

You can:
| Name | D(1A) dopamine receptor |
|---|---|
| Species | Sus scrofa (Pig) |
| Gene | DRD1 |
| Synonym | Dopamine D1 receptor |
| Disease | N/A for non-human GPCRs |
| Length | 446 |
| Amino acid sequence | MRTLNTSTMDGTGLVVERDFSFRILTACFLSLLILSTLLGNTLVCAAVIRFRHLRSKVTNFFVISLAVSDLLVAVLVMPWKAVAEIAGFWPFGSFCNIWVAFDIMCSTASILNLCVISVDRYWAISSPFRYERKMTPKAAFILISVAWTLSVLISFIPVQLSWHKAKPTSPSDGNVTSLGKTTHNCDSSLSRTYAISSSLISFYIPVAIMIVTYTRIYRIAQKQIRRISALERAAVHAKNCQTTAGNGNPAECSQPESSFKMSFKRETKVLKTLSVIMGVFVCCWLPFFILNCMVPFCGSGETKPFCIDSITFDVFVWFGWANSSLNPIIYAFNADFRKAFSTLLGCYRLCPTSTNAIETVSINNNGAVVFSSHHEPRGSISKDCNLVYLIPHAVGSSEDLKKEEAGGIASPLEKLSPALSVILDYDTDVSLEKIQPITQNGQHPT |
| UniProt | P50130 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5067 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 126 | 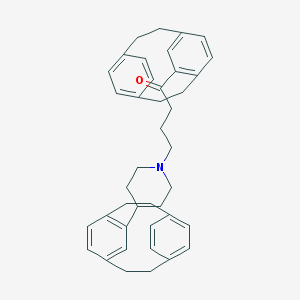 CHEMBL1256170 CHEMBL1256170 | C41H45NO | 567.817 | 2 / 0 | 9.2 | No |
| 633 | 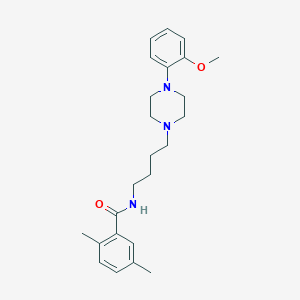 CHEMBL1258494 CHEMBL1258494 | C24H33N3O2 | 395.547 | 4 / 1 | 4.2 | Yes |
| 1584 | 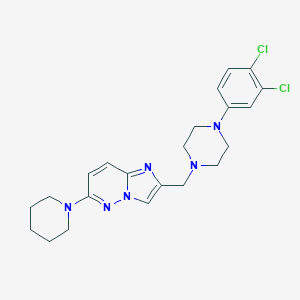 CHEMBL207543 CHEMBL207543 | C22H26Cl2N6 | 445.392 | 5 / 0 | 4.3 | Yes |
| 441750 | 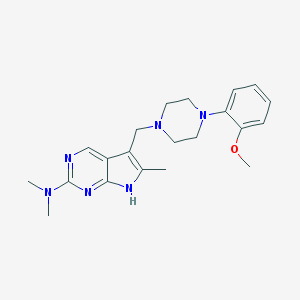 CHEMBL558838 CHEMBL558838 | C21H28N6O | 380.496 | 6 / 1 | 2.9 | Yes |
| 5033 | 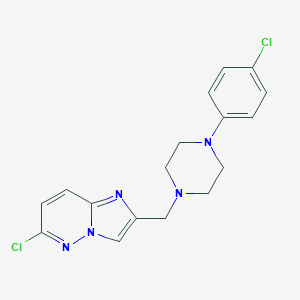 CHEMBL211135 CHEMBL211135 | C17H17Cl2N5 | 362.258 | 4 / 0 | 3.4 | Yes |
| 5238 | 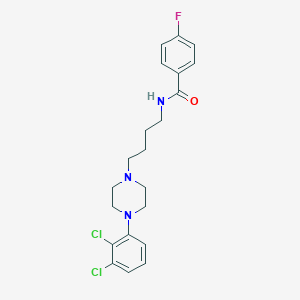 CHEMBL196744 CHEMBL196744 | C21H24Cl2FN3O | 424.341 | 4 / 1 | 4.9 | Yes |
