

You can:
| Name | Muscarinic acetylcholine receptor M4 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Chrm4 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 4 Chrm-4 HM3 M4 receptor |
| Disease | N/A for non-human GPCRs |
| Length | 478 |
| Amino acid sequence | MXNFTPVNGSSANQSVRLVTAAHNHLETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQLQTVNNYFLFSLGCADLIIGAFSMNLYTLYIIKGYWPLGAVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPARRTTKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTVPDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVHKHRPEGPKEKKAKTLAFLKSPLMKPSIKKPPPGGASREELRNGKLEEAPPPALPPPPRPVPDKDTSNESSSGSATQNTKERPPTELSTAEATTPALPAPTLQPRTLNPASKWSKIQIVTKQTGNECVTAIEIVPATPAGMRPAANVARKFASIARNQVRKKRQMAARERKVTRTIFAILLAFILTWTPYNVMVLVNTFCQSCIPERVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLCQYRNIGTAR |
| UniProt | P08485 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL317 |
| IUPHAR | 16 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 459244 | 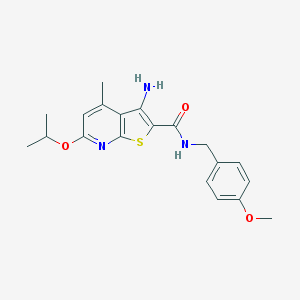 VU0152123 VU0152123 | C20H23N3O3S | 385.482 | 6 / 2 | 4.6 | Yes |
| 459257 | 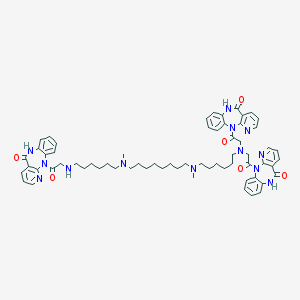 CHEMBL1202000 CHEMBL1202000 | C64H77N13O6 | 1124.41 | 13 / 4 | 7.7 | No |
| 459260 | 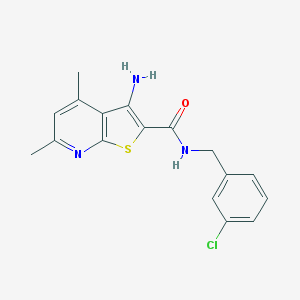 VU0402472-1 VU0402472-1 | C17H16ClN3OS | 345.845 | 4 / 2 | 4.6 | Yes |
| 6615 | 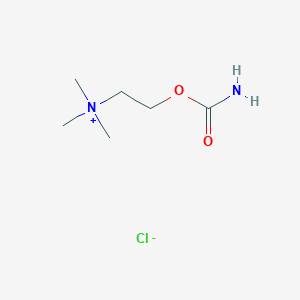 carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 18367 | 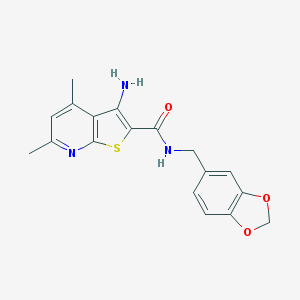 VU0152099 VU0152099 | C18H17N3O3S | 355.412 | 6 / 2 | 3.8 | Yes |
