

You can:
| Name | Metabotropic glutamate receptor 4 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Grm4 |
| Synonym | glutamate receptor GPRC1D mGlu4 receptor mGluR4 |
| Disease | N/A for non-human GPCRs |
| Length | 912 |
| Amino acid sequence | MSGKGGWAWWWARLPLCLLLSLYAPWVPSSLGKPKGHPHMNSIRIDGDITLGGLFPVHGRGSEGKACGELKKEKGIHRLEAMLFALDRINNDPDLLPNITLGARILDTCSRDTHALEQSLTFVQALIEKDGTEVRCGSGGPPIITKPERVVGVIGASGSSVSIMVANILRLFKIPQISYASTAPDLSDNSRYDFFSRVVPSDTYQAQAMVDIVRALKWNYVSTLASEGSYGESGVEAFIQKSRENGGVCIAQSVKIPREPKTGEFDKIIKRLLETSNARGIIIFANEDDIRRVLEAARRANQTGHFFWMGSDSWGSKSAPVLRLEEVAEGAVTILPKRMSVRGFDRYFSSRTLDNNRRNIWFAEFWEDNFHCKLSRHALKKGSHIKKCTNRERIGQDSAYEQEGKVQFVIDAVYAMGHALHAMHRDLCPGRVGLCPRMDPVDGTQLLKYIRNVNFSGIAGNPVTFNENGDAPGRYDIYQYQLRNGSAEYKVIGSWTDHLHLRIERMQWPGSGQQLPRSICSLPCQPGERKKTVKGMACCWHCEPCTGYQYQVDRYTCKTCPYDMRPTENRTSCQPIPIVKLEWDSPWAVLPLFLAVVGIAATLFVVVTFVRYNDTPIVKASGRELSYVLLAGIFLCYATTFLMIAEPDLGTCSLRRIFLGLGMSISYAALLTKTNRIYRIFEQGKRSVSAPRFISPASQLAITFILISLQLLGICVWFVVDPSHSVVDFQDQRTLDPRFARGVLKCDISDLSLICLLGYSMLLMVTCTVYAIKTRGVPETFNEAKPIGFTMYTTCIVWLAFIPIFFGTSQSADKLYIQTTTLTVSVSLSASVSLGMLYMPKVYIILFHPEQNVPKRKRSLKAVVTAATMSNKFTQKGNFRPNGEAKSELCENLETPALATKQTYVTYTNHAI |
| UniProt | P31423 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2787 |
| IUPHAR | 292 |
| DrugBank | N/A |
| Name | L-AP4 |
|---|---|
| Molecular formula | C4H10NO5P |
| IUPAC name | (2S)-2-amino-4-phosphonobutanoic acid |
| Molecular weight | 183.1 |
| Hydrogen bond acceptor | 6 |
| Hydrogen bond donor | 4 |
| XlogP | -5.5 |
| Synonyms | (S)-2-amino-4-phosphonobutanoic acid ACM23052815 CCG-37553 FCH919870 L-(+)-2-Amino-4-phosphono-butanoic acid [ Show all ] |
| Inchi Key | DDOQBQRIEWHWBT-VKHMYHEASA-N |
| Inchi ID | InChI=1S/C4H10NO5P/c5-3(4(6)7)1-2-11(8,9)10/h3H,1-2,5H2,(H,6,7)(H2,8,9,10)/t3-/m0/s1 |
| PubChem CID | 179394 |
| ChEMBL | CHEMBL33567 |
| IUPHAR | 1410, 1412 |
| BindingDB | 50007548 |
| DrugBank | N/A |
Structure | 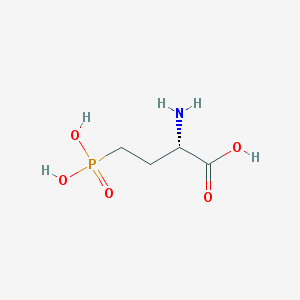 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| EC10 | 10.0 nM | MedChemComm, (2014) 5:10:1548 | ChEMBL |
| EC50 | 30.0 nM | PMID26814576, PMID25958247 | BindingDB,ChEMBL |
| EC50 | 80.0 nM | PMID17722902 | BindingDB,ChEMBL |
| EC50 | 96.0 nM | PMID20218620 | BindingDB,ChEMBL |
| EC50 | 130.0 nM | PMID22750138, PMID20218620 | BindingDB,ChEMBL |
| EC50 | 200.0 nM | PMID15801843 | BindingDB,ChEMBL |
| EC50 | 320.0 nM | PMID14711315, PMID15801843 | BindingDB,ChEMBL |
| EC50 | 500.0 nM | PMID7738999 | BindingDB,ChEMBL |
| EC50 | 510.0 nM | PMID17350267, PMID16213710 | BindingDB,ChEMBL |
| EC50 | 910.0 nM | PMID9357538 | BindingDB,ChEMBL |
| EC50 | 1200.0 nM | PMID7738999 | ChEMBL |
| EC50 | >100000.0 nM | PMID15801843 | ChEMBL |
| EC50 | 129000.0 nM | PMID15801843 | BindingDB,ChEMBL |
| IC50 | 398.107 - 1000.0 nM | PMID8463825, PMID9144638, PMID8719808, PMID10187777 | IUPHAR |
| Kd | 501.187 nM | PMID10187777 | IUPHAR |
| Ki | 160.0 nM | PMID16213710, PMID17350267 | BindingDB,ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417