| 8375 | 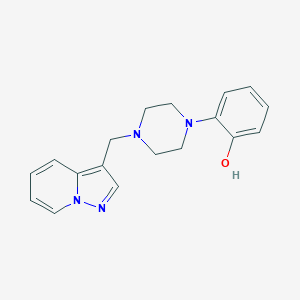 CHEMBL272602 CHEMBL272602 | C18H20N4O | 308.385 | 4 / 1 | 2.1 | Yes |
| 8939 | 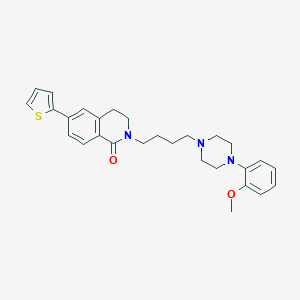 CHEMBL1771110 CHEMBL1771110 | C28H33N3O2S | 475.651 | 5 / 0 | 5.1 | No |
| 8994 |  CHEMBL1928132 CHEMBL1928132 | C62H92N6O13 | 1129.45 | 17 / 2 | 5.1 | No |
| 442128 |  CHEMBL3318840 CHEMBL3318840 | C58H72Cl2F2N2O6 | 1002.12 | 10 / 0 | 14.6 | No |
| 17030 |  CHEMBL240376 CHEMBL240376 | C25H31N3O2 | 405.542 | 4 / 1 | 4.1 | Yes |
| 18844 |  CHEMBL182379 CHEMBL182379 | C24H28IN3O3 | 533.41 | 5 / 1 | 4.9 | No |
| 19402 | 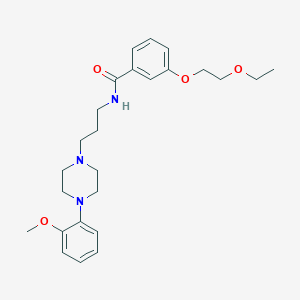 CHEMBL1928119 CHEMBL1928119 | C25H35N3O4 | 441.572 | 6 / 1 | 3.3 | Yes |
| 19693 | 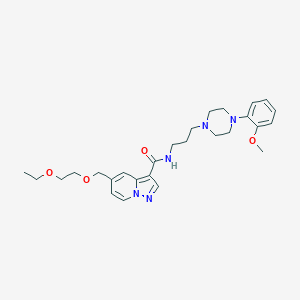 CHEMBL1928135 CHEMBL1928135 | C27H37N5O4 | 495.624 | 7 / 1 | 2.2 | Yes |
| 22054 | 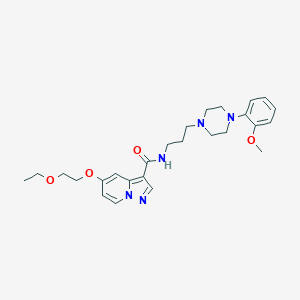 CHEMBL1928133 CHEMBL1928133 | C26H35N5O4 | 481.597 | 7 / 1 | 2.5 | Yes |
| 23144 | 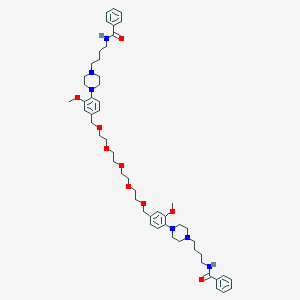 CHEMBL1928248 CHEMBL1928248 | C54H76N6O9 | 953.235 | 13 / 2 | 5.7 | No |
| 23242 | 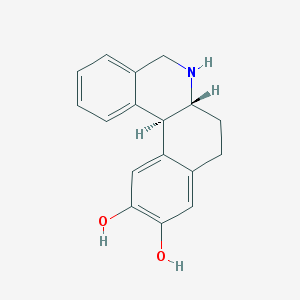 DIHYDREXIDINE DIHYDREXIDINE | C17H17NO2 | 267.328 | 3 / 3 | 2.5 | Yes |
| 23512 | 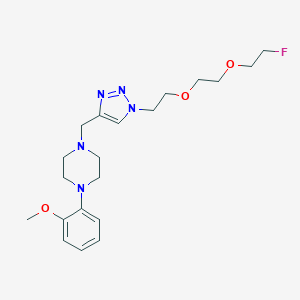 CHEMBL2443000 CHEMBL2443000 | C20H30FN5O3 | 407.49 | 8 / 0 | 1.1 | Yes |
| 24112 | 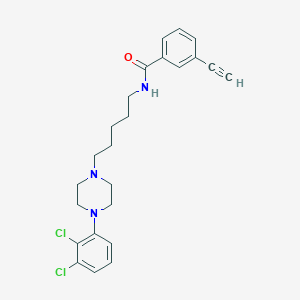 CHEMBL239733 CHEMBL239733 | C24H27Cl2N3O | 444.4 | 3 / 1 | 5.4 | No |
| 24261 | 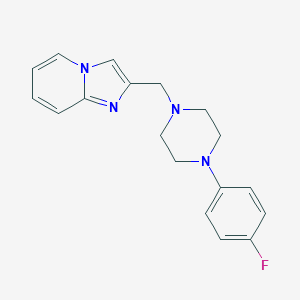 CHEMBL380054 CHEMBL380054 | C18H19FN4 | 310.376 | 4 / 0 | 3.3 | Yes |