| 18946 | 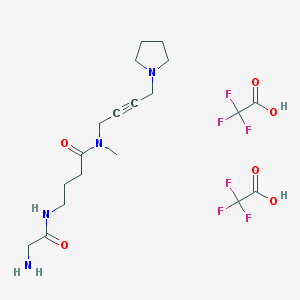 CHEMBL1202133 CHEMBL1202133 | C19H28F6N4O6 | 522.445 | 14 / 4 | N/A | No |
| 19116 | 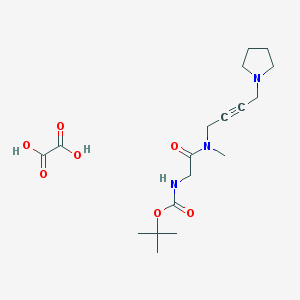 CHEMBL1202047 CHEMBL1202047 | C18H29N3O7 | 399.444 | 8 / 3 | N/A | N/A |
| 20538 | 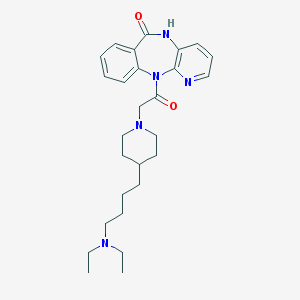 AQ-RA 741 AQ-RA 741 | C27H37N5O2 | 463.626 | 5 / 1 | 3.1 | Yes |
| 536500 |  CHEMBL3974708 CHEMBL3974708 | C17H18N4O3S2 | 390.476 | 7 / 2 | 2.2 | Yes |
| 21406 |  CHEMBL68240 CHEMBL68240 | C12H15NO2 | 205.257 | 3 / 1 | 1.2 | Yes |
| 22823 |  CHEMBL68771 CHEMBL68771 | C20H24N6O2 | 380.452 | 6 / 2 | -0.8 | Yes |
| 459412 |  tropicamide tropicamide | C17H20N2O2 | 284.359 | 3 / 1 | 1.5 | Yes |
| 27436 |  CHEMBL305403 CHEMBL305403 | C28H31N7O3 | 513.602 | 7 / 3 | -0.1 | No |
| 30958 |  CHEMBL303678 CHEMBL303678 | C20H21N5O3 | 379.42 | 5 / 1 | -0.4 | Yes |
| 33723 |  CHEMBL304112 CHEMBL304112 | C30H30N6O4 | 538.608 | 7 / 1 | 1.5 | No |
| 36720 |  CHEMBL1202058 CHEMBL1202058 | C18H21ClN2O5 | 380.825 | 6 / 2 | N/A | N/A |
| 37695 |  CHEMBL1916225 CHEMBL1916225 | C21H30FN3O3 | 391.487 | 5 / 1 | 3.1 | Yes |
| 37953 |  CHEMBL68709 CHEMBL68709 | C25H23N5O3 | 441.491 | 5 / 1 | 1.2 | Yes |
| 522660 |  CHEMBL1357034 CHEMBL1357034 | C20H23N5O3S2 | 445.556 | 8 / 2 | 2.7 | Yes |
| 459584 |  VU0402471-1 VU0402471-1 | C17H16FN3OS | 329.393 | 5 / 2 | 4.0 | Yes |
| 459586 | 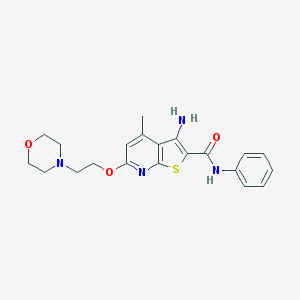 VU0359455 VU0359455 | C21H24N4O3S | 412.508 | 7 / 2 | 3.6 | Yes |
