

You can:
| Name | Metabotropic glutamate receptor 4 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm4 |
| Synonym | glutamate receptor GPRC1D mGlu4 receptor mGluR4 |
| Disease | N/A for non-human GPCRs |
| Length | 912 |
| Amino acid sequence | MSGKGGWAWWWARLPLCLLLSLYAPWVPSSLGKPKGHPHMNSIRIDGDITLGGLFPVHGRGSEGKACGELKKEKGIHRLEAMLFALDRINNDPDLLPNITLGARILDTCSRDTHALEQSLTFVQALIEKDGTEVRCGSGGPPIITKPERVVGVIGASGSSVSIMVANILRLFKIPQISYASTAPDLSDNSRYDFFSRVVPSDTYQAQAMVDIVRALKWNYVSTLASEGSYGESGVEAFIQKSRENGGVCIAQSVKIPREPKTGEFDKIIKRLLETSNARGIIIFANEDDIRRVLEAARRANQTGHFFWMGSDSWGSKSAPVLRLEEVAEGAVTILPKRMSVRGFDRYFSSRTLDNNRRNIWFAEFWEDNFHCKLSRHALKKGSHIKKCTNRERIGQDSAYEQEGKVQFVIDAVYAMGHALHAMHRDLCPGRVGLCPRMDPVDGTQLLKYIRNVNFSGIAGNPVTFNENGDAPGRYDIYQYQLRNGSAEYKVIGSWTDHLHLRIERMQWPGSGQQLPRSICSLPCQPGERKKTVKGMACCWHCEPCTGYQYQVDRYTCKTCPYDMRPTENRTSCQPIPIVKLEWDSPWAVLPLFLAVVGIAATLFVVVTFVRYNDTPIVKASGRELSYVLLAGIFLCYATTFLMIAEPDLGTCSLRRIFLGLGMSISYAALLTKTNRIYRIFEQGKRSVSAPRFISPASQLAITFILISLQLLGICVWFVVDPSHSVVDFQDQRTLDPRFARGVLKCDISDLSLICLLGYSMLLMVTCTVYAIKTRGVPETFNEAKPIGFTMYTTCIVWLAFIPIFFGTSQSADKLYIQTTTLTVSVSLSASVSLGMLYMPKVYIILFHPEQNVPKRKRSLKAVVTAATMSNKFTQKGNFRPNGEAKSELCENLETPALATKQTYVTYTNHAI |
| UniProt | P31423 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2787 |
| IUPHAR | 292 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 557353 | 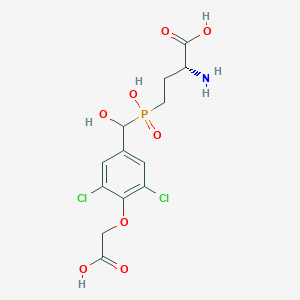 US9212196, Derivative 9 US9212196, Derivative 9 | C13H16Cl2NO8P | 416.144 | 9 / 5 | -2.6 | Yes |
| 535969 | 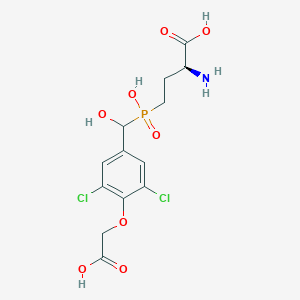 CHEMBL3974450 CHEMBL3974450 | C13H16Cl2NO8P | 416.144 | 9 / 5 | -2.6 | Yes |