| 31571 | 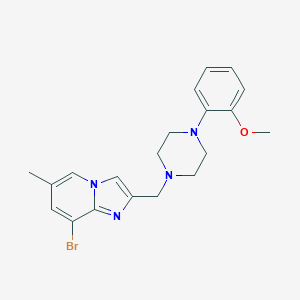 CHEMBL211539 CHEMBL211539 | C20H23BrN4O | 415.335 | 4 / 0 | 4.3 | Yes |
| 34184 | 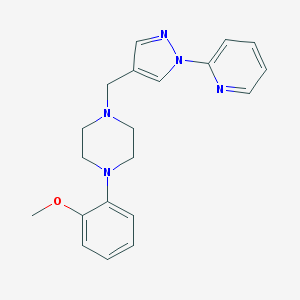 CHEMBL210404 CHEMBL210404 | C20H23N5O | 349.438 | 5 / 0 | 2.6 | Yes |
| 443081 | 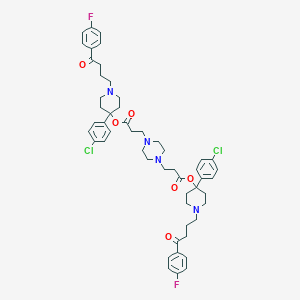 CHEMBL3318846 CHEMBL3318846 | C52H60Cl2F2N4O6 | 945.971 | 12 / 0 | 8.3 | No |
| 36125 | 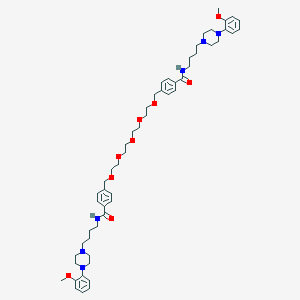 CHEMBL1928126 CHEMBL1928126 | C54H76N6O9 | 953.235 | 13 / 2 | 5.7 | No |
| 36461 | 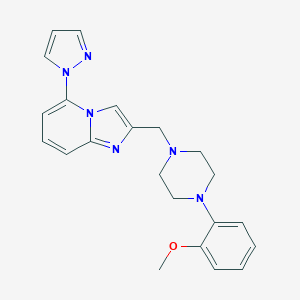 CHEMBL212160 CHEMBL212160 | C22H24N6O | 388.475 | 5 / 0 | 3.5 | Yes |
| 37348 | 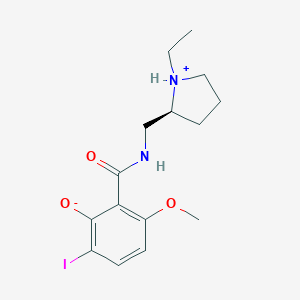 CID 29982233 CID 29982233 | C15H21IN2O3 | 404.248 | 3 / 2 | 2.9 | Yes |
| 37446 | 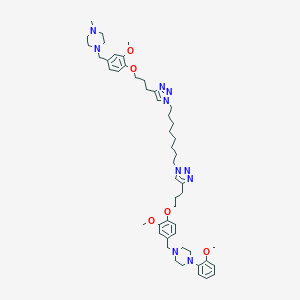 CHEMBL1803054 CHEMBL1803054 | C50H72N10O5 | 893.191 | 13 / 0 | 7.2 | No |
| 37567 | 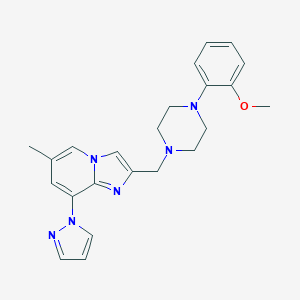 CHEMBL210551 CHEMBL210551 | C23H26N6O | 402.502 | 5 / 0 | 3.5 | Yes |
| 39369 | 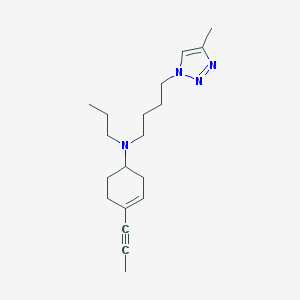 CHEMBL484203 CHEMBL484203 | C19H30N4 | 314.477 | 3 / 0 | 3.6 | Yes |
| 39771 | 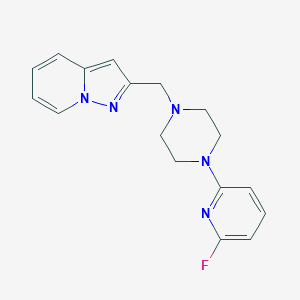 CHEMBL405292 CHEMBL405292 | C17H18FN5 | 311.364 | 5 / 0 | 2.2 | Yes |
| 39864 | 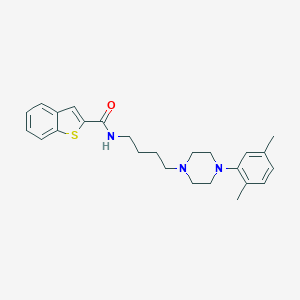 CHEMBL1258383 CHEMBL1258383 | C25H31N3OS | 421.603 | 4 / 1 | 5.6 | No |
| 443425 | 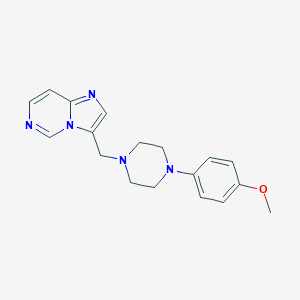 CHEMBL560125 CHEMBL560125 | C18H21N5O | 323.4 | 5 / 0 | 2.5 | Yes |