| 459597 | 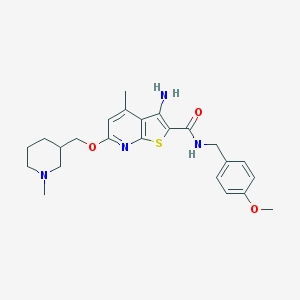 VU0152121 VU0152121 | C24H30N4O3S | 454.589 | 7 / 2 | 4.6 | Yes |
| 44166 | 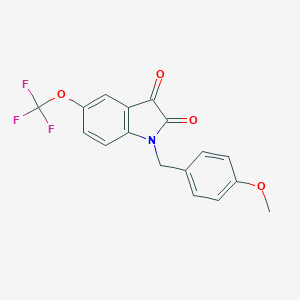 VU 0238429 VU 0238429 | C17H12F3NO4 | 351.281 | 7 / 0 | 3.5 | Yes |
| 45149 | 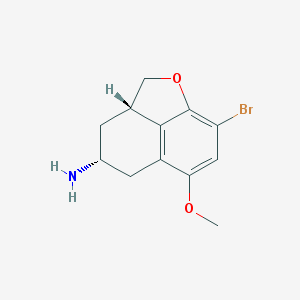 CHEMBL445106 CHEMBL445106 | C12H14BrNO2 | 284.153 | 3 / 1 | 1.9 | Yes |
| 45941 | 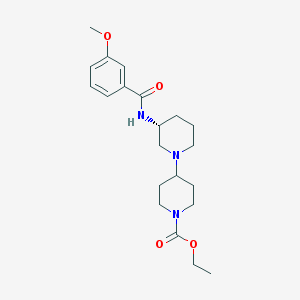 CHEMBL1916228 CHEMBL1916228 | C21H31N3O4 | 389.496 | 5 / 1 | 2.6 | Yes |
| 459646 | 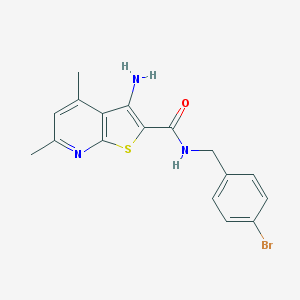 VU0402479-1 VU0402479-1 | C17H16BrN3OS | 390.299 | 4 / 2 | 4.6 | Yes |
| 459674 | 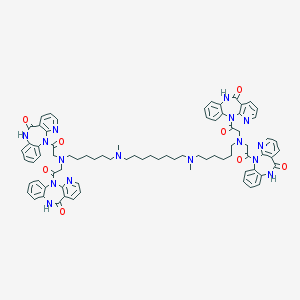 CHEMBL1202002 CHEMBL1202002 | C78H86N16O8 | 1375.65 | 16 / 4 | 9.0 | No |
| 459678 | 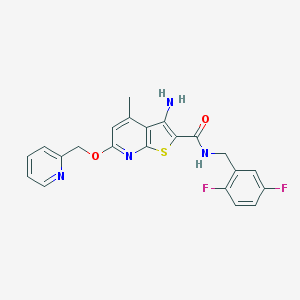 VU0359480 VU0359480 | C22H18F2N4O2S | 440.469 | 8 / 2 | 4.5 | Yes |
| 51901 |  CHEMBL112244 CHEMBL112244 | C36H59N7O2 | 621.915 | 7 / 3 | 4.3 | No |
| 52290 |  TBPB TBPB | C25H32N4O | 404.558 | 3 / 1 | 4.4 | Yes |
| 537357 |  CHEMBL3966208 CHEMBL3966208 | C19H22N4O3S2 | 418.53 | 7 / 2 | 3.0 | Yes |
| 57553 |  CHEMBL1202001 CHEMBL1202001 | C58H76N10O20 | 1233.3 | 26 / 12 | N/A | No |
| 57880 | 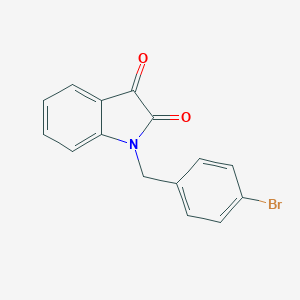 1-(4-Bromobenzyl)indole-2,3-dione 1-(4-Bromobenzyl)indole-2,3-dione | C15H10BrNO2 | 316.154 | 2 / 0 | 2.8 | Yes |
| 459759 | 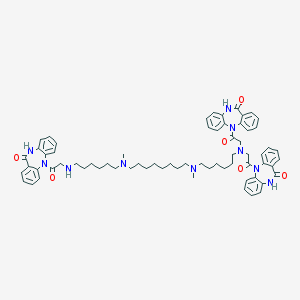 CHEMBL1202003 CHEMBL1202003 | C67H80N10O6 | 1121.44 | 10 / 4 | 9.9 | No |