| 535990 | 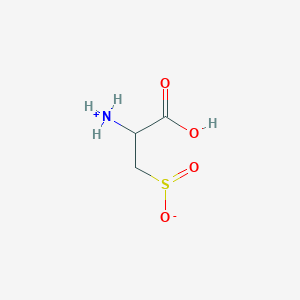 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 6714 | 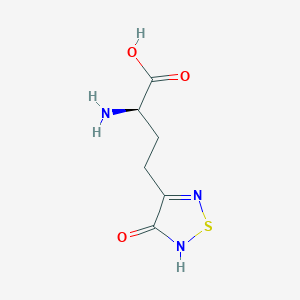 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 6717 | 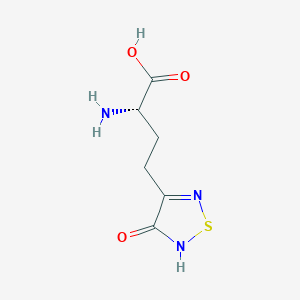 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 8297 | 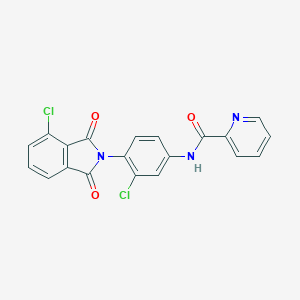 VU0405623-1 VU0405623-1 | C20H11Cl2N3O3 | 412.226 | 4 / 1 | 3.8 | Yes |
| 442080 | 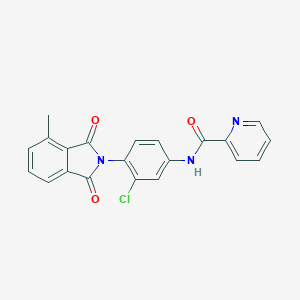 CHEMBL3357575 CHEMBL3357575 | C21H14ClN3O3 | 391.811 | 4 / 1 | 3.5 | Yes |
| 9584 | 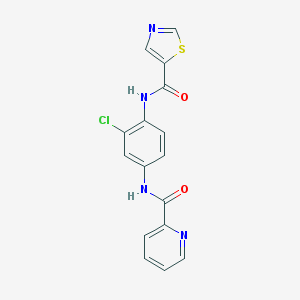 CHEMBL1909433 CHEMBL1909433 | C16H11ClN4O2S | 358.8 | 5 / 2 | 2.8 | Yes |
| 10198 | 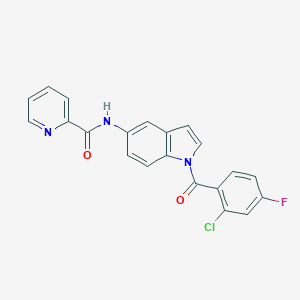 CHEMBL1672256 CHEMBL1672256 | C21H13ClFN3O2 | 393.802 | 4 / 1 | 4.4 | Yes |
| 557605 | 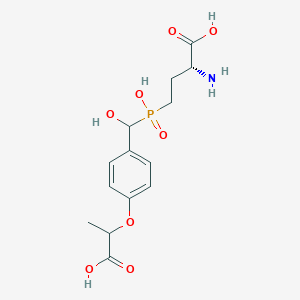 US9212196, Derivative 29 US9212196, Derivative 29 | C14H20NO8P | 361.287 | 9 / 5 | -3.4 | Yes |
| 536280 | 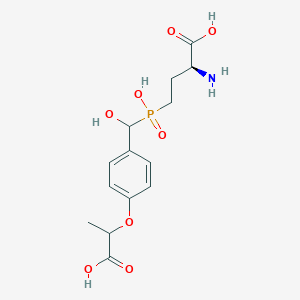 CHEMBL3967637 CHEMBL3967637 | C14H20NO8P | 361.287 | 9 / 5 | -3.4 | Yes |
| 521773 | 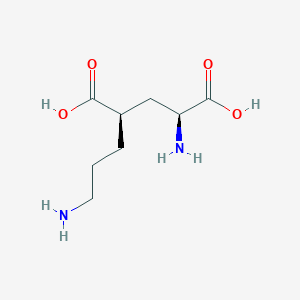 CHEMBL2312685 CHEMBL2312685 | C8H16N2O4 | 204.226 | 6 / 4 | -6.8 | Yes |
| 12905 | 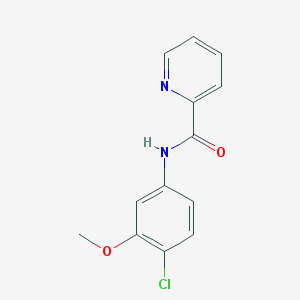 VU 0361737 VU 0361737 | C13H11ClN2O2 | 262.693 | 3 / 1 | 2.6 | Yes |
| 13245 | 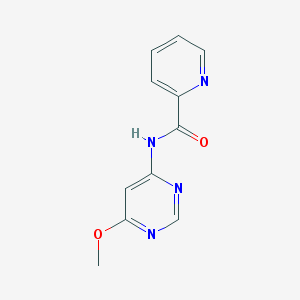 N-(6-methoxypyrimidin-4-yl)picolinamide N-(6-methoxypyrimidin-4-yl)picolinamide | C11H10N4O2 | 230.227 | 5 / 1 | 0.9 | Yes |
| 13320 | 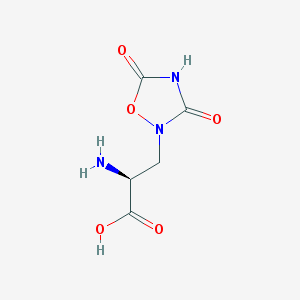 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 557654 | 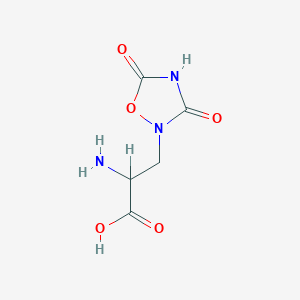 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid 2-amino-3-(3,5-dioxo-1,2,4-oxadiazolidin-2-yl)propanoic acid | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 13334 | 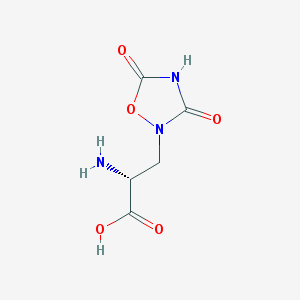 CHEMBL168279 CHEMBL168279 | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 15043 | 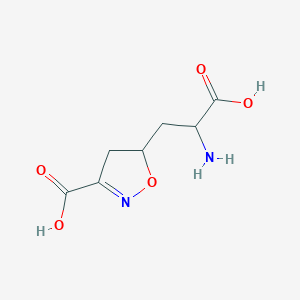 CHEMBL197629 CHEMBL197629 | C7H10N2O5 | 202.166 | 7 / 3 | -3.4 | Yes |