| 45132 | 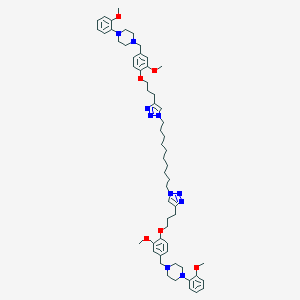 CHEMBL1803025 CHEMBL1803025 | C58H80N10O6 | 1013.34 | 14 / 0 | 10.0 | No |
| 443563 | 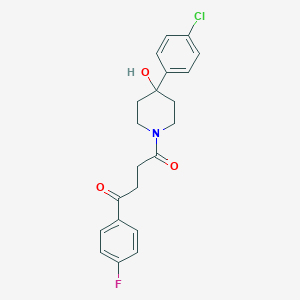 CHEMBL3318852 CHEMBL3318852 | C21H21ClFNO3 | 389.851 | 4 / 1 | 2.9 | Yes |
| 47680 | 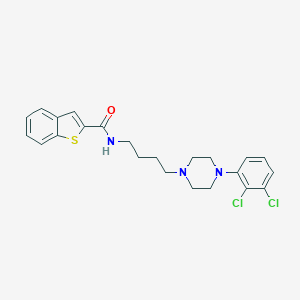 fauc-365 fauc-365 | C23H25Cl2N3OS | 462.433 | 4 / 1 | 6.2 | No |
| 47684 | 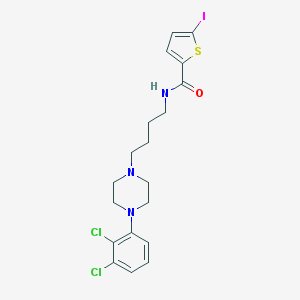 CHEMBL362101 CHEMBL362101 | C19H22Cl2IN3OS | 538.269 | 4 / 1 | 5.5 | No |
| 47912 |  CHEMBL391256 CHEMBL391256 | C24H29N3O2 | 391.515 | 4 / 1 | 3.8 | Yes |
| 48189 |  CHEMBL241212 CHEMBL241212 | C24H29N3O2 | 391.515 | 4 / 1 | 3.8 | Yes |
| 49831 |  CHEMBL1803049 CHEMBL1803049 | C28H39N5O3 | 493.652 | 7 / 0 | 4.6 | Yes |
| 50494 |  CHEMBL1928124 CHEMBL1928124 | C27H39N3O4 | 469.626 | 6 / 1 | 3.4 | Yes |
| 51696 | 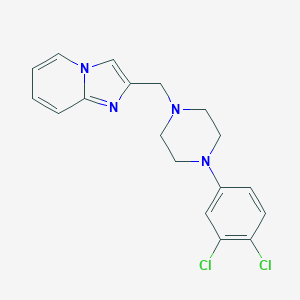 CHEMBL378680 CHEMBL378680 | C18H18Cl2N4 | 361.27 | 3 / 0 | 4.5 | Yes |
| 53507 | 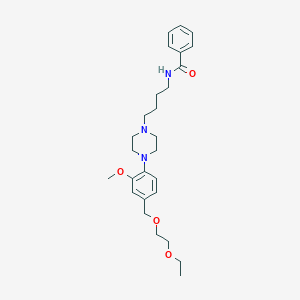 CHEMBL1928247 CHEMBL1928247 | C27H39N3O4 | 469.626 | 6 / 1 | 3.4 | Yes |
| 443899 | 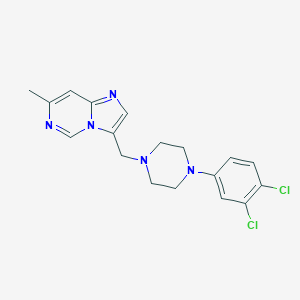 CHEMBL562591 CHEMBL562591 | C18H19Cl2N5 | 376.285 | 4 / 0 | 4.2 | Yes |
| 56549 | 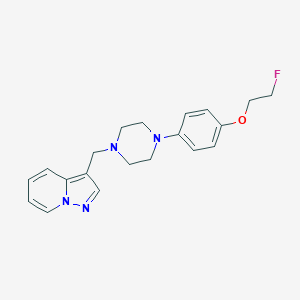 CHEMBL272873 CHEMBL272873 | C20H23FN4O | 354.429 | 5 / 0 | 2.8 | Yes |
| 61842 | 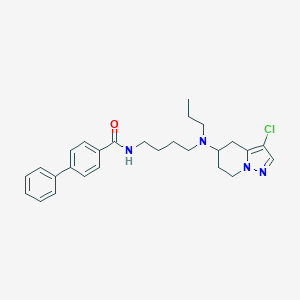 CHEMBL1765630 CHEMBL1765630 | C27H33ClN4O | 465.038 | 3 / 1 | 5.5 | No |
| 65392 | 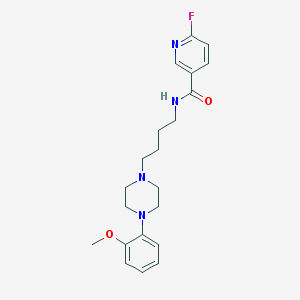 CHEMBL198993 CHEMBL198993 | C21H27FN4O2 | 386.471 | 6 / 1 | 2.9 | Yes |
