

You can:
| Name | Alpha-1B adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRA1B |
| Synonym | alpha1B-adrenoceptor alpha1B-adrenergic receptor Alpha-1B adrenoreceptor Alpha-1B adrenoceptor alpha 1B-adrenoreceptor [ Show all ] |
| Disease | Psychiatric disorder Hypertension Exogenous obesity Attention deficit hyperactivity disorder |
| Length | 520 |
| Amino acid sequence | MNPDLDTGHNTSAPAHWGELKNANFTGPNQTSSNSTLPQLDITRAISVGLVLGAFILFAIVGNILVILSVACNRHLRTPTNYFIVNLAMADLLLSFTVLPFSAALEVLGYWVLGRIFCDIWAAVDVLCCTASILSLCAISIDRYIGVRYSLQYPTLVTRRKAILALLSVWVLSTVISIGPLLGWKEPAPNDDKECGVTEEPFYALFSSLGSFYIPLAVILVMYCRVYIVAKRTTKNLEAGVMKEMSNSKELTLRIHSKNFHEDTLSSTKAKGHNPRSSIAVKLFKFSREKKAAKTLGIVVGMFILCWLPFFIALPLGSLFSTLKPPDAVFKVVFWLGYFNSCLNPIIYPCSSKEFKRAFVRILGCQCRGRGRRRRRRRRRLGGCAYTYRPWTRGGSLERSQSRKDSLDDSGSCLSGSQRTLPSASPSPGYLGRGAPPPVELCAFPEWKAPGALLSLPAPEPPGRRGRHDSGPLFTFKLLTEPESPGTDGGASNGGCEAAADVANGQPGFKSNMPLAPGQF |
| UniProt | P35368 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T29500 |
| ChEMBL | CHEMBL232 |
| IUPHAR | 23 |
| DrugBank | BE0000575 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 62 | 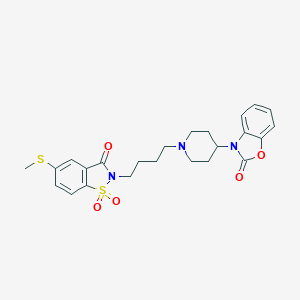 CHEMBL309518 CHEMBL309518 | C24H27N3O5S2 | 501.616 | 7 / 0 | 3.5 | No |
| 77 | 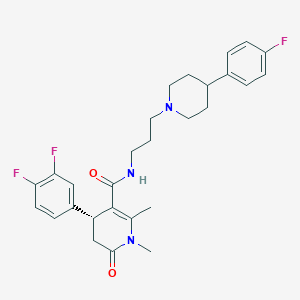 CHEMBL287791 CHEMBL287791 | C28H32F3N3O2 | 499.578 | 6 / 1 | 3.9 | Yes |
| 547908 | 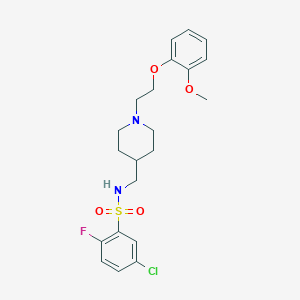 CHEMBL3937911 CHEMBL3937911 | C21H26ClFN2O4S | 456.957 | 7 / 1 | 4.0 | Yes |
| 907 | 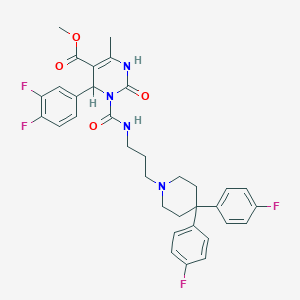 CHEMBL342126 CHEMBL342126 | C34H34F4N4O4 | 638.664 | 9 / 2 | 6.1 | No |