| 16024 | 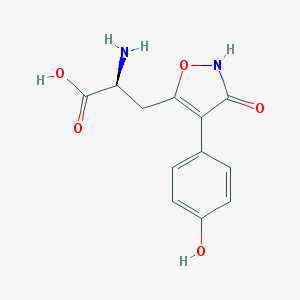 CHEMBL367027 CHEMBL367027 | C12H12N2O5 | 264.237 | 6 / 4 | -2.3 | Yes |
| 522217 | 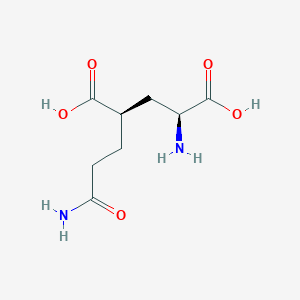 CHEMBL479804 CHEMBL479804 | C8H14N2O5 | 218.209 | 6 / 4 | -3.3 | Yes |
| 22859 | 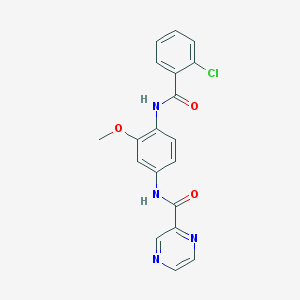 CHEMBL1672235 CHEMBL1672235 | C19H15ClN4O3 | 382.804 | 5 / 2 | 2.4 | Yes |
| 536571 | 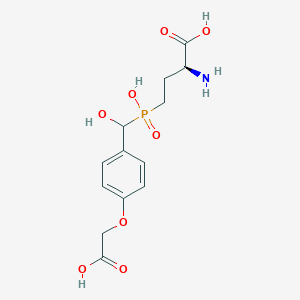 LSP4-2022 LSP4-2022 | C13H18NO8P | 347.26 | 9 / 5 | -3.8 | Yes |
| 522260 | 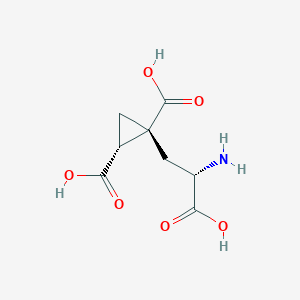 CHEMBL3786667 CHEMBL3786667 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522269 | 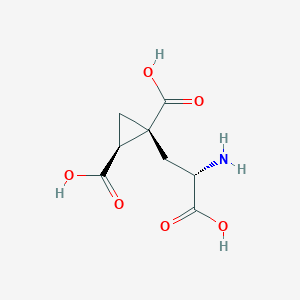 CHEMBL3787264 CHEMBL3787264 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522275 | 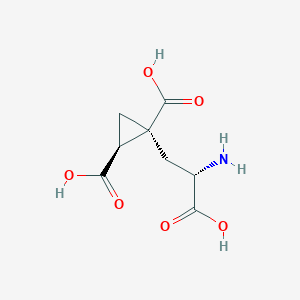 CHEMBL3786026 CHEMBL3786026 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 522288 | 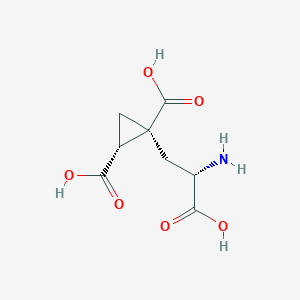 CHEMBL3786938 CHEMBL3786938 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 25660 | 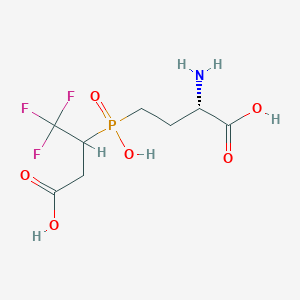 CHEMBL1093010 CHEMBL1093010 | C8H13F3NO6P | 307.162 | 10 / 4 | -3.7 | Yes |
| 467015 | 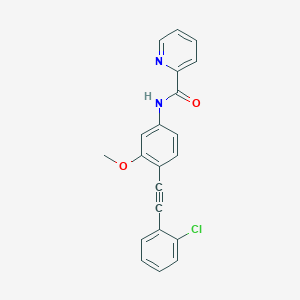 CHEMBL3596586 CHEMBL3596586 | C21H15ClN2O2 | 362.813 | 3 / 1 | 4.6 | Yes |
| 33521 | 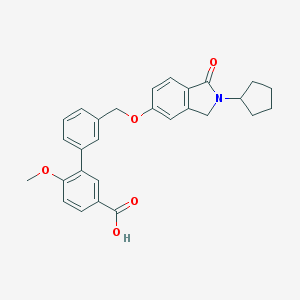 CHEMBL2179631 CHEMBL2179631 | C28H27NO5 | 457.526 | 5 / 1 | 4.8 | Yes |
| 34971 | 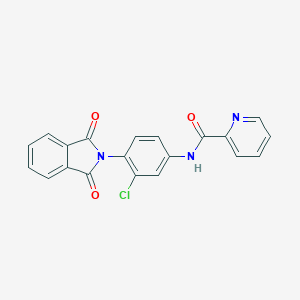 VU0405622-1 VU0405622-1 | C20H12ClN3O3 | 377.784 | 4 / 1 | 3.2 | Yes |
| 36689 | 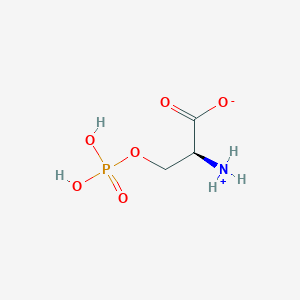 CID 57689797 CID 57689797 | C3H8NO6P | 185.072 | 6 / 3 | -4.5 | Yes |
| 37461 | 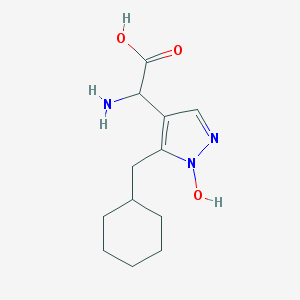 CHEMBL242344 CHEMBL242344 | C12H19N3O3 | 253.302 | 5 / 3 | -0.3 | Yes |
