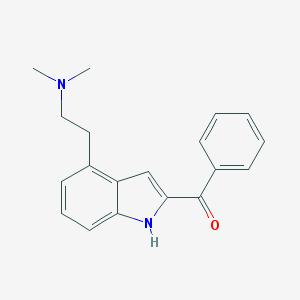 CHEMBL124382 CHEMBL124382 | C19H20N2O | 292.382 | 2 / 1 | 4.0 | Yes |
| 8827 | 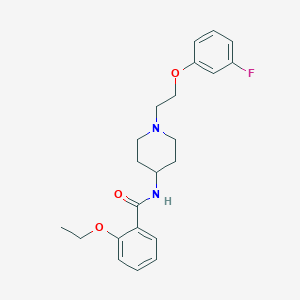 CHEMBL2159463 CHEMBL2159463 | C22H27FN2O3 | 386.467 | 5 / 1 | 3.9 | Yes |
| 8895 | 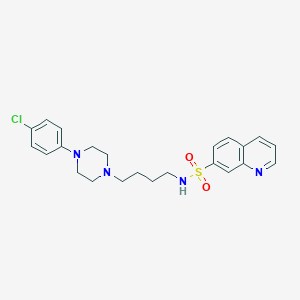 CHEMBL1949969 CHEMBL1949969 | C23H27ClN4O2S | 459.005 | 6 / 1 | 4.1 | Yes |
| 8953 | 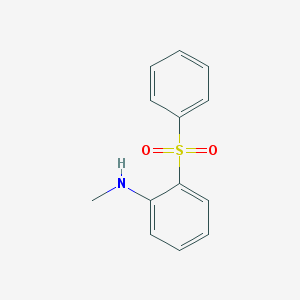 2-(Methylamino)diphenyl sulfone 2-(Methylamino)diphenyl sulfone | C13H13NO2S | 247.312 | 3 / 1 | 3.3 | Yes |
| 9289 | 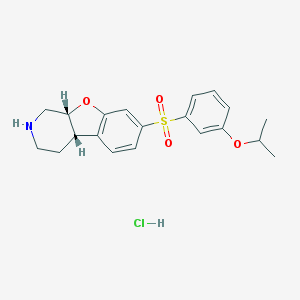 CHEMBL1949768 CHEMBL1949768 | C20H24ClNO4S | 409.925 | 5 / 2 | N/A | N/A |
| 9349 | 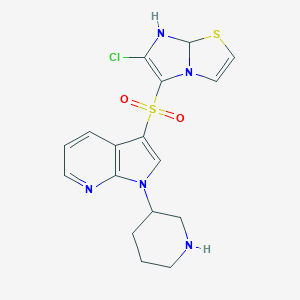 CID 44433196 CID 44433196 | C17H18ClN5O2S2 | 423.934 | 7 / 2 | 2.3 | Yes |
| 9362 | 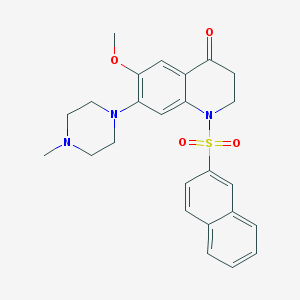 CHEMBL1642121 CHEMBL1642121 | C25H27N3O4S | 465.568 | 7 / 0 | 3.3 | Yes |
| 9544 | 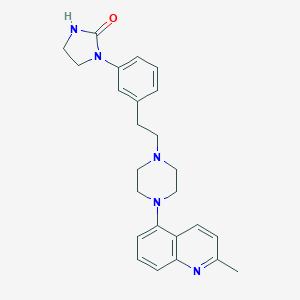 GSK163090 GSK163090 | C25H29N5O | 415.541 | 4 / 1 | 3.4 | Yes |
| 9635 | 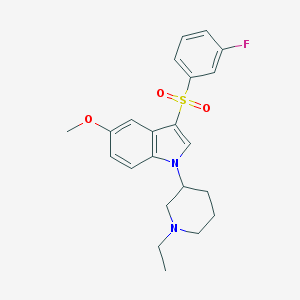 CHEMBL1081793 CHEMBL1081793 | C22H25FN2O3S | 416.511 | 5 / 0 | 3.7 | Yes |
| 9996 | 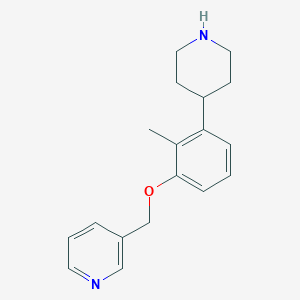 tolylpiperidine, 9g tolylpiperidine, 9g | C18H22N2O | 282.387 | 3 / 1 | 2.8 | Yes |
| 10070 | 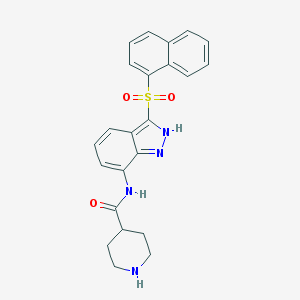 CHEMBL1642876 CHEMBL1642876 | C23H22N4O3S | 434.514 | 5 / 3 | 3.1 | Yes |
| 10123 | 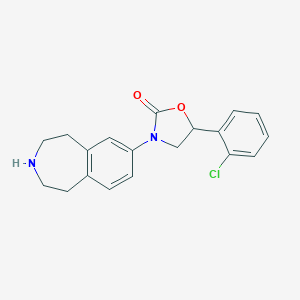 CHEMBL3260801 CHEMBL3260801 | C19H19ClN2O2 | 342.823 | 3 / 1 | 3.5 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417