| 1936 | 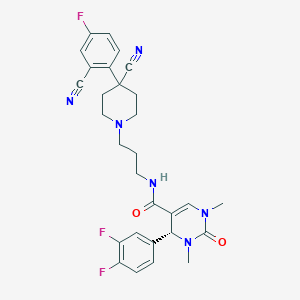 CHEMBL93843 CHEMBL93843 | C29H29F3N6O2 | 550.586 | 8 / 1 | 2.5 | No |
| 2009 | 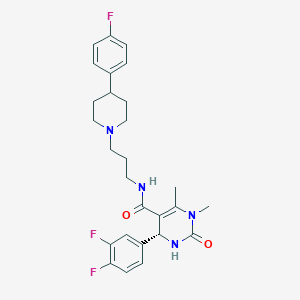 CHEMBL90421 CHEMBL90421 | C27H31F3N4O2 | 500.566 | 6 / 2 | 3.6 | No |
| 2013 | 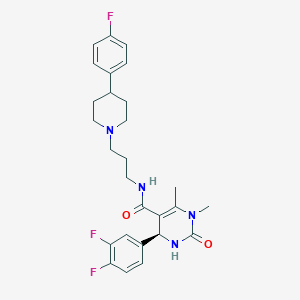 CHEMBL328268 CHEMBL328268 | C27H31F3N4O2 | 500.566 | 6 / 2 | 3.6 | No |
| 2185 | 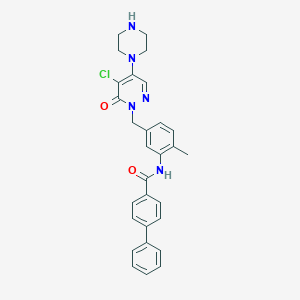 CHEMBL458002 CHEMBL458002 | C29H28ClN5O2 | 514.026 | 5 / 2 | 4.3 | No |
| 2282 | 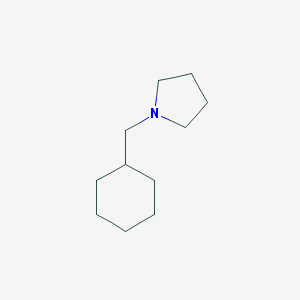 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 3038 | 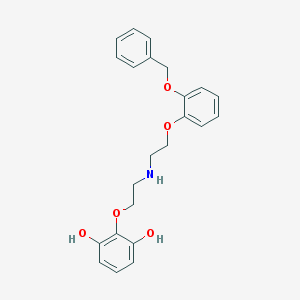 CHEMBL1255085 CHEMBL1255085 | C23H25NO5 | 395.455 | 6 / 3 | 3.9 | Yes |
| 3140 | 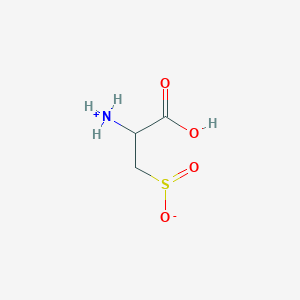 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 441853 | 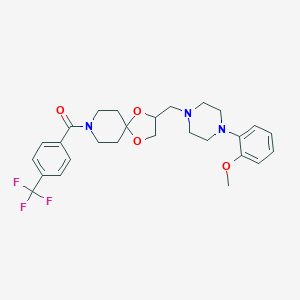 CHEMBL3342860 CHEMBL3342860 | C27H32F3N3O4 | 519.565 | 9 / 0 | 3.9 | No |
| 4498 | 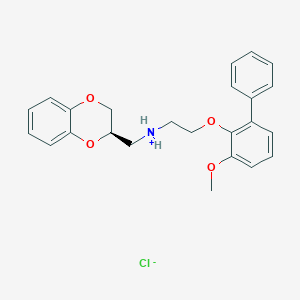 CID 44417700 CID 44417700 | C24H26ClNO4 | 427.925 | 5 / 1 | N/A | N/A |
| 4502 | 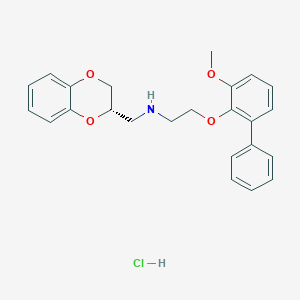 CHEMBL215151 CHEMBL215151 | C24H26ClNO4 | 427.925 | 5 / 2 | N/A | N/A |
| 4952 | 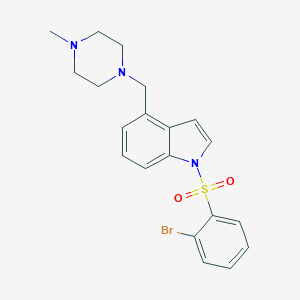 5HT6-ligand-1 5HT6-ligand-1 | C20H22BrN3O2S | 448.379 | 4 / 0 | 3.6 | Yes |
| 5062 | 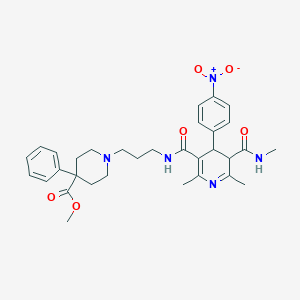 BDBM50068818 BDBM50068818 | C32H39N5O6 | 589.693 | 8 / 2 | 2.9 | No |
| 5252 | 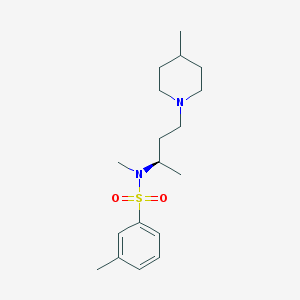 SB-258719 SB-258719 | C18H30N2O2S | 338.51 | 4 / 0 | 3.5 | Yes |