| 444276 | 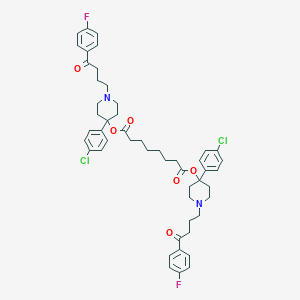 CHEMBL3318835 CHEMBL3318835 | C50H56Cl2F2N2O6 | 889.903 | 10 / 0 | 10.2 | No |
| 65731 | 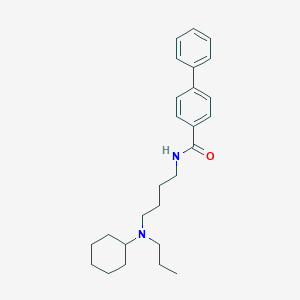 CHEMBL485227 CHEMBL485227 | C26H36N2O | 392.587 | 2 / 1 | 6.3 | No |
| 66164 | 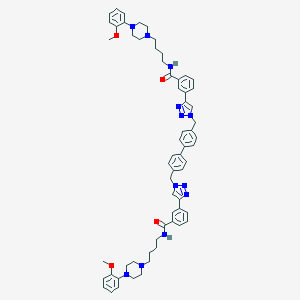 CHEMBL574569 CHEMBL574569 | C62H70N12O4 | 1047.32 | 12 / 2 | 8.8 | No |
| 444305 | 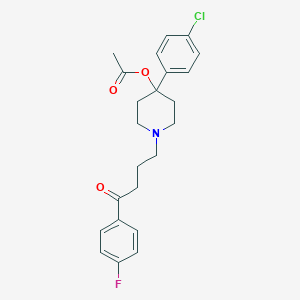 CHEMBL3318845 CHEMBL3318845 | C23H25ClFNO3 | 417.905 | 5 / 0 | 3.8 | Yes |
| 66874 | 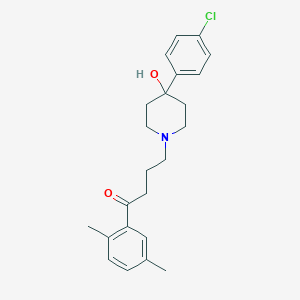 CHEMBL1257811 CHEMBL1257811 | C23H28ClNO2 | 385.932 | 3 / 1 | 3.9 | Yes |
| 68622 | 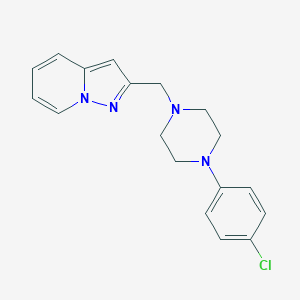 FAUC 213 FAUC 213 | C18H19ClN4 | 326.828 | 3 / 0 | 3.2 | Yes |
| 71180 |  CHEMBL1803030 CHEMBL1803030 | C26H35N5O3 | 465.598 | 7 / 0 | 3.7 | Yes |
| 72150 |  CHEMBL391042 CHEMBL391042 | C25H31N3O2 | 405.542 | 4 / 1 | 4.1 | Yes |
| 444668 |  CHEMBL3318831 CHEMBL3318831 | C46H48Cl2F2N2O6 | 833.795 | 10 / 0 | 8.4 | No |
| 444684 |  CHEMBL550920 CHEMBL550920 | C20H25N5O | 351.454 | 5 / 0 | 3.3 | Yes |
| 76902 | 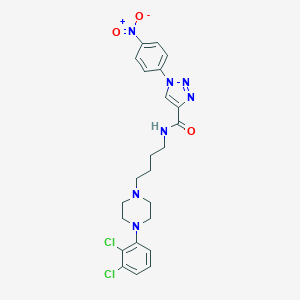 CHEMBL573792 CHEMBL573792 | C23H25Cl2N7O3 | 518.399 | 7 / 1 | 4.4 | No |
| 77830 | 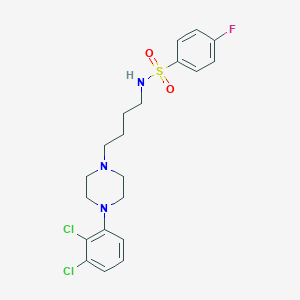 CHEMBL195247 CHEMBL195247 | C20H24Cl2FN3O2S | 460.389 | 6 / 1 | 4.6 | Yes |
| 80456 | 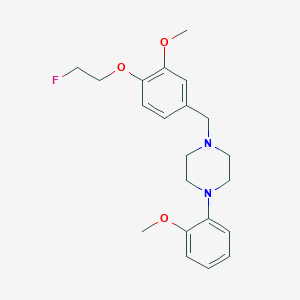 CHEMBL256492 CHEMBL256492 | C21H27FN2O3 | 374.456 | 6 / 0 | 3.6 | Yes |