| 59353 | 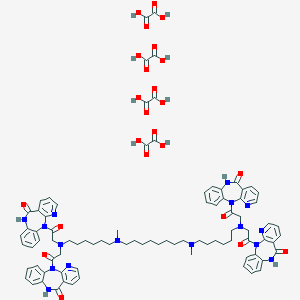 CHEMBL1202002 CHEMBL1202002 | C86H94N16O24 | 1735.79 | 32 / 12 | N/A | No |
| 459778 | 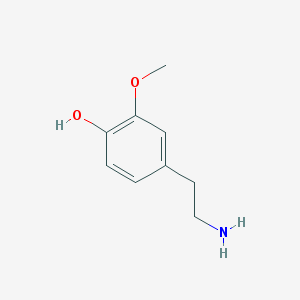 4-(2-Aminoethyl)-2-methoxyphenol 4-(2-Aminoethyl)-2-methoxyphenol | C9H13NO2 | 167.208 | 3 / 2 | 0.2 | Yes |
| 459817 |  VU0402481-1 VU0402481-1 | C17H15F2N3OS | 347.384 | 6 / 2 | 4.1 | Yes |
| 444856 |  CHEMBL3329755 CHEMBL3329755 | C23H25F3N4O3S | 494.533 | 8 / 1 | 3.4 | Yes |
| 459962 |  Himbacine Himbacine | C22H35NO2 | 345.527 | 3 / 0 | 5.0 | Yes |
| 82153 |  Himbacine Himbacine | C22H35NO2 | 345.527 | 3 / 0 | 5.0 | Yes |
| 84305 | 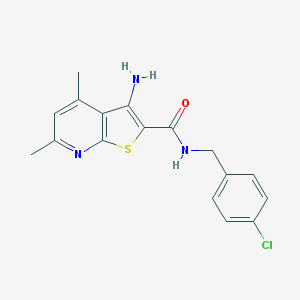 633283-39-3 633283-39-3 | C17H16ClN3OS | 345.845 | 4 / 2 | 4.6 | Yes |
| 548904 | 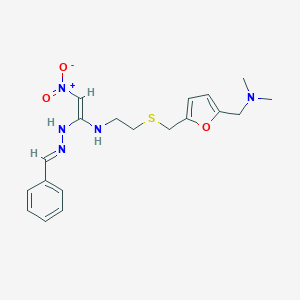 CHEMBL3923597 CHEMBL3923597 | C19H25N5O3S | 403.501 | 8 / 2 | 1.9 | Yes |
| 548925 | 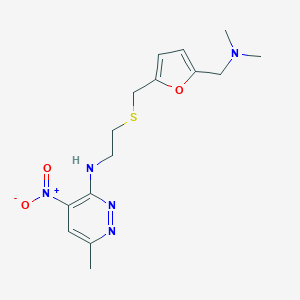 CHEMBL3921095 CHEMBL3921095 | C15H21N5O3S | 351.425 | 8 / 1 | 2.1 | Yes |
| 91128 | 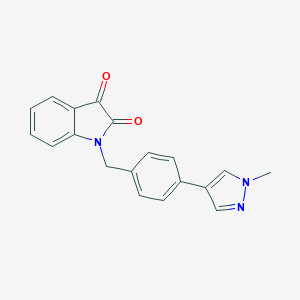 CHEMBL1084941 CHEMBL1084941 | C19H15N3O2 | 317.348 | 3 / 0 | 2.3 | Yes |
| 97341 | 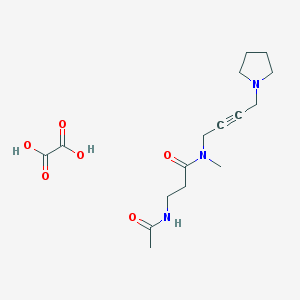 CHEMBL1202043 CHEMBL1202043 | C16H25N3O6 | 355.391 | 7 / 3 | N/A | N/A |
| 99126 | 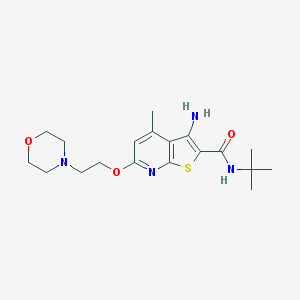 VU0359460 VU0359460 | C19H28N4O3S | 392.518 | 7 / 2 | 3.0 | Yes |
| 99151 | 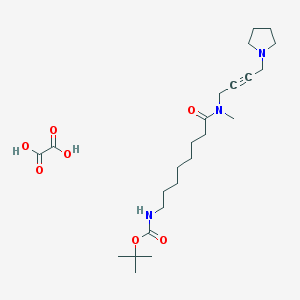 CHEMBL1202052 CHEMBL1202052 | C24H41N3O7 | 483.606 | 8 / 3 | N/A | N/A |
| 460117 | 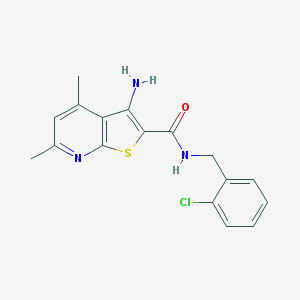 VU0402473-1 VU0402473-1 | C17H16ClN3OS | 345.845 | 4 / 2 | 4.6 | Yes |