| 5683 | 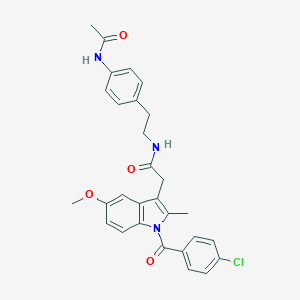 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 6456 | 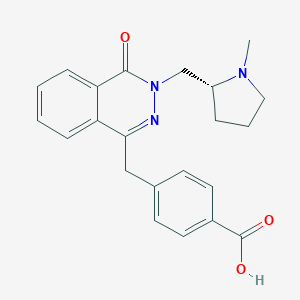 CHEMBL1767144 CHEMBL1767144 | C22H23N3O3 | 377.444 | 5 / 1 | 0.6 | Yes |
| 463711 | 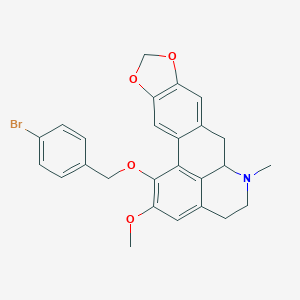 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7236 | 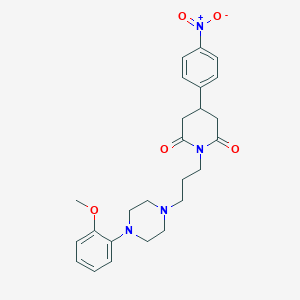 CHEMBL1796042 CHEMBL1796042 | C25H30N4O5 | 466.538 | 7 / 0 | 2.7 | Yes |
| 7403 | 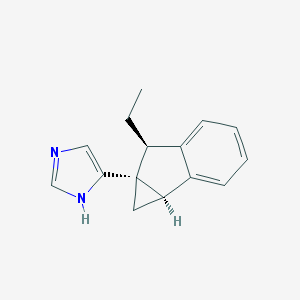 CHEMBL1256610 CHEMBL1256610 | C15H16N2 | 224.307 | 1 / 1 | 2.9 | Yes |
| 442019 | 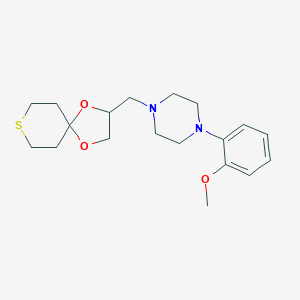 CHEMBL3342852 CHEMBL3342852 | C19H28N2O3S | 364.504 | 6 / 0 | 2.5 | Yes |
| 521684 | 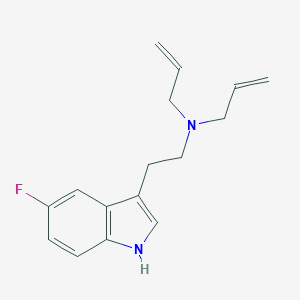 CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8644 | 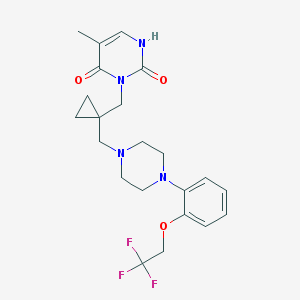 CHEMBL55629 CHEMBL55629 | C22H27F3N4O3 | 452.478 | 8 / 1 | 2.9 | Yes |
| 536201 | 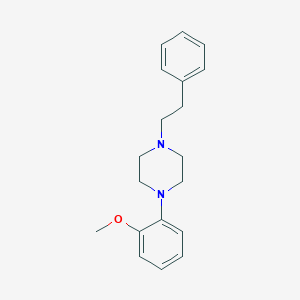 CHEMBL27021 CHEMBL27021 | C19H24N2O | 296.414 | 3 / 0 | 3.8 | Yes |
| 8778 | 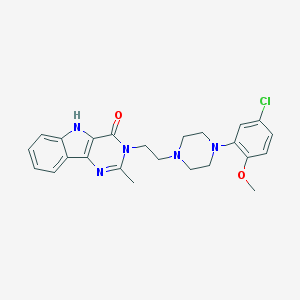 CHEMBL220515 CHEMBL220515 | C24H26ClN5O2 | 451.955 | 5 / 1 | 3.6 | Yes |
| 9877 | 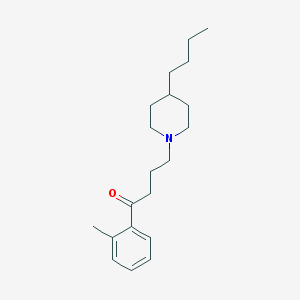 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 10045 | 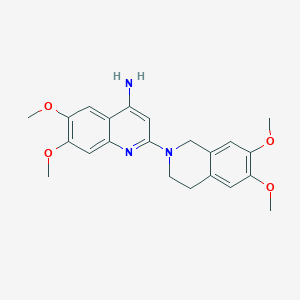 Abanoquil Abanoquil | C22H25N3O4 | 395.459 | 7 / 1 | 3.4 | Yes |
| 10266 | 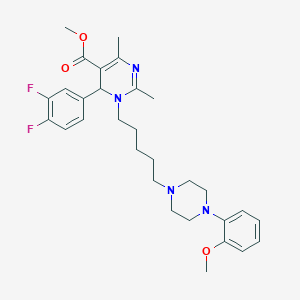 CHEMBL145587 CHEMBL145587 | C30H38F2N4O3 | 540.656 | 8 / 0 | 4.3 | No |
| 10729 | 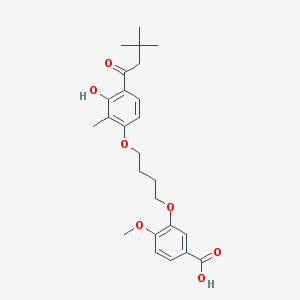 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10780 | 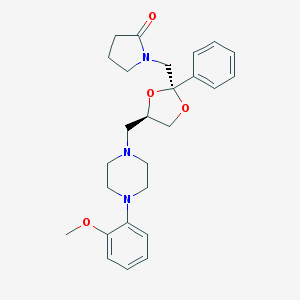 CHEMBL1242445 CHEMBL1242445 | C26H33N3O4 | 451.567 | 6 / 0 | 2.4 | Yes |