| 558420 | 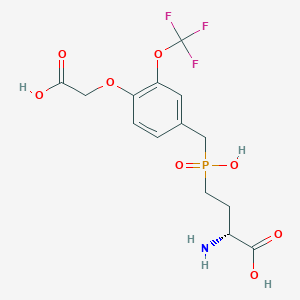 US9212196, Derivative 32 US9212196, Derivative 32 | C14H17F3NO8P | 415.258 | 12 / 4 | -2.1 | No |
| 536936 | 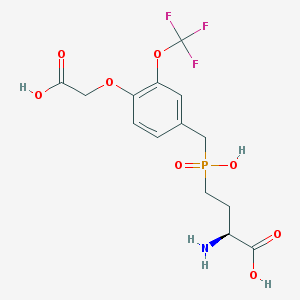 CHEMBL3910941 CHEMBL3910941 | C14H17F3NO8P | 415.258 | 12 / 4 | -2.1 | No |
| 536962 | 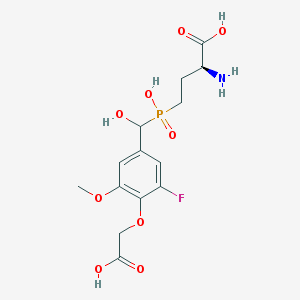 CHEMBL3898023 CHEMBL3898023 | C14H19FNO9P | 395.276 | 11 / 5 | -3.8 | No |
| 558443 | 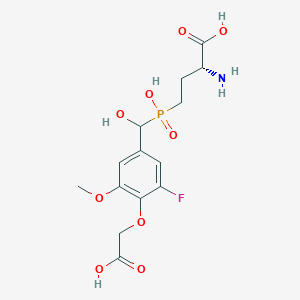 US9212196, Derivative 6 US9212196, Derivative 6 | C14H19FNO9P | 395.276 | 11 / 5 | -3.8 | No |
| 40144 | 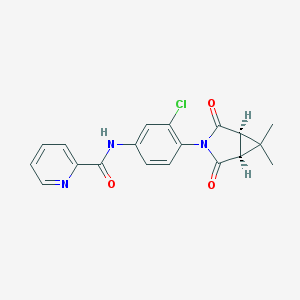 CHEMBL1711049 CHEMBL1711049 | C19H16ClN3O3 | 369.805 | 4 / 1 | 2.5 | Yes |
| 537065 | 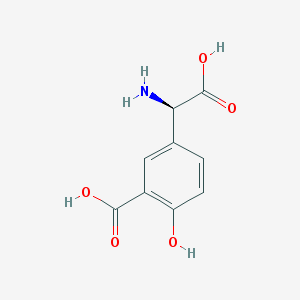 (R)-3-Carboxy-4-hydroxyphenylglycine (R)-3-Carboxy-4-hydroxyphenylglycine | C9H9NO5 | 211.173 | 6 / 4 | -1.2 | Yes |
| 537066 | 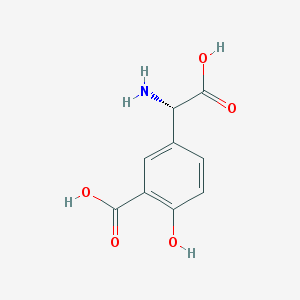 (S)-3C4HPG (S)-3C4HPG | C9H9NO5 | 211.173 | 6 / 4 | -1.2 | Yes |
| 42847 | 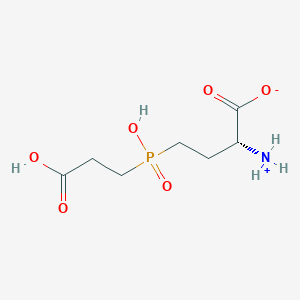 MolPort-000-743-361 MolPort-000-743-361 | C7H14NO6P | 239.164 | 6 / 3 | -4.2 | Yes |
| 42851 | 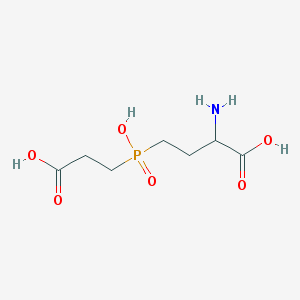 AC1MWWXC AC1MWWXC | C7H14NO6P | 239.164 | 7 / 4 | -4.8 | Yes |
| 42856 | 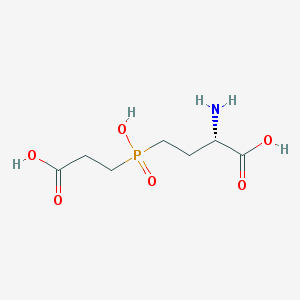 CHEMBL1089515 CHEMBL1089515 | C7H14NO6P | 239.164 | 7 / 4 | -4.8 | Yes |
| 537084 | 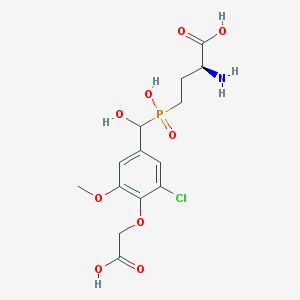 CHEMBL3969216 CHEMBL3969216 | C14H19ClNO9P | 411.728 | 10 / 5 | -3.2 | Yes |
| 558600 | 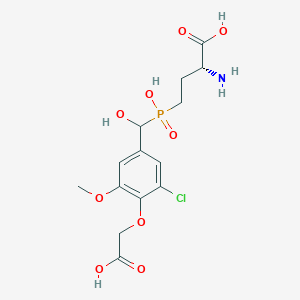 US9212196, Derivative 7 US9212196, Derivative 7 | C14H19ClNO9P | 411.728 | 10 / 5 | -3.2 | Yes |
