

You can:
| Name | Neuropeptide Y receptor type 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NPY5R |
| Synonym | NPYY5-R NPY5-R NPY-Y5 receptor neuropeptide Y receptor type 5 food intake receptor [ Show all ] |
| Disease | Eating disorder; Obesity Eating disorder; Obesity; Diabetes Major depressive disorder Obesity |
| Length | 445 |
| Amino acid sequence | MDLELDEYYNKTLATENNTAATRNSDFPVWDDYKSSVDDLQYFLIGLYTFVSLLGFMGNLLILMALMKKRNQKTTVNFLIGNLAFSDILVVLFCSPFTLTSVLLDQWMFGKVMCHIMPFLQCVSVLVSTLILISIAIVRYHMIKHPISNNLTANHGYFLIATVWTLGFAICSPLPVFHSLVELQETFGSALLSSRYLCVESWPSDSYRIAFTISLLLVQYILPLVCLTVSHTSVCRSISCGLSNKENRLEENEMINLTLHPSKKSGPQVKLSGSHKWSYSFIKKHRRRYSKKTACVLPAPERPSQENHSRILPENFGSVRSQLSSSSKFIPGVPTCFEIKPEENSDVHELRVKRSVTRIKKRSRSVFYRLTILILVFAVSWMPLHLFHVVTDFNDNLISNRHFKLVYCICHLLGMMSCCLNPILYGFLNNGIKADLVSLIHCLHM |
| UniProt | Q15761 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q15761 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q15761. |
| BioLiP | N/A |
| Therapeutic Target Database | T20331 |
| ChEMBL | CHEMBL4561 |
| IUPHAR | 308 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 336 | 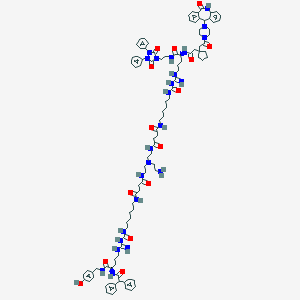 BDBM50442547 BDBM50442547 | C104H138N24O15 | 1964.4 | 20 / 19 | 6.9 | No |
| 868 | 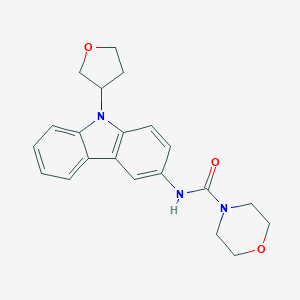 CHEMBL324028 CHEMBL324028 | C21H23N3O3 | 365.433 | 3 / 1 | 2.1 | Yes |
| 1668 | 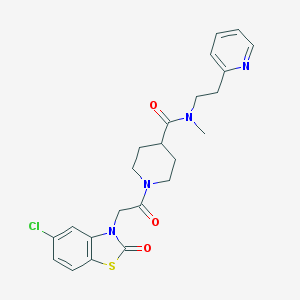 CHEMBL16994 CHEMBL16994 | C23H25ClN4O3S | 472.988 | 5 / 0 | 2.9 | Yes |