| 10786 | 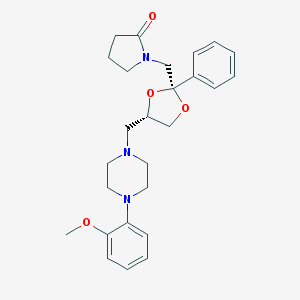 CHEMBL1242444 CHEMBL1242444 | C26H33N3O4 | 451.567 | 6 / 0 | 2.4 | Yes |
| 11278 | 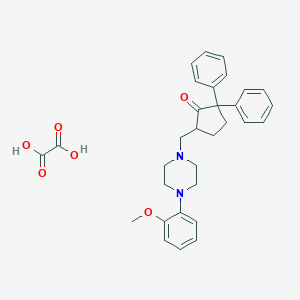 CHEMBL1946783 CHEMBL1946783 | C31H34N2O6 | 530.621 | 8 / 2 | N/A | No |
| 11356 | 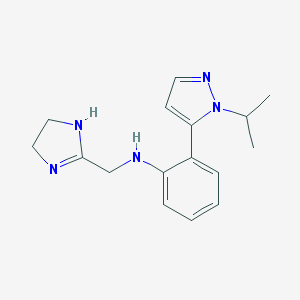 CHEMBL18303 CHEMBL18303 | C16H21N5 | 283.379 | 3 / 2 | 1.6 | Yes |
| 11532 | 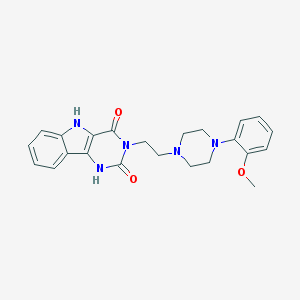 3-MPPI 3-MPPI | C23H25N5O3 | 419.485 | 5 / 2 | 2.7 | Yes |
| 12105 | 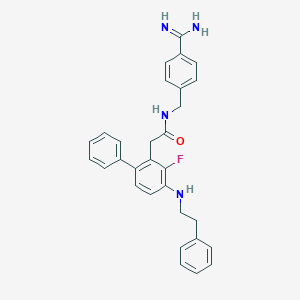 CHEMBL343173 CHEMBL343173 | C30H29FN4O | 480.587 | 4 / 4 | 5.3 | No |
| 12493 | 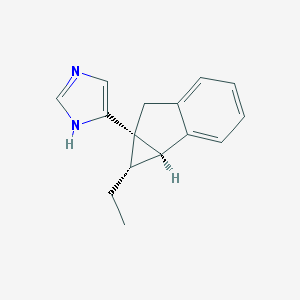 CHEMBL1255723 CHEMBL1255723 | C15H16N2 | 224.307 | 1 / 1 | 3.1 | Yes |
| 12629 | 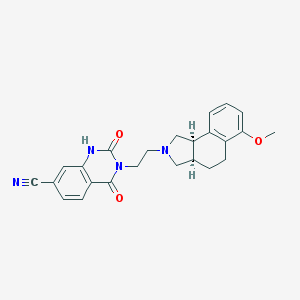 CHEMBL62033 CHEMBL62033 | C24H24N4O3 | 416.481 | 5 / 1 | 2.5 | Yes |
| 12638 | 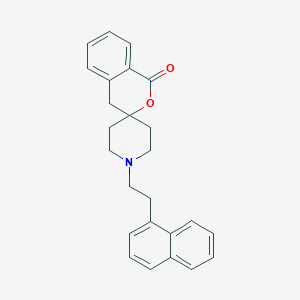 CHEMBL98168 CHEMBL98168 | C25H25NO2 | 371.48 | 3 / 0 | 5.4 | No |
| 12810 | 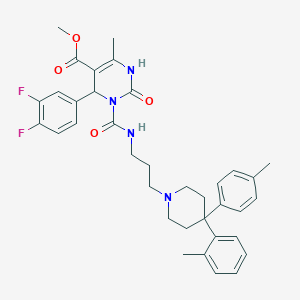 CHEMBL145894 CHEMBL145894 | C36H40F2N4O4 | 630.737 | 7 / 2 | 6.6 | No |
| 13157 | 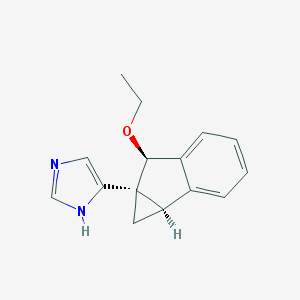 CHEMBL1256565 CHEMBL1256565 | C15H16N2O | 240.306 | 2 / 1 | 1.9 | Yes |
| 13393 | 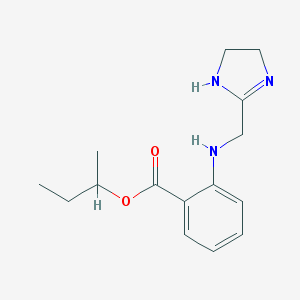 CHEMBL97859 CHEMBL97859 | C15H21N3O2 | 275.352 | 4 / 2 | 2.7 | Yes |
| 13645 | 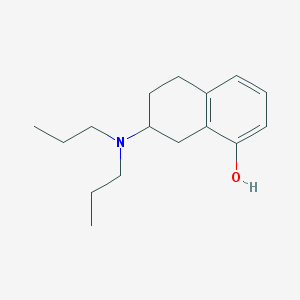 8-OH-Dpat 8-OH-Dpat | C16H25NO | 247.382 | 2 / 1 | 4.1 | Yes |
| 13977 | 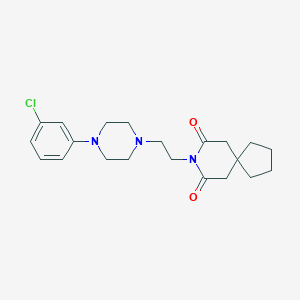 CHEMBL57364 CHEMBL57364 | C21H28ClN3O2 | 389.924 | 4 / 0 | 3.5 | Yes |