| 519954 | 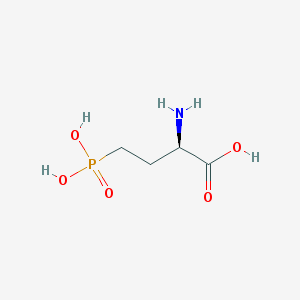 D-AP4 D-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 537399 | 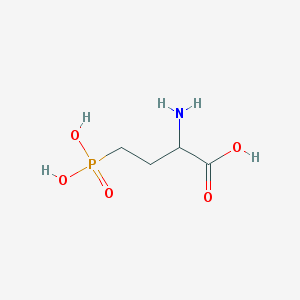 2-amino-4-phosphonobutanoic acid 2-amino-4-phosphonobutanoic acid | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 57285 | 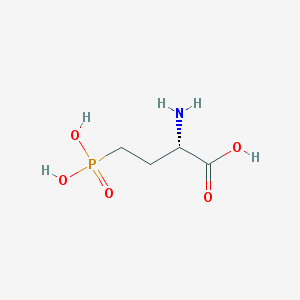 L-AP4 L-AP4 | C4H10NO5P | 183.1 | 6 / 4 | -5.5 | Yes |
| 58689 | 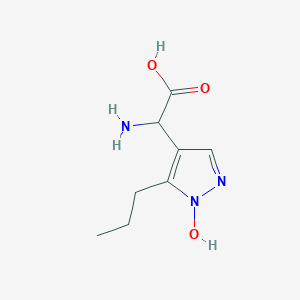 CHEMBL389585 CHEMBL389585 | C8H13N3O3 | 199.21 | 5 / 3 | -2.1 | Yes |
| 62847 | 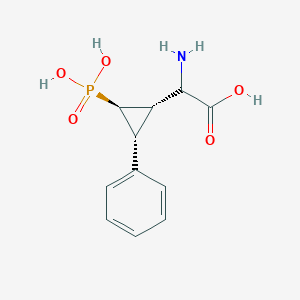 CHEMBL234138 CHEMBL234138 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 62851 | 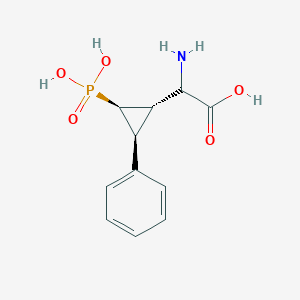 CHEMBL392419 CHEMBL392419 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 62861 | 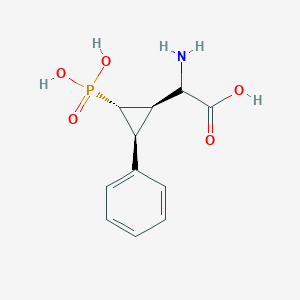 CHEMBL396319 CHEMBL396319 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 62864 | 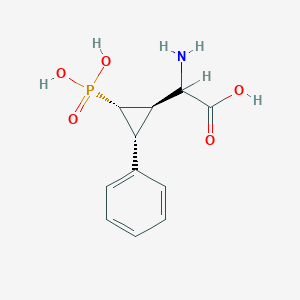 CHEMBL392420 CHEMBL392420 | C11H14NO5P | 271.209 | 6 / 4 | -3.3 | Yes |
| 537498 | 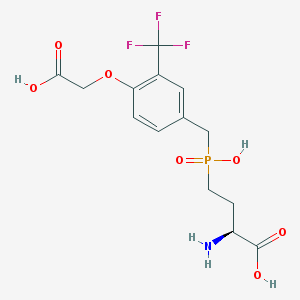 CHEMBL3980977 CHEMBL3980977 | C14H17F3NO7P | 399.259 | 11 / 4 | -2.4 | No |
| 559220 | 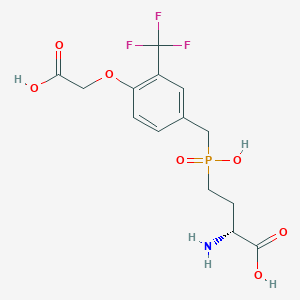 US9212196, Derivative 33 US9212196, Derivative 33 | C14H17F3NO7P | 399.259 | 11 / 4 | -2.4 | No |
| 537526 | 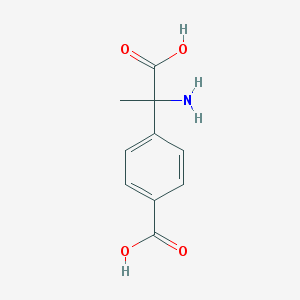 (RS)-MCPG (RS)-MCPG | C10H11NO4 | 209.201 | 5 / 3 | -2.0 | Yes |
| 537608 | 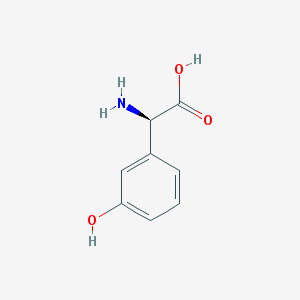 25698-27-5 25698-27-5 | C8H9NO3 | 167.164 | 4 / 3 | -2.1 | Yes |
| 537609 |  CID 40428795 CID 40428795 | C8H9NO3 | 167.164 | 3 / 2 | -1.4 | Yes |
| 537631 |  CHEMBL3895389 CHEMBL3895389 | C14H20NO8P | 361.287 | 9 / 4 | -3.3 | Yes |
| 559389 |  US9212196, Derivative 31 US9212196, Derivative 31 | C14H20NO8P | 361.287 | 9 / 4 | -3.3 | Yes |
| 68260 |  SCHEMBL1897912 SCHEMBL1897912 | C16H12ClN3O3 | 329.74 | 4 / 1 | 1.4 | Yes |