| 80735 | 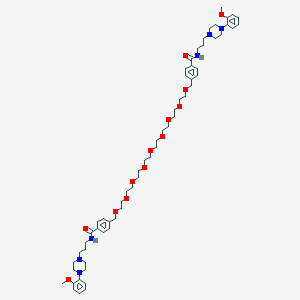 CHEMBL1928131 CHEMBL1928131 | C60H88N6O13 | 1101.39 | 17 / 2 | 4.4 | No |
| 80856 |  CHEMBL240106 CHEMBL240106 | C17H23BrN2O3 | 383.286 | 4 / 1 | 3.1 | Yes |
| 81396 |  CHEMBL2443010 CHEMBL2443010 | C18H26FN5O2 | 363.437 | 7 / 0 | 1.3 | Yes |
| 82398 |  CHEMBL1258036 CHEMBL1258036 | C30H33FN2O | 456.605 | 4 / 0 | 6.1 | No |
| 83258 |  CHEMBL241438 CHEMBL241438 | C31H35N3O2 | 481.64 | 4 / 1 | 5.9 | No |
| 87001 | 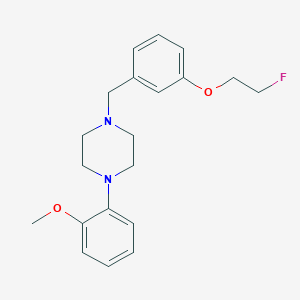 CHEMBL270408 CHEMBL270408 | C20H25FN2O2 | 344.43 | 5 / 0 | 3.6 | Yes |
| 87066 | 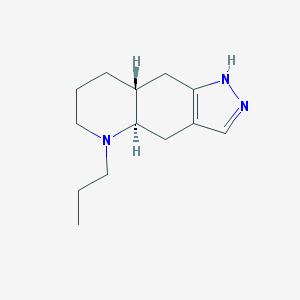 QUINPIROLE QUINPIROLE | C13H21N3 | 219.332 | 2 / 1 | 2.3 | Yes |
| 91499 | 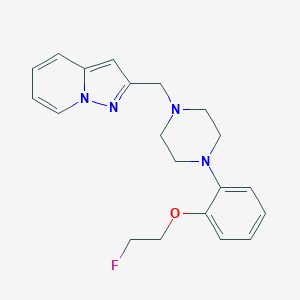 CHEMBL405260 CHEMBL405260 | C20H23FN4O | 354.429 | 5 / 0 | 2.8 | Yes |
| 445410 | 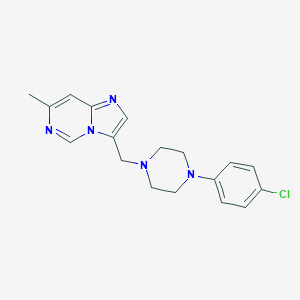 CHEMBL549575 CHEMBL549575 | C18H20ClN5 | 341.843 | 4 / 0 | 3.6 | Yes |
| 93546 | 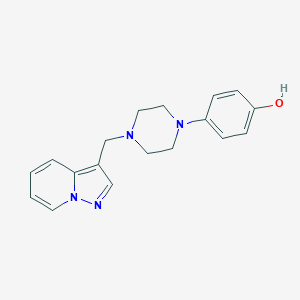 CHEMBL407818 CHEMBL407818 | C18H20N4O | 308.385 | 4 / 1 | 2.1 | Yes |
| 95126 | 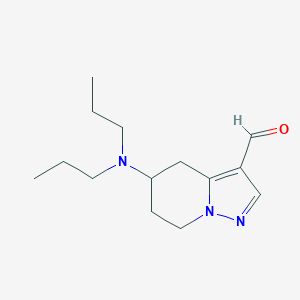 CHEMBL198174 CHEMBL198174 | C14H23N3O | 249.358 | 3 / 0 | 1.9 | Yes |
| 96273 | 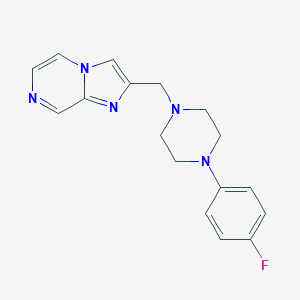 CHEMBL377542 CHEMBL377542 | C17H18FN5 | 311.364 | 5 / 0 | 2.3 | Yes |
| 97845 | 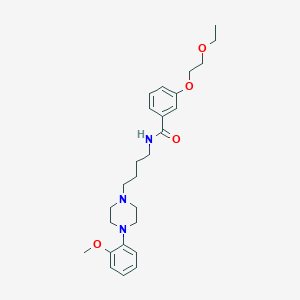 CHEMBL1928120 CHEMBL1928120 | C26H37N3O4 | 455.599 | 6 / 1 | 3.7 | Yes |
| 98176 |  CHEMBL241424 CHEMBL241424 | C30H33N3O2 | 467.613 | 4 / 1 | 5.5 | No |
| 101428 |  CHEMBL484202 CHEMBL484202 | C29H34N4 | 438.619 | 3 / 0 | 5.9 | No |