| 14038 | 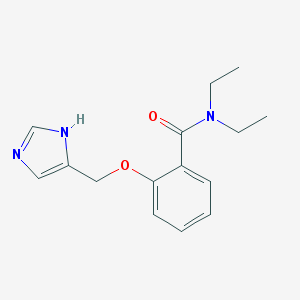 CHEMBL326837 CHEMBL326837 | C15H19N3O2 | 273.336 | 3 / 1 | 1.8 | Yes |
| 548048 | 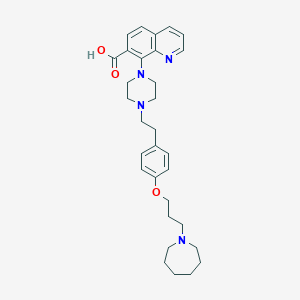 CHEMBL3925977 CHEMBL3925977 | C31H40N4O3 | 516.686 | 7 / 1 | 2.9 | No |
| 14686 | 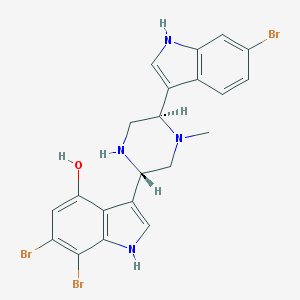 CHEMBL302882 CHEMBL302882 | C21H19Br3N4O | 583.122 | 3 / 4 | 4.6 | No |
| 14904 | 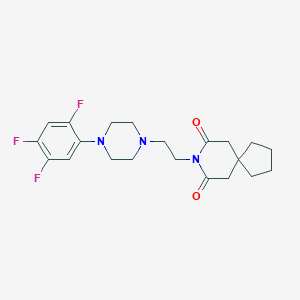 CHEMBL193620 CHEMBL193620 | C21H26F3N3O2 | 409.453 | 7 / 0 | 3.1 | Yes |
| 15288 | 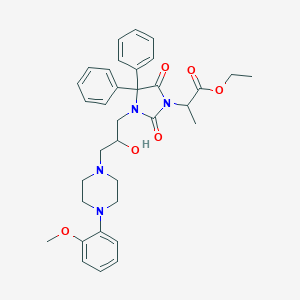 CHEMBL1256344 CHEMBL1256344 | C34H40N4O6 | 600.716 | 8 / 1 | 4.2 | No |
| 15298 | 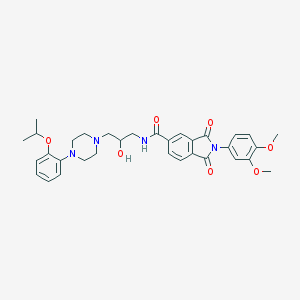 CHEMBL70095 CHEMBL70095 | C33H38N4O7 | 602.688 | 9 / 2 | 3.5 | No |
| 15324 | 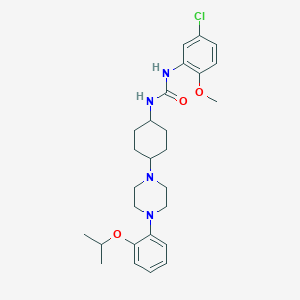 CHEMBL271913 CHEMBL271913 | C27H37ClN4O3 | 501.068 | 5 / 2 | 5.3 | No |
| 15341 | 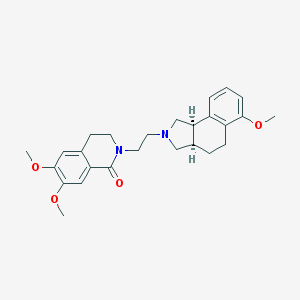 CHEMBL291305 CHEMBL291305 | C26H32N2O4 | 436.552 | 5 / 0 | 3.5 | Yes |
| 15478 | 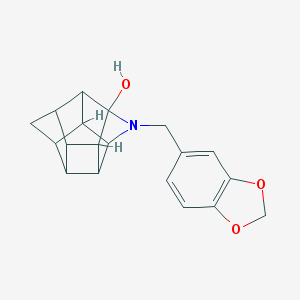 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15791 | 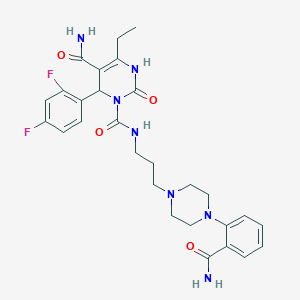 CHEMBL142718 CHEMBL142718 | C28H33F2N7O4 | 569.614 | 8 / 4 | 1.9 | No |
| 16119 | 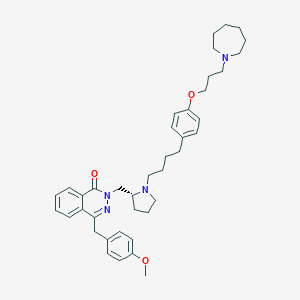 CHEMBL1767165 CHEMBL1767165 | C40H52N4O3 | 636.881 | 6 / 0 | 7.6 | No |
| 16532 | 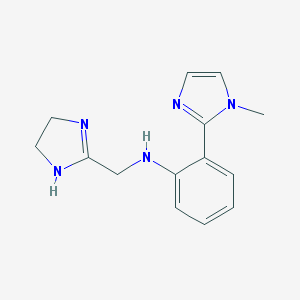 CHEMBL18256 CHEMBL18256 | C14H17N5 | 255.325 | 3 / 2 | 0.7 | Yes |
| 16690 | 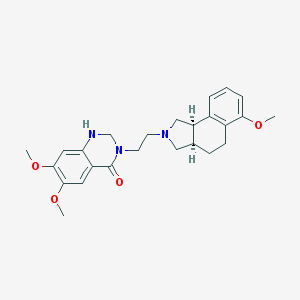 CHEMBL303988 CHEMBL303988 | C25H31N3O4 | 437.54 | 6 / 1 | 3.4 | Yes |