| 460146 | 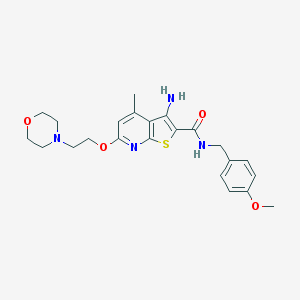 VU0152119 VU0152119 | C23H28N4O4S | 456.561 | 8 / 2 | 3.5 | Yes |
| 460147 | 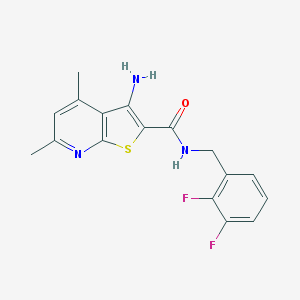 VU0402483-1 VU0402483-1 | C17H15F2N3OS | 347.384 | 6 / 2 | 4.1 | Yes |
| 103512 | 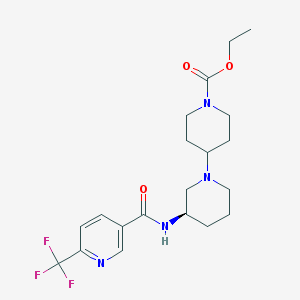 CHEMBL2315680 CHEMBL2315680 | C20H27F3N4O3 | 428.456 | 8 / 1 | 2.5 | Yes |
| 107434 | 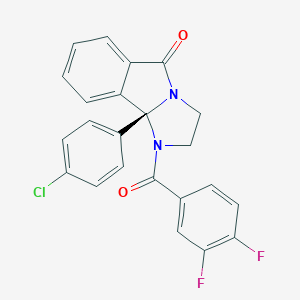 ml375 ml375 | C23H15ClF2N2O2 | 424.832 | 4 / 0 | 4.3 | Yes |
| 112441 | 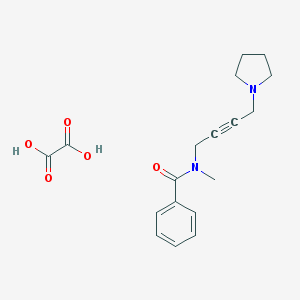 CHEMBL1202057 CHEMBL1202057 | C18H22N2O5 | 346.383 | 6 / 2 | N/A | N/A |
| 115348 | 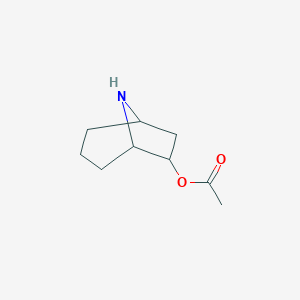 CHEMBL64000 CHEMBL64000 | C9H15NO2 | 169.224 | 3 / 1 | 0.9 | Yes |
| 116436 | 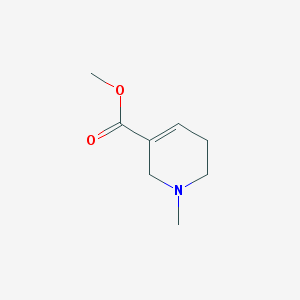 arecoline arecoline | C8H13NO2 | 155.197 | 3 / 0 | 0.3 | Yes |
| 460280 | 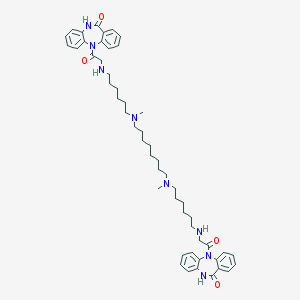 CHEMBL1202004 CHEMBL1202004 | C52H70N8O4 | 871.184 | 8 / 4 | 7.9 | No |
| 460297 | 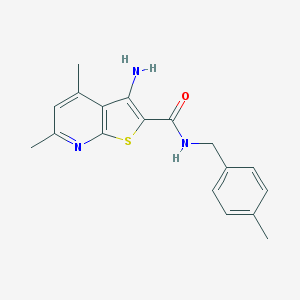 VU0402477-1 VU0402477-1 | C18H19N3OS | 325.43 | 4 / 2 | 4.3 | Yes |
| 549354 | 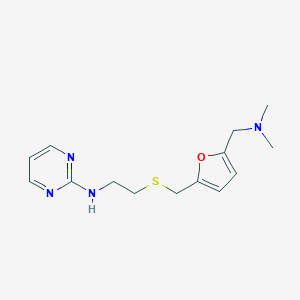 CHEMBL3963436 CHEMBL3963436 | C14H20N4OS | 292.401 | 6 / 1 | 1.6 | Yes |
| 122439 | 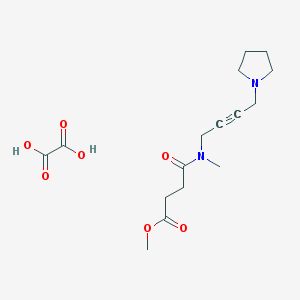 CHEMBL1202040 CHEMBL1202040 | C16H24N2O7 | 356.375 | 8 / 2 | N/A | N/A |
| 124120 | 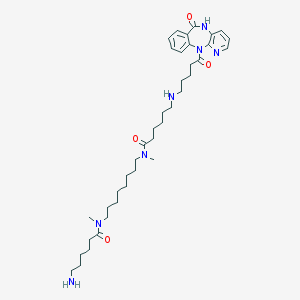 CHEMBL134692 CHEMBL134692 | C39H61N7O4 | 691.962 | 7 / 3 | 3.0 | No |