| 1793 | 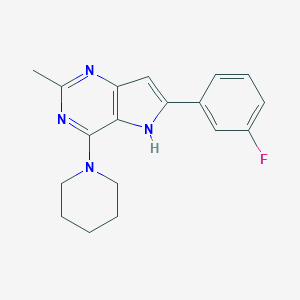 CHEMBL343817 CHEMBL343817 | C18H19FN4 | 310.376 | 4 / 1 | 3.9 | Yes |
| 4018 | 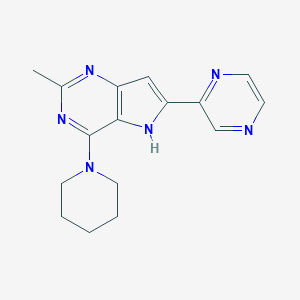 CHEMBL423720 CHEMBL423720 | C16H18N6 | 294.362 | 5 / 1 | 1.7 | Yes |
| 4374 | 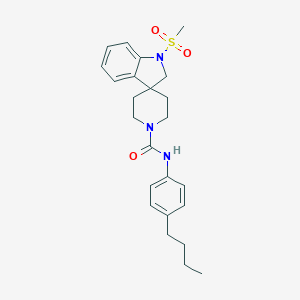 CHEMBL523330 CHEMBL523330 | C24H31N3O3S | 441.59 | 4 / 1 | 4.2 | Yes |
| 4904 | 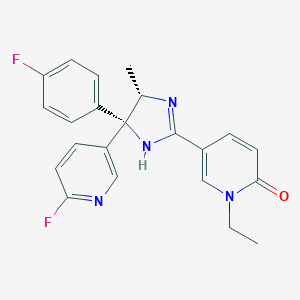 CHEMBL1784127 CHEMBL1784127 | C22H20F2N4O | 394.426 | 5 / 1 | 2.3 | Yes |
| 5227 | 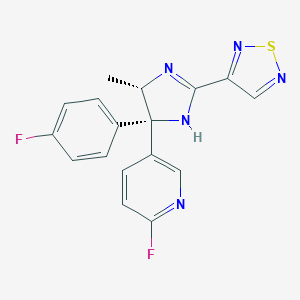 CHEMBL523945 CHEMBL523945 | C17H13F2N5S | 357.383 | 7 / 1 | 2.5 | Yes |
| 5571 |  CHEMBL522183 CHEMBL522183 | C23H21F2N3 | 377.439 | 4 / 1 | 4.2 | Yes |
| 5728 |  CHEMBL495625 CHEMBL495625 | C24H22N4O4 | 430.464 | 6 / 1 | 2.8 | Yes |
| 6536 |  CHEMBL117400 CHEMBL117400 | C29H33FN4O3S | 536.666 | 7 / 2 | 4.0 | No |
| 6678 |  CHEMBL498396 CHEMBL498396 | C19H16FN3O2 | 337.354 | 5 / 1 | 3.5 | Yes |
| 6726 |  CHEMBL7147 CHEMBL7147 | C15H14N4 | 250.305 | 2 / 2 | 2.2 | Yes |
| 7177 |  CHEMBL136974 CHEMBL136974 | C19H19F3N4 | 360.384 | 6 / 1 | 4.7 | Yes |
| 7566 |  CHEMBL269561 CHEMBL269561 | C17H14FN3 | 279.318 | 3 / 1 | 3.1 | Yes |
| 7644 |  CHEMBL494174 CHEMBL494174 | C21H17F2N3 | 349.385 | 4 / 2 | 3.4 | Yes |
| 7914 |  CHEMBL349328 CHEMBL349328 | C38H49IN4O2 | 720.74 | 5 / 0 | 8.2 | No |
| 8570 |  CHEMBL326736 CHEMBL326736 | C14H11N3 | 221.263 | 2 / 1 | 2.3 | Yes |
| 8598 |  CHEMBL331319 CHEMBL331319 | C28H32F2N4O3S | 542.646 | 8 / 3 | 3.7 | No |
| 8639 |  CHEMBL523715 CHEMBL523715 | C24H25N5O3S | 463.556 | 6 / 1 | 2.2 | Yes |