| 16888 |  Glemanserin Glemanserin | C20H25NO | 295.426 | 2 / 1 | 3.8 | Yes |
| 16925 | 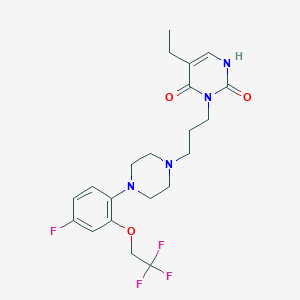 CHEMBL53675 CHEMBL53675 | C21H26F4N4O3 | 458.458 | 9 / 1 | 3.2 | Yes |
| 17202 | 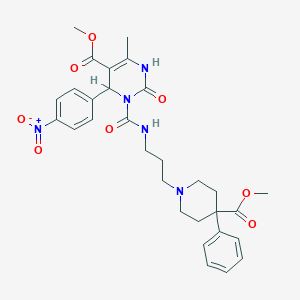 CHEMBL142383 CHEMBL142383 | C30H35N5O8 | 593.637 | 9 / 2 | 3.4 | No |
| 17875 |  CHEMBL267336 CHEMBL267336 | C26H29F4N3O5S | 571.588 | 12 / 1 | 4.0 | No |
| 17883 |  Bmy-7378 Bmy-7378 | C22H31N3O3 | 385.508 | 5 / 0 | 2.8 | Yes |
| 18691 |  SK&F-106686 SK&F-106686 | C14H16ClNS | 265.799 | 2 / 0 | 4.3 | Yes |
| 19144 |  CHEMBL1255003 CHEMBL1255003 | C25H29NO3 | 391.511 | 4 / 1 | 5.3 | No |
| 19339 | 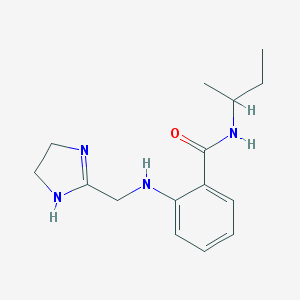 CHEMBL432192 CHEMBL432192 | C15H22N4O | 274.368 | 3 / 3 | 2.1 | Yes |
| 536470 | 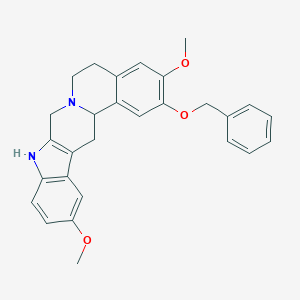 CHEMBL3957222 CHEMBL3957222 | C28H28N2O3 | 440.543 | 4 / 1 | 4.9 | Yes |
| 20050 | 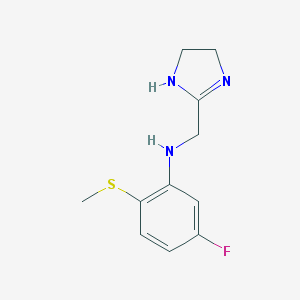 CHEMBL72768 CHEMBL72768 | C11H14FN3S | 239.312 | 4 / 2 | 1.5 | Yes |
| 20199 | 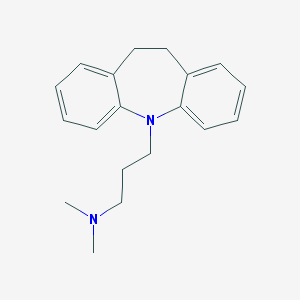 imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 21144 | 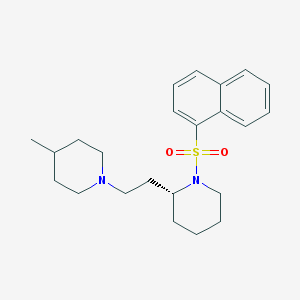 SCHEMBL3825498 SCHEMBL3825498 | C23H32N2O2S | 400.581 | 4 / 0 | 4.9 | Yes |
| 21599 | 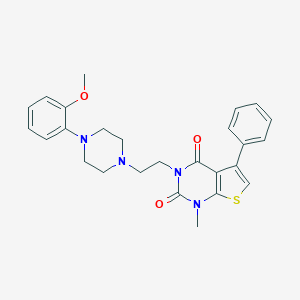 CHEMBL14213 CHEMBL14213 | C26H28N4O3S | 476.595 | 6 / 0 | 4.1 | Yes |
| 21831 | 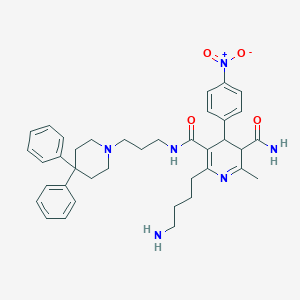 BDBM50081825 BDBM50081825 | C38H46N6O4 | 650.824 | 7 / 3 | 4.3 | No |
| 21896 | 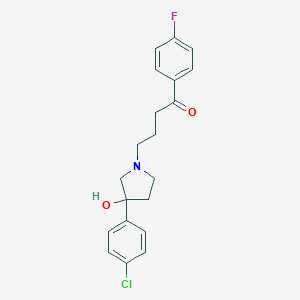 CHEMBL150161 CHEMBL150161 | C20H21ClFNO2 | 361.841 | 4 / 1 | 2.9 | Yes |
