

You can:
| Name | CHEMBL2397392 |
|---|---|
| Molecular formula | C23H30N4O |
| IUPAC name | N-[4-[[(1R)-4-ethynylcyclohex-3-en-1-yl]-propylamino]butyl]pyrazolo[1,5-a]pyridine-3-carboxamide |
| Molecular weight | 378.52 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 1 |
| XlogP | 3.4 |
| Synonyms | BDBM50436538 |
| Inchi Key | IDKIRFAIRCIGSS-FQEVSTJZSA-N |
| Inchi ID | InChI=1S/C23H30N4O/c1-3-15-26(20-12-10-19(4-2)11-13-20)16-8-6-14-24-23(28)21-18-25-27-17-7-5-9-22(21)27/h2,5,7,9-10,17-18,20H,3,6,8,11-16H2,1H3,(H,24,28)/t20-/m0/s1 |
| PubChem CID | 71734029 |
| ChEMBL | CHEMBL2397392 |
| IUPHAR | N/A |
| BindingDB | 50436538 |
| DrugBank | N/A |
Structure | 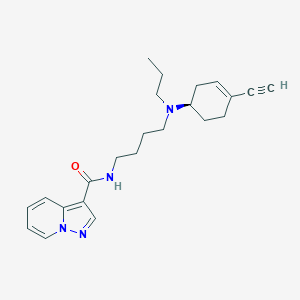 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
You can:
| GLASS ID | Name | UniProt | Gene | Species | Length |
|---|---|---|---|---|---|
| 130542 | D(1A) dopamine receptor | P50130 | DRD1 | Sus scrofa (Pig) | 446 |
| 130540 | D(2) dopamine receptor | P14416 | DRD2 | Homo sapiens (Human) | 443 |
| 130541 | D(3) dopamine receptor | P35462 | DRD3 | Homo sapiens (Human) | 400 |
| 525291 | D(4) dopamine receptor | P21917 | DRD4 | Homo sapiens (Human) | 467 |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417