| 101537 |  UNII-UGT5535REQ UNII-UGT5535REQ | C17H18ClNO | 287.787 | 2 / 1 | 4.0 | Yes |
| 445731 |  CHEMBL562125 CHEMBL562125 | C19H21Cl2N5 | 390.312 | 4 / 0 | 4.6 | Yes |
| 445770 | 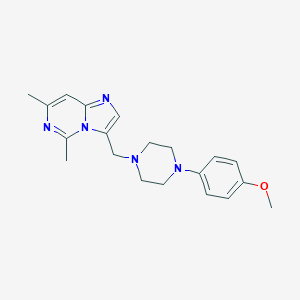 CHEMBL562064 CHEMBL562064 | C20H25N5O | 351.454 | 5 / 0 | 3.3 | Yes |
| 104570 | 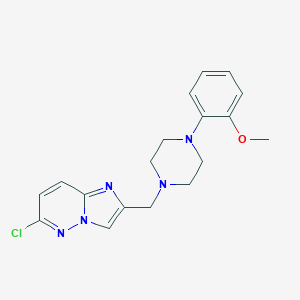 CHEMBL378515 CHEMBL378515 | C18H20ClN5O | 357.842 | 5 / 0 | 2.7 | Yes |
| 105784 | 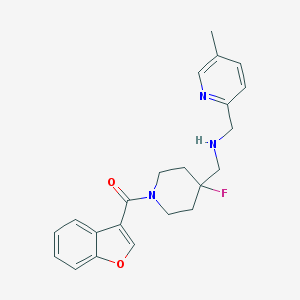 CHEMBL1258999 CHEMBL1258999 | C22H24FN3O2 | 381.451 | 5 / 1 | 2.9 | Yes |
| 445880 | 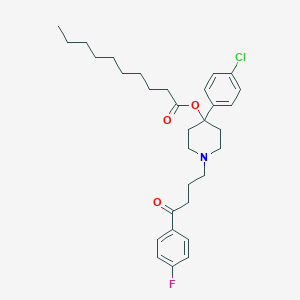 HALOPERIDOL DECANOATE HALOPERIDOL DECANOATE | C31H41ClFNO3 | 530.121 | 5 / 0 | 7.9 | No |
| 106709 | 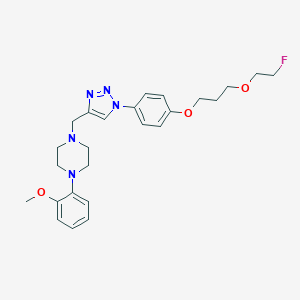 CHEMBL2443007 CHEMBL2443007 | C25H32FN5O3 | 469.561 | 8 / 0 | 3.3 | Yes |
| 111033 | 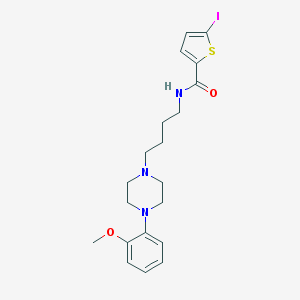 CHEMBL182652 CHEMBL182652 | C20H26IN3O2S | 499.411 | 5 / 1 | 4.2 | Yes |
| 113615 | 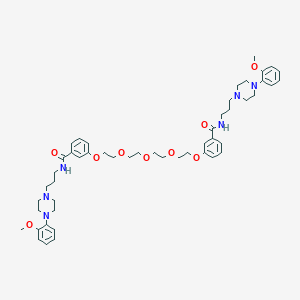 CHEMBL1928121 CHEMBL1928121 | C50H68N6O9 | 897.127 | 13 / 2 | 5.6 | No |
| 114441 | 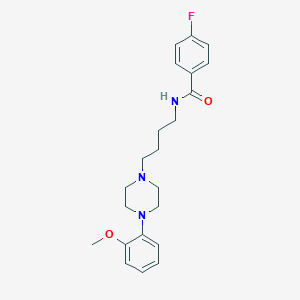 CHEMBL194727 CHEMBL194727 | C22H28FN3O2 | 385.483 | 5 / 1 | 3.6 | Yes |
| 115693 |  CHEMBL1258382 CHEMBL1258382 | C33H37N3OS | 523.739 | 4 / 1 | 7.4 | No |
| 117373 |  CHEMBL2397478 CHEMBL2397478 | C32H47N5O3 | 549.76 | 6 / 1 | 6.0 | No |
| 117375 |  CHEMBL2397477 CHEMBL2397477 | C32H47N5O3 | 549.76 | 6 / 1 | 6.0 | No |
| 118723 |  CHEMBL1928123 CHEMBL1928123 | C26H37N3O4 | 455.599 | 6 / 1 | 3.0 | Yes |