| 22204 | 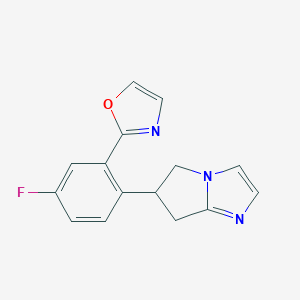 CHEMBL501404 CHEMBL501404 | C15H12FN3O | 269.279 | 4 / 0 | 1.9 | Yes |
| 22422 | 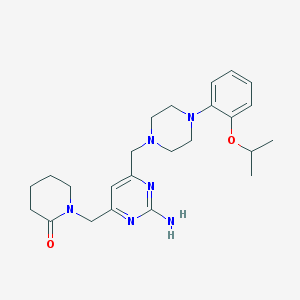 CHEMBL75808 CHEMBL75808 | C24H34N6O2 | 438.576 | 7 / 1 | 1.8 | Yes |
| 22957 | 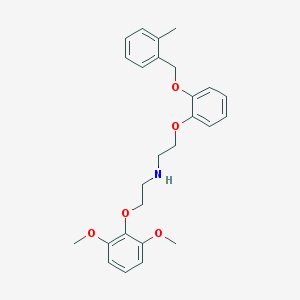 CHEMBL382972 CHEMBL382972 | C26H31NO5 | 437.536 | 6 / 1 | 4.9 | Yes |
| 23421 | 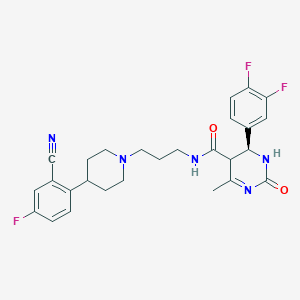 BDBM50090015 BDBM50090015 | C27H28F3N5O2 | 511.549 | 7 / 2 | 3.2 | No |
| 23438 | 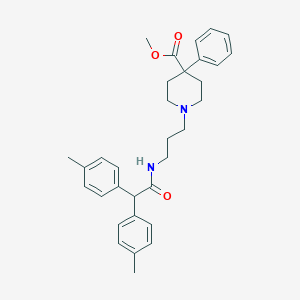 CHEMBL41677 CHEMBL41677 | C32H38N2O3 | 498.667 | 4 / 1 | 5.8 | No |
| 442605 | 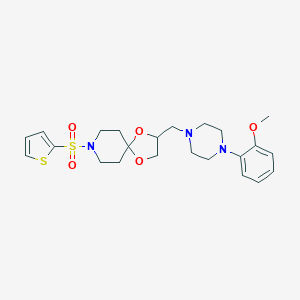 CHEMBL3342872 CHEMBL3342872 | C23H31N3O5S2 | 493.637 | 9 / 0 | 2.7 | Yes |
| 24429 | 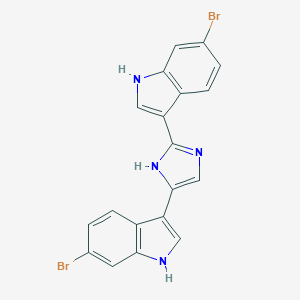 Nortopsentin A Nortopsentin A | C19H12Br2N4 | 456.141 | 1 / 3 | 5.0 | Yes |
| 25053 | 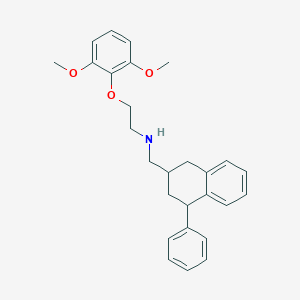 CHEMBL42231 CHEMBL42231 | C27H31NO3 | 417.549 | 4 / 1 | 5.5 | No |
| 25060 | 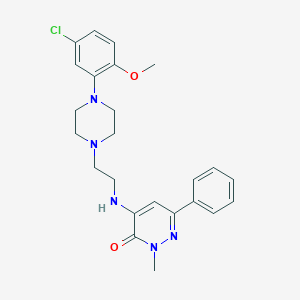 CHEMBL81486 CHEMBL81486 | C24H28ClN5O2 | 453.971 | 6 / 1 | 3.9 | Yes |
| 26057 | 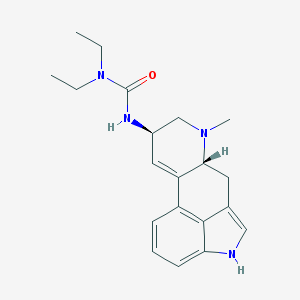 S-(-)-Lisuride S-(-)-Lisuride | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26086 | 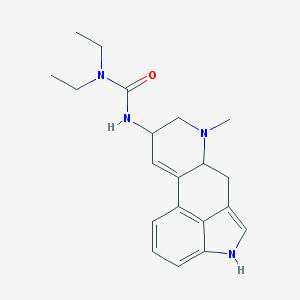 AC1L1H1T AC1L1H1T | C20H26N4O | 338.455 | 2 / 2 | 2.7 | Yes |
| 26297 | 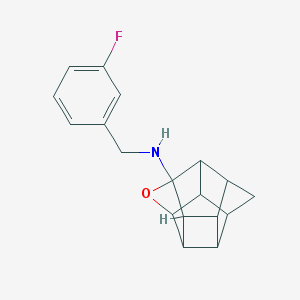 CHEMBL600053 CHEMBL600053 | C18H18FNO | 283.346 | 3 / 1 | 2.5 | Yes |
| 26480 |  CHEMBL342347 CHEMBL342347 | C31H39F2N5O3 | 567.682 | 8 / 1 | 3.7 | No |
| 26483 |  CHEMBL358313 CHEMBL358313 | C31H39F2N5O3 | 567.682 | 8 / 1 | 3.7 | No |