| 8737 |  CHEMBL75827 CHEMBL75827 | C23H19F3N4O2S | 472.486 | 8 / 2 | 5.0 | Yes |
| 8871 |  CID 73351118 CID 73351118 | C184H287N53O53S2 | 4153.76 | 61 / 58 | -13.6 | No |
| 8983 |  CHEMBL137786 CHEMBL137786 | C19H18N4O | 318.38 | 4 / 2 | 3.8 | Yes |
| 9041 |  CHEMBL133032 CHEMBL133032 | C18H19N3O | 293.37 | 4 / 0 | 4.1 | Yes |
| 9163 |  CHEMBL350379 CHEMBL350379 | C28H35N3O2S | 477.667 | 5 / 2 | 4.9 | Yes |
| 9239 |  CID 76335489 CID 76335489 | C177H272N52O55 | 4008.43 | 62 / 60 | -15.6 | No |
| 9479 |  BDBM50261952 BDBM50261952 | C23H25BrO4 | 445.353 | 4 / 0 | 4.1 | Yes |
| 10097 |  CHEMBL6938 CHEMBL6938 | C19H18N2O | 290.366 | 2 / 1 | 4.0 | Yes |
| 10606 |  CHEMBL288355 CHEMBL288355 | C28H33N5O3 | 487.604 | 4 / 5 | 2.9 | Yes |
| 11411 |  CHEMBL1912066 CHEMBL1912066 | C19H24N4O4S3 | 468.605 | 8 / 2 | 3.1 | Yes |
| 11437 |  CHEMBL42747 CHEMBL42747 | C30H35N5O3 | 513.642 | 4 / 4 | 3.1 | No |
| 11938 |  CHEMBL120117 CHEMBL120117 | C15H10F3N3 | 289.261 | 5 / 1 | 3.2 | Yes |
| 12045 | 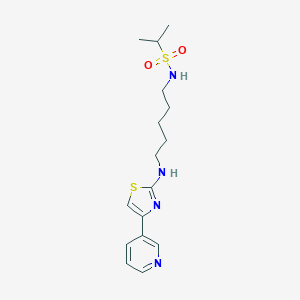 CHEMBL1909718 CHEMBL1909718 | C16H24N4O2S2 | 368.514 | 7 / 2 | 2.8 | Yes |
| 12145 | 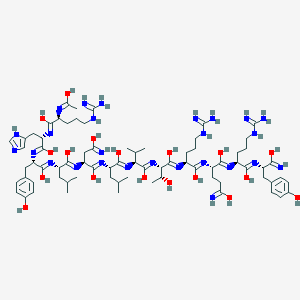 Ac-Peptide YY(25-36) Ac-Peptide YY(25-36) | C74H118N26O18 | 1659.92 | 37 / 31 | 4.7 | No |
| 13103 | 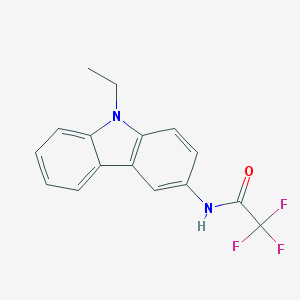 CHEMBL61879 CHEMBL61879 | C16H13F3N2O | 306.288 | 4 / 1 | 3.9 | Yes |
| 13872 | 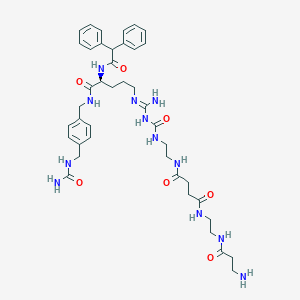 CHEMBL2440903 CHEMBL2440903 | C41H56N12O7 | 828.976 | 9 / 11 | -1.2 | No |