| 119243 | 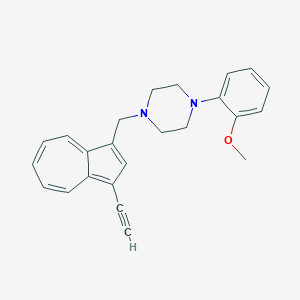 CHEMBL2207632 CHEMBL2207632 | C24H24N2O | 356.469 | 3 / 0 | 4.6 | Yes |
| 120525 | 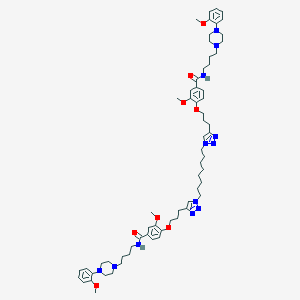 CHEMBL1916550 CHEMBL1916550 | C64H90N12O8 | 1155.5 | 16 / 2 | 9.3 | No |
| 122080 | 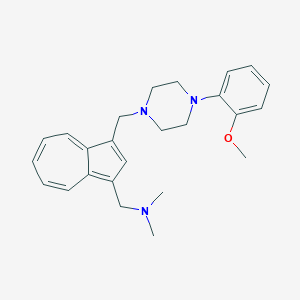 CHEMBL2207637 CHEMBL2207637 | C25H31N3O | 389.543 | 4 / 0 | 4.1 | Yes |
| 124933 | 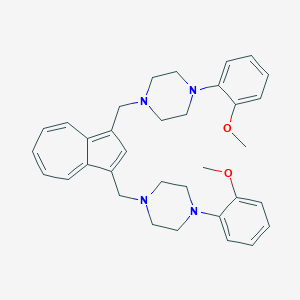 CHEMBL2207641 CHEMBL2207641 | C34H40N4O2 | 536.72 | 6 / 0 | 5.7 | No |
| 126076 | 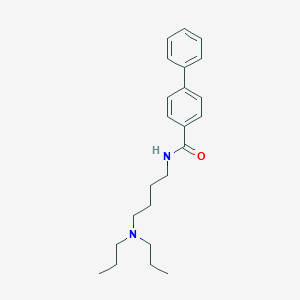 CHEMBL484204 CHEMBL484204 | C23H32N2O | 352.522 | 2 / 1 | 5.0 | Yes |
| 128139 | 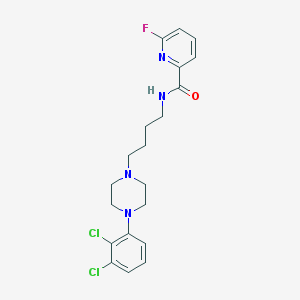 CHEMBL197195 CHEMBL197195 | C20H23Cl2FN4O | 425.329 | 5 / 1 | 4.5 | Yes |
| 130490 | 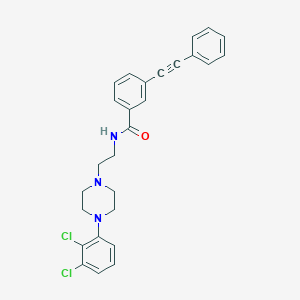 CHEMBL241211 CHEMBL241211 | C27H25Cl2N3O | 478.417 | 3 / 1 | 6.1 | No |
| 130542 | 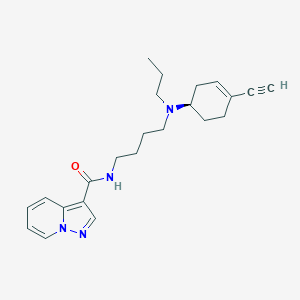 CHEMBL2397392 CHEMBL2397392 | C23H30N4O | 378.52 | 3 / 1 | 3.4 | Yes |
| 130544 | 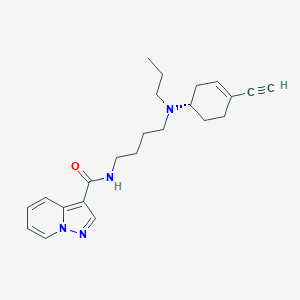 CHEMBL2397476 CHEMBL2397476 | C23H30N4O | 378.52 | 3 / 1 | 3.4 | Yes |
| 134924 | 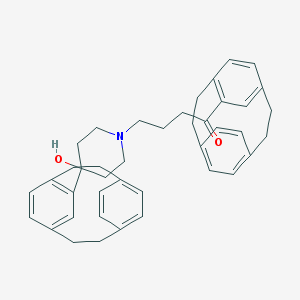 CHEMBL1256169 CHEMBL1256169 | C41H45NO2 | 583.816 | 3 / 1 | 8.1 | No |
| 137700 | 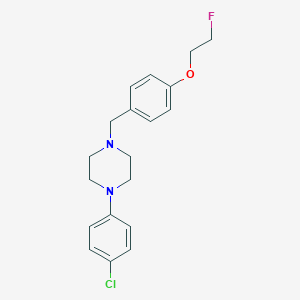 CHEMBL272073 CHEMBL272073 | C19H22ClFN2O | 348.846 | 4 / 0 | 4.3 | Yes |
| 137873 | 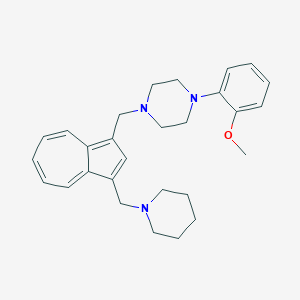 CHEMBL2207638 CHEMBL2207638 | C28H35N3O | 429.608 | 4 / 0 | 5.0 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417