| 125944 | 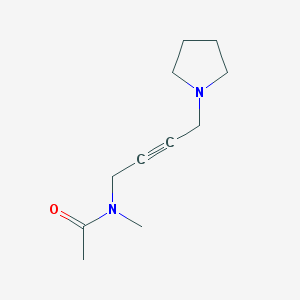 Oxotremorine 1 Oxotremorine 1 | C11H18N2O | 194.278 | 2 / 0 | 0.3 | Yes |
| 126436 | 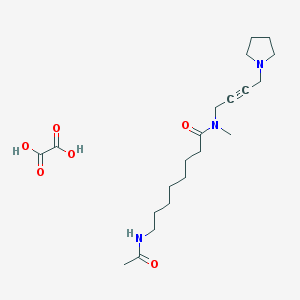 CHEMBL1202051 CHEMBL1202051 | C21H35N3O6 | 425.526 | 7 / 3 | N/A | N/A |
| 127081 | 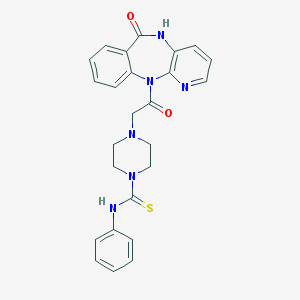 CHEMBL420739 CHEMBL420739 | C25H24N6O2S | 472.567 | 5 / 2 | 1.5 | Yes |
| 127097 | 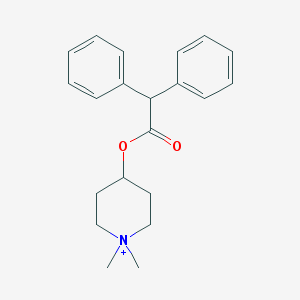 4-Diphenylacetoxy-1,1-dimethylpiperidinium 4-Diphenylacetoxy-1,1-dimethylpiperidinium | C21H26NO2+ | 324.444 | 2 / 0 | 4.0 | Yes |
| 132155 |  CHEMBL1202039 CHEMBL1202039 | C17H28N4O6 | 384.433 | 8 / 4 | N/A | N/A |
| 133381 |  CHEMBL404874 CHEMBL404874 | C18H28ClN3O | 337.892 | 3 / 1 | 3.7 | Yes |
| 134883 |  CHEMBL1916229 CHEMBL1916229 | C20H27F2N3O3 | 395.451 | 6 / 1 | 2.8 | Yes |
| 135535 |  CHEMBL271704 CHEMBL271704 | C22H26ClN3O | 383.92 | 3 / 1 | 3.9 | Yes |
| 136725 |  CHEMBL270580 CHEMBL270580 | C23H30ClN3O2 | 415.962 | 4 / 1 | 4.9 | Yes |
| 460519 |  VU0402476-1 VU0402476-1 | C16H16N4OS | 312.391 | 5 / 2 | 2.9 | Yes |
| 143614 |  CHEMBL255523 CHEMBL255523 | C20H30ClN3O | 363.93 | 3 / 1 | 4.5 | Yes |
| 549775 |  CHEMBL3927243 CHEMBL3927243 | C24H27Cl2N3O2S | 492.459 | 5 / 2 | 5.3 | No |
| 153067 |  CHEMBL1916230 CHEMBL1916230 | C21H31N3O3 | 373.497 | 4 / 1 | 3.0 | Yes |
| 539985 |  CHEMBL3942363 CHEMBL3942363 | C17H15F3N4O3S2 | 444.447 | 10 / 2 | 3.2 | Yes |
| 460630 |  VU0152117 VU0152117 | C22H28N4O3S | 428.551 | 7 / 2 | 4.2 | Yes |
| 159780 |  JTTPGMRULUVVEK-UHFFFAOYSA-N JTTPGMRULUVVEK-UHFFFAOYSA-N | C13H20N2O3 | 252.314 | 4 / 1 | -2.5 | Yes |