| 26557 |  Rauwolscine Rauwolscine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26566 |  corynanthine corynanthine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26590 | 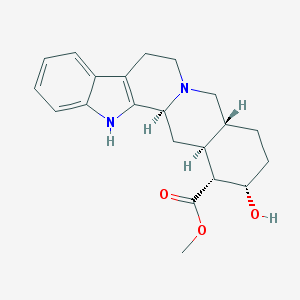 Yohimbine Yohimbine | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 26646 | 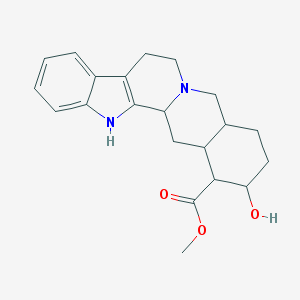 CHEBI:48565 CHEBI:48565 | C21H26N2O3 | 354.45 | 4 / 2 | 2.9 | Yes |
| 27039 | 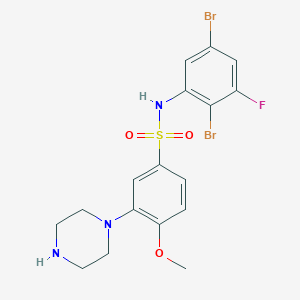 SB-357134 SB-357134 | C17H18Br2FN3O3S | 523.215 | 7 / 2 | 3.4 | No |
| 27075 | 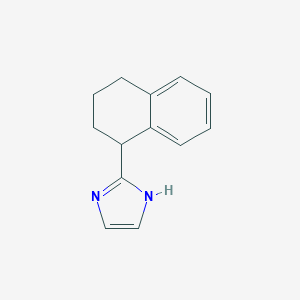 CHEMBL258007 CHEMBL258007 | C13H14N2 | 198.269 | 1 / 1 | 2.7 | Yes |
| 27194 | 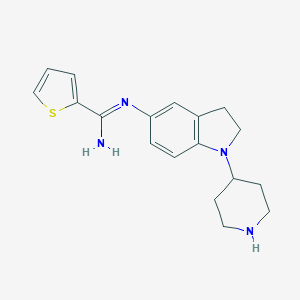 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27512 | 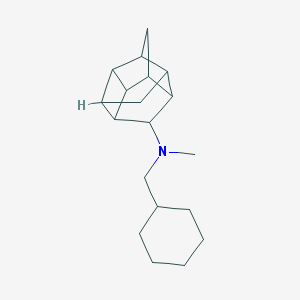 CHEMBL2432063 CHEMBL2432063 | C19H29N | 271.448 | 1 / 0 | 4.7 | Yes |
| 27581 | 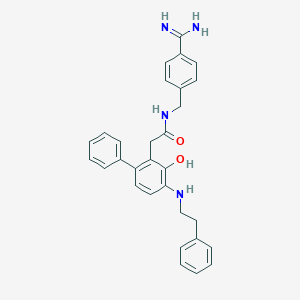 Substituted Benzene Analog 51 Substituted Benzene Analog 51 | C30H30N4O2 | 478.596 | 4 / 5 | 4.8 | Yes |
| 28151 | 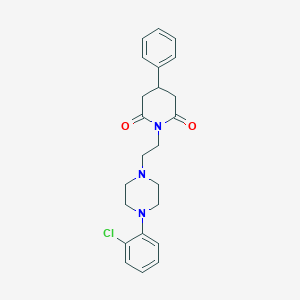 CHEMBL1796029 CHEMBL1796029 | C23H26ClN3O2 | 411.93 | 4 / 0 | 3.2 | Yes |
| 29370 | 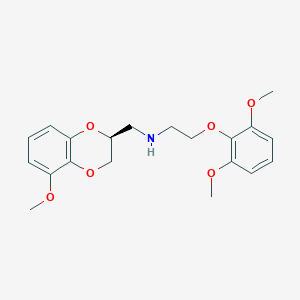 CHEMBL2153559 CHEMBL2153559 | C20H25NO6 | 375.421 | 7 / 1 | 2.9 | Yes |
| 29374 | 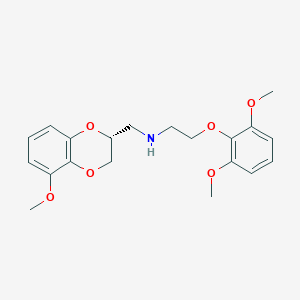 CHEMBL2153560 CHEMBL2153560 | C20H25NO6 | 375.421 | 7 / 1 | 2.9 | Yes |
| 29435 | 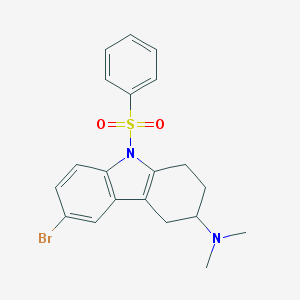 CHEMBL2165524 CHEMBL2165524 | C20H21BrN2O2S | 433.364 | 3 / 0 | 4.7 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417