| 559478 |  US9212196, Derivative 19 US9212196, Derivative 19 | C15H22NO9P | 391.313 | 10 / 5 | -3.5 | Yes |
| 537686 | 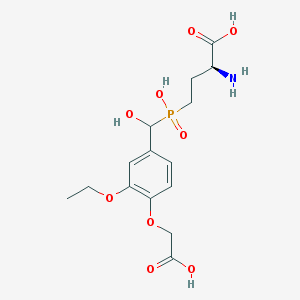 CHEMBL3941586 CHEMBL3941586 | C15H22NO9P | 391.313 | 10 / 5 | -3.5 | Yes |
| 70059 | 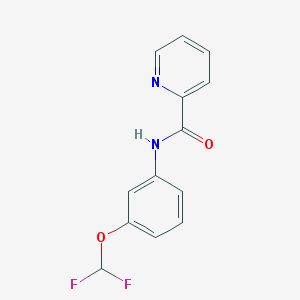 CHEMBL541754 CHEMBL541754 | C13H10F2N2O2 | 264.232 | 5 / 1 | 2.9 | Yes |
| 76546 | 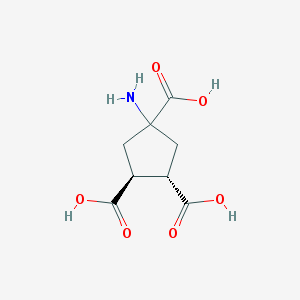 CHEMBL329236 CHEMBL329236 | C8H11NO6 | 217.177 | 7 / 4 | -4.0 | Yes |
| 82270 | 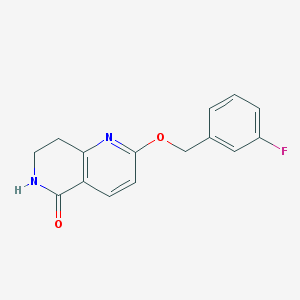 CHEMBL3298273 CHEMBL3298273 | C15H13FN2O2 | 272.279 | 4 / 1 | 2.1 | Yes |
| 84450 | 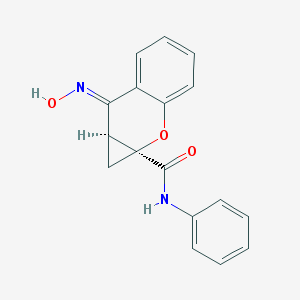 CHEMBL507522 CHEMBL507522 | C17H14N2O3 | 294.31 | 4 / 2 | 2.6 | Yes |
| 84930 | 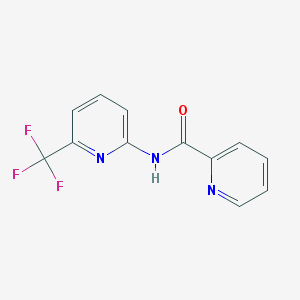 CHEMBL549669 CHEMBL549669 | C12H8F3N3O | 267.211 | 6 / 1 | 2.2 | Yes |
| 85067 | 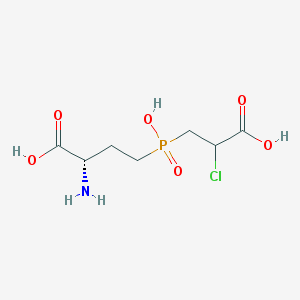 CHEMBL1076865 CHEMBL1076865 | C7H13ClNO6P | 273.606 | 7 / 4 | -4.1 | Yes |
| 85076 | 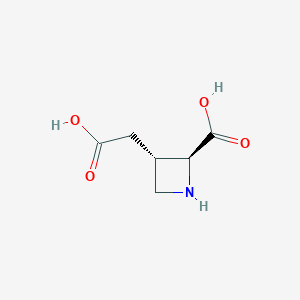 (2S,3S)-3-(carboxymethyl)azetidine-2-carboxylic acid (2S,3S)-3-(carboxymethyl)azetidine-2-carboxylic acid | C6H9NO4 | 159.141 | 5 / 3 | -3.3 | Yes |
| 89646 | 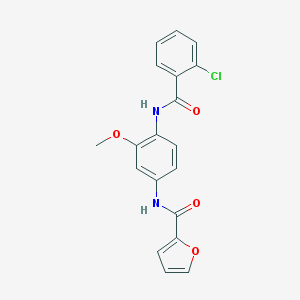 CHEMBL1385271 CHEMBL1385271 | C19H15ClN2O4 | 370.789 | 4 / 2 | 3.6 | Yes |
| 524194 | 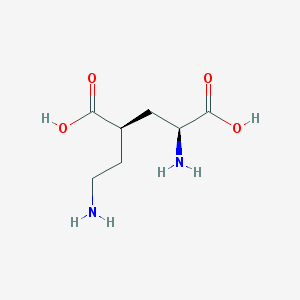 CHEMBL2312684 CHEMBL2312684 | C7H14N2O4 | 190.199 | 6 / 4 | -6.3 | Yes |
| 98488 | 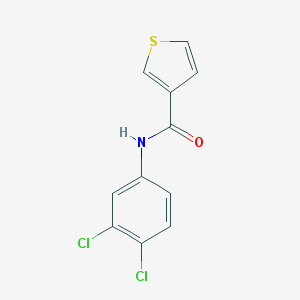 N-(3,4-dichlorophenyl)thiophene-3-carboxamide N-(3,4-dichlorophenyl)thiophene-3-carboxamide | C11H7Cl2NOS | 272.143 | 2 / 1 | 4.1 | Yes |
| 100372 | 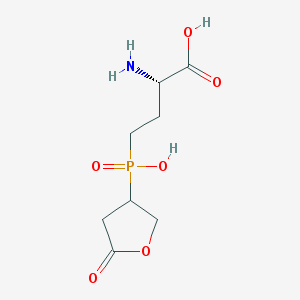 SCHEMBL8239107 SCHEMBL8239107 | C8H14NO6P | 251.175 | 7 / 3 | -4.7 | Yes |
| 102752 | 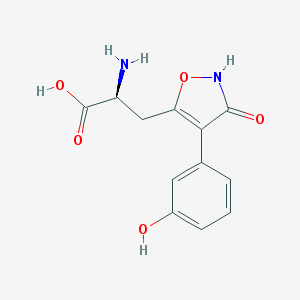 CHEMBL368556 CHEMBL368556 | C12H12N2O5 | 264.237 | 6 / 4 | -2.3 | Yes |