| 460765 | 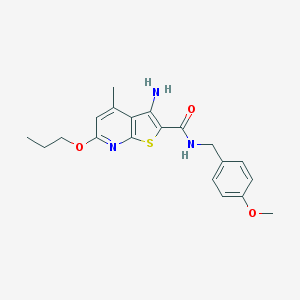 VU0152122 VU0152122 | C20H23N3O3S | 385.482 | 6 / 2 | 4.7 | Yes |
| 540455 | 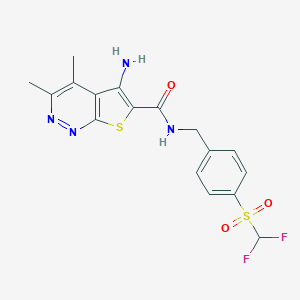 CHEMBL3963788 CHEMBL3963788 | C17H16F2N4O3S2 | 426.457 | 9 / 2 | 3.1 | Yes |
| 175878 | 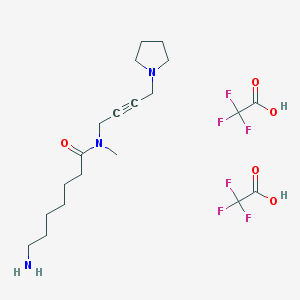 CHEMBL1202131 CHEMBL1202131 | C20H31F6N3O5 | 507.474 | 13 / 3 | N/A | No |
| 178616 | 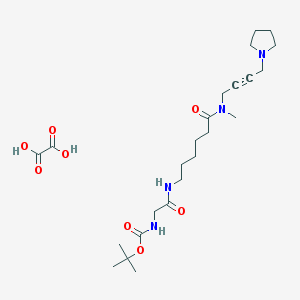 CHEMBL1202054 CHEMBL1202054 | C24H40N4O8 | 512.604 | 9 / 4 | N/A | No |
| 460859 | 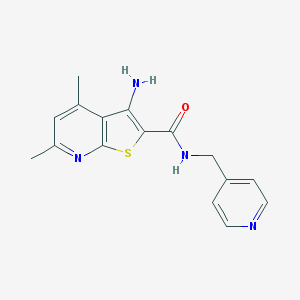 VU0402475-1 VU0402475-1 | C16H16N4OS | 312.391 | 5 / 2 | 2.9 | Yes |
| 540761 | 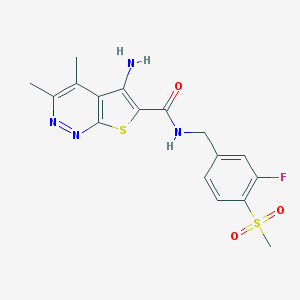 CHEMBL3896591 CHEMBL3896591 | C17H17FN4O3S2 | 408.466 | 8 / 2 | 2.3 | Yes |
| 550238 | 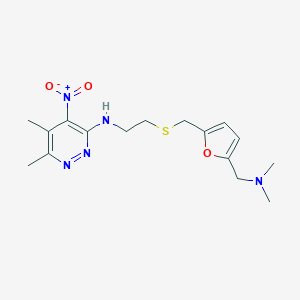 CHEMBL3964368 CHEMBL3964368 | C16H23N5O3S | 365.452 | 8 / 1 | 2.4 | Yes |
| 550257 | 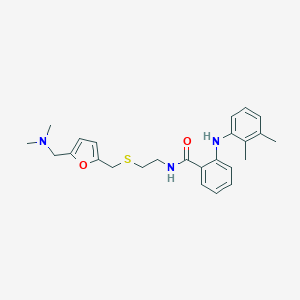 CHEMBL3937756 CHEMBL3937756 | C25H31N3O2S | 437.602 | 5 / 2 | 6.0 | No |
| 460912 | 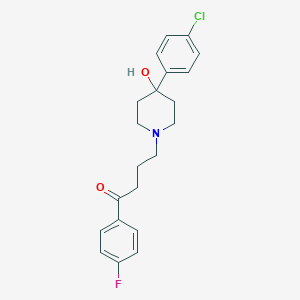 haloperidol haloperidol | C21H23ClFNO2 | 375.868 | 4 / 1 | 3.2 | Yes |
| 194895 | 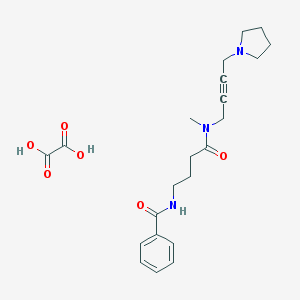 CHEMBL1202049 CHEMBL1202049 | C22H29N3O6 | 431.489 | 7 / 3 | N/A | N/A |
| 195466 | 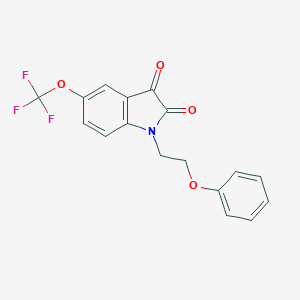 CHEMBL2381694 CHEMBL2381694 | C17H12F3NO4 | 351.281 | 7 / 0 | 3.7 | Yes |
| 196396 | 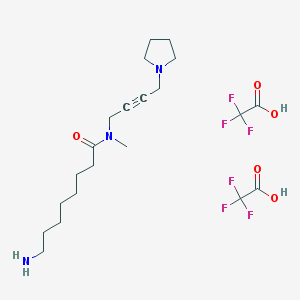 CHEMBL1202134 CHEMBL1202134 | C21H33F6N3O5 | 521.501 | 13 / 3 | N/A | No |