| 102897 | 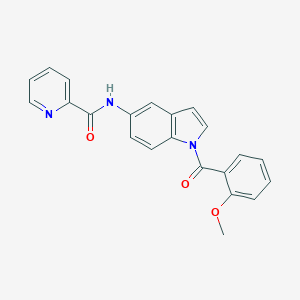 CHEMBL1672257 CHEMBL1672257 | C22H17N3O3 | 371.396 | 4 / 1 | 3.6 | Yes |
| 108031 | 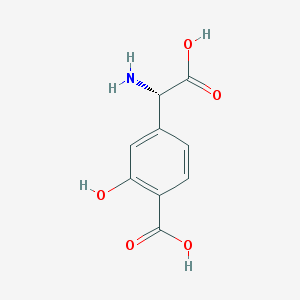 (S)-4-CARBOXY-3-HYDROXYPHENYLGLYCINE (S)-4-CARBOXY-3-HYDROXYPHENYLGLYCINE | C9H9NO5 | 211.173 | 6 / 4 | -1.0 | Yes |
| 109300 | 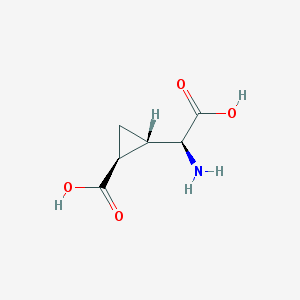 (2S,1'S,2'S)-2-(carboxycyclopropyl)glycine (2S,1'S,2'S)-2-(carboxycyclopropyl)glycine | C6H9NO4 | 159.141 | 5 / 3 | -3.4 | Yes |
| 109311 | 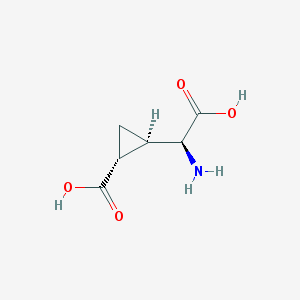 CCG-II CCG-II | C6H9NO4 | 159.141 | 5 / 3 | -3.4 | Yes |
| 109321 | 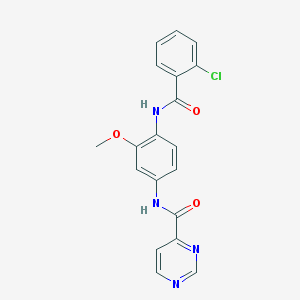 CHEMBL1672236 CHEMBL1672236 | C19H15ClN4O3 | 382.804 | 5 / 2 | 2.8 | Yes |
| 446116 | 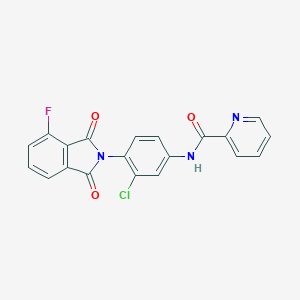 CHEMBL3357574 CHEMBL3357574 | C20H11ClFN3O3 | 395.774 | 5 / 1 | 3.3 | Yes |
| 538736 | 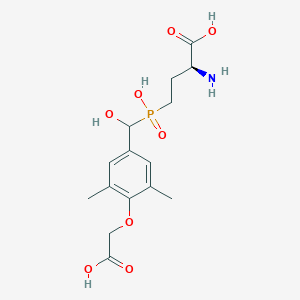 CHEMBL3912541 CHEMBL3912541 | C15H22NO8P | 375.314 | 9 / 5 | -3.1 | Yes |
| 560800 | 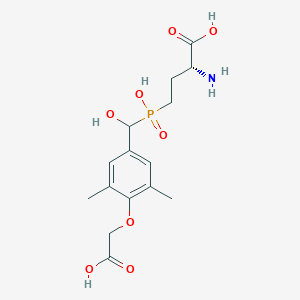 US9212196, Derivative 10 US9212196, Derivative 10 | C15H22NO8P | 375.314 | 9 / 5 | -3.1 | Yes |
| 117648 | 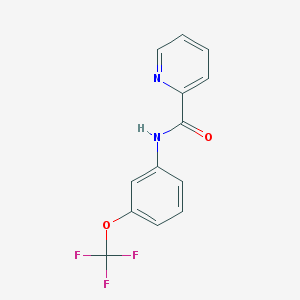 CHEMBL559271 CHEMBL559271 | C13H9F3N2O2 | 282.222 | 6 / 1 | 3.2 | Yes |
| 538880 | 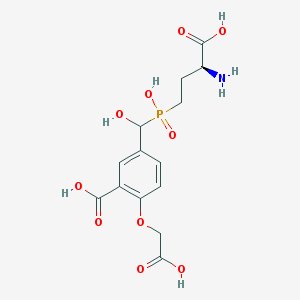 CHEMBL3927149 CHEMBL3927149 | C14H18NO10P | 391.269 | 11 / 6 | -4.3 | No |
| 560944 | 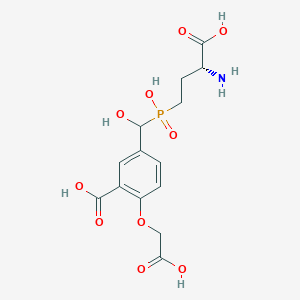 US9212196, Derivative 23 US9212196, Derivative 23 | C14H18NO10P | 391.269 | 11 / 6 | -4.3 | No |
| 538898 | 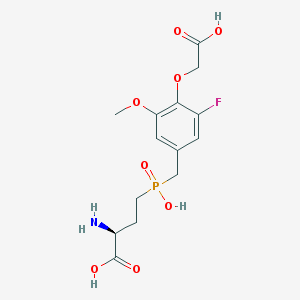 CHEMBL3913188 CHEMBL3913188 | C14H19FNO8P | 379.277 | 10 / 4 | -3.2 | Yes |
| 560962 | 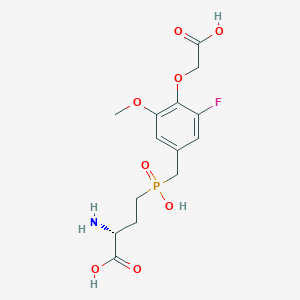 US9212196, Derivative 34 US9212196, Derivative 34 | C14H19FNO8P | 379.277 | 10 / 4 | -3.2 | Yes |
| 538965 | 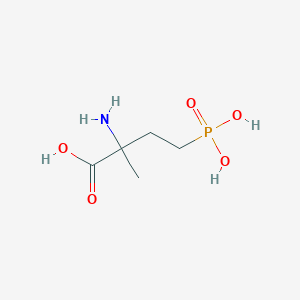 4-Phosphonoisovaline 4-Phosphonoisovaline | C5H12NO5P | 197.127 | 6 / 4 | -4.6 | Yes |
| 553878 | 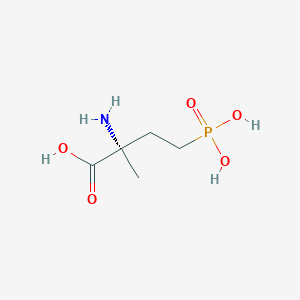 MAP4 MAP4 | C5H12NO5P | 197.127 | 6 / 4 | -4.6 | Yes |
| 120218 | 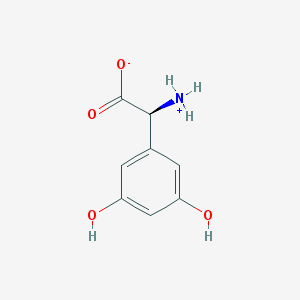 CHEBI:75204 CHEBI:75204 | C8H9NO4 | 183.163 | 4 / 3 | -1.8 | Yes |
| 128449 | 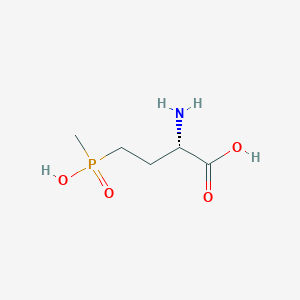 L-PHOSPHINOTHRICIN L-PHOSPHINOTHRICIN | C5H12NO4P | 181.128 | 5 / 3 | -5.0 | Yes |
| 130323 | 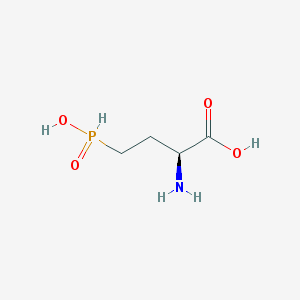 demethylphosphinothricin demethylphosphinothricin | C4H10NO4P | 167.101 | 5 / 3 | -5.1 | Yes |
| 130722 | 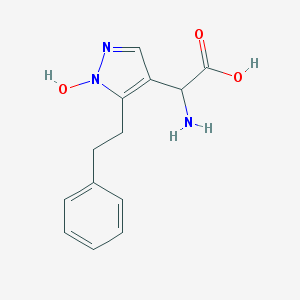 CHEMBL397048 CHEMBL397048 | C13H15N3O3 | 261.281 | 5 / 3 | -1.1 | Yes |
| 553949 | 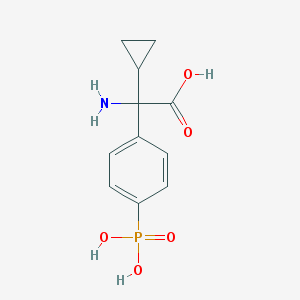 cppg cppg | C11H14NO5P | 271.209 | 6 / 4 | -3.1 | Yes |
| 135009 | 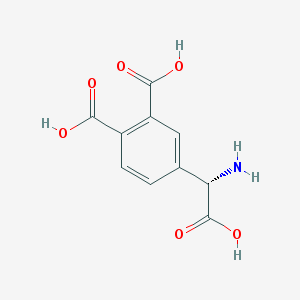 (S)-3,4-DCPG (S)-3,4-DCPG | C10H9NO6 | 239.183 | 7 / 4 | -2.7 | Yes |
| 561522 | 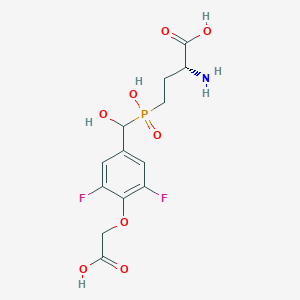 US9212196, Derivative 28 US9212196, Derivative 28 | C13H16F2NO8P | 383.241 | 11 / 5 | -3.6 | No |
| 539422 | 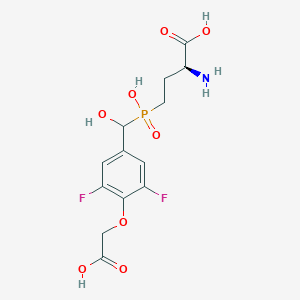 CHEMBL3896442 CHEMBL3896442 | C13H16F2NO8P | 383.241 | 11 / 5 | -3.6 | No |
| 136965 | 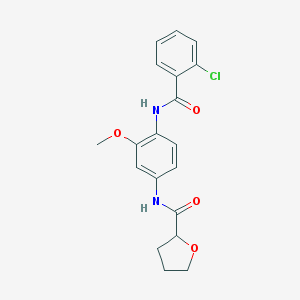 CHEMBL1672231 CHEMBL1672231 | C19H19ClN2O4 | 374.821 | 4 / 2 | 2.9 | Yes |
| 539452 | 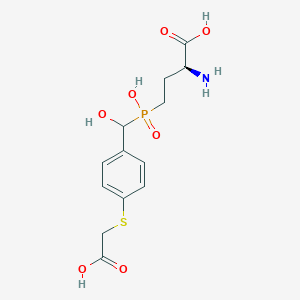 CHEMBL3905370 CHEMBL3905370 | C13H18NO7PS | 363.321 | 9 / 5 | -3.3 | Yes |
| 561552 | 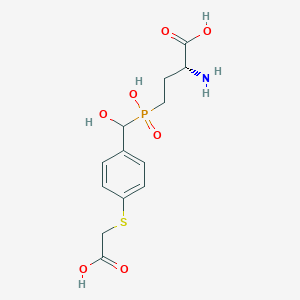 US9212196, Derivative 26 US9212196, Derivative 26 | C13H18NO7PS | 363.321 | 9 / 5 | -3.3 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417