| 196757 | 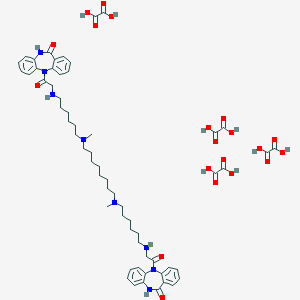 CHEMBL1202004 CHEMBL1202004 | C60H78N8O20 | 1231.32 | 24 / 12 | N/A | No |
| 201849 | 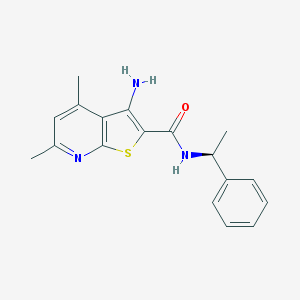 VU10007 VU10007 | C18H19N3OS | 325.43 | 4 / 2 | 4.3 | Yes |
| 202858 | 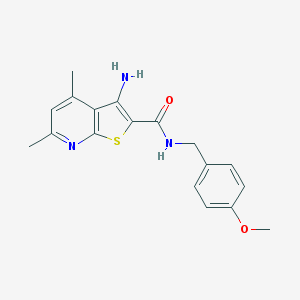 VU0152100 VU0152100 | C18H19N3O2S | 341.429 | 5 / 2 | 3.9 | Yes |
| 205997 | 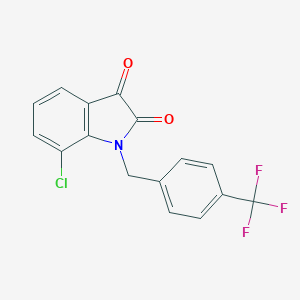 CHEMBL503735 CHEMBL503735 | C16H9ClF3NO2 | 339.698 | 5 / 0 | 3.9 | Yes |
| 461030 | 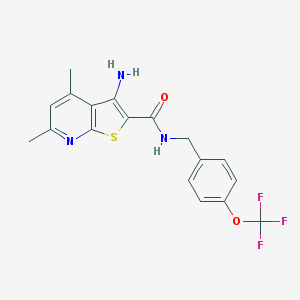 VU0402480-1 VU0402480-1 | C18H16F3N3O2S | 395.4 | 8 / 2 | 5.1 | No |
| 209339 | 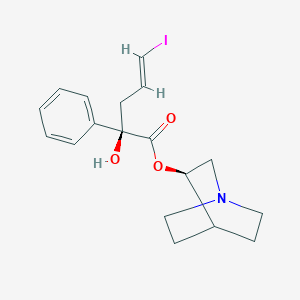 CHEMBL127587 CHEMBL127587 | C18H22INO3 | 427.282 | 4 / 1 | 3.2 | Yes |
| 541506 | 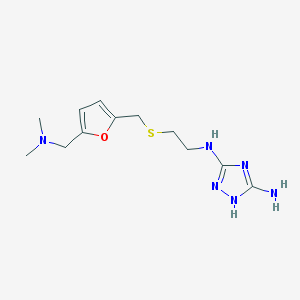 CHEMBL3926121 CHEMBL3926121 | C12H20N6OS | 296.393 | 7 / 3 | 1.1 | Yes |
| 550534 | 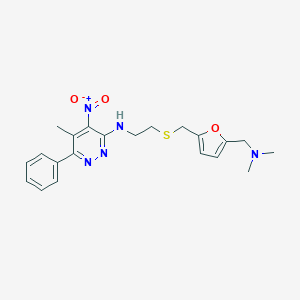 CHEMBL3961411 CHEMBL3961411 | C21H25N5O3S | 427.523 | 8 / 1 | 3.7 | Yes |
| 219022 | 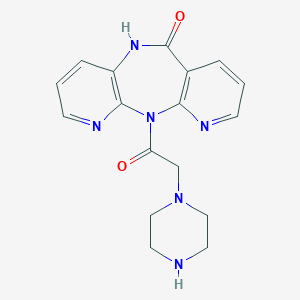 CHEMBL2112217 CHEMBL2112217 | C17H18N6O2 | 338.371 | 6 / 2 | -0.3 | Yes |
| 222335 | 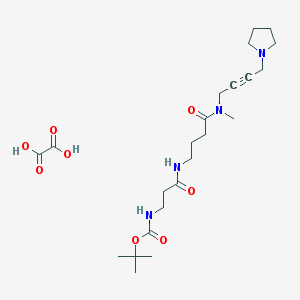 CHEMBL1202037 CHEMBL1202037 | C23H38N4O8 | 498.577 | 9 / 4 | N/A | N/A |
| 541813 | 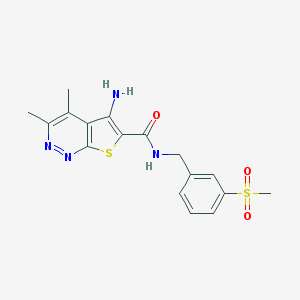 CHEMBL3894866 CHEMBL3894866 | C17H18N4O3S2 | 390.476 | 7 / 2 | 2.2 | Yes |
| 224015 | 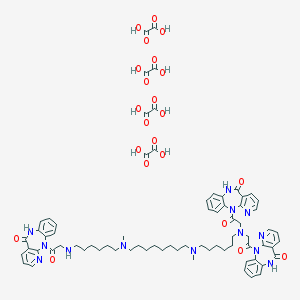 CHEMBL1202000 CHEMBL1202000 | C72H85N13O22 | 1484.54 | 29 / 12 | N/A | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417