| 14162 |  CID 44577739 CID 44577739 | C27H34O4 | 422.565 | 4 / 1 | 5.8 | No |
| 14627 |  CHEMBL360671 CHEMBL360671 | C22H23BrF3N5O3S | 574.417 | 11 / 1 | 5.6 | No |
| 14631 |  D0P9KP D0P9KP | C180H279N53O54 | 4049.53 | 61 / 61 | -14.9 | No |
| 15271 |  CHEMBL304896 CHEMBL304896 | C22H19N3O2 | 357.413 | 3 / 1 | 2.7 | Yes |
| 15355 | 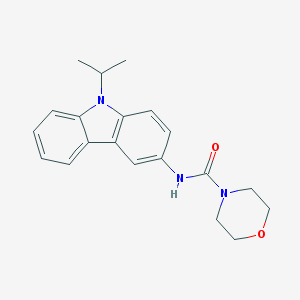 CHEMBL419951 CHEMBL419951 | C20H23N3O2 | 337.423 | 2 / 1 | 2.9 | Yes |
| 16306 | 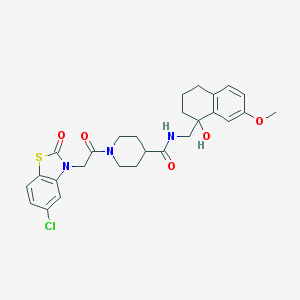 CHEMBL349486 CHEMBL349486 | C27H30ClN3O5S | 544.063 | 6 / 2 | 3.2 | No |
| 16607 | 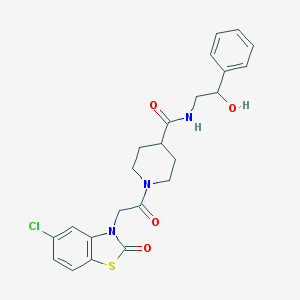 CHEMBL276322 CHEMBL276322 | C23H24ClN3O4S | 473.972 | 5 / 2 | 2.6 | Yes |
| 19109 | 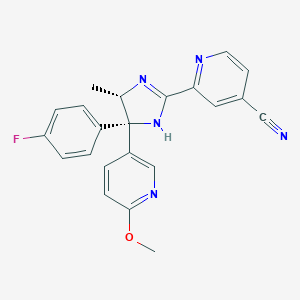 CHEMBL506163 CHEMBL506163 | C22H18FN5O | 387.418 | 6 / 1 | 2.6 | Yes |
| 19161 | 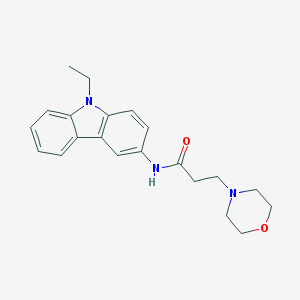 CHEMBL59680 CHEMBL59680 | C21H25N3O2 | 351.45 | 3 / 1 | 2.4 | Yes |
| 19347 | 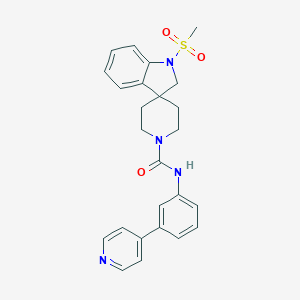 CHEMBL492375 CHEMBL492375 | C25H26N4O3S | 462.568 | 5 / 1 | 2.8 | Yes |
| 19358 | 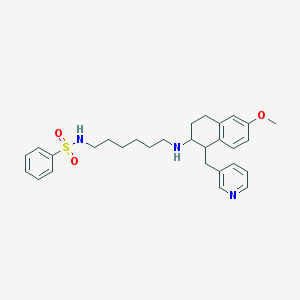 CHEMBL118931 CHEMBL118931 | C29H37N3O3S | 507.693 | 6 / 2 | 4.9 | No |
| 19395 | 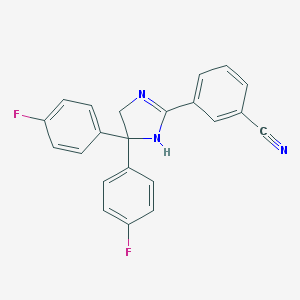 CHEMBL523819 CHEMBL523819 | C22H15F2N3 | 359.38 | 4 / 1 | 3.8 | Yes |
| 19999 |  CHEMBL164586 CHEMBL164586 | C29H38N4O2S | 506.709 | 6 / 3 | 4.6 | No |
| 20251 |  CHEMBL76512 CHEMBL76512 | C18H12Cl2F3N3O | 414.209 | 5 / 1 | 5.6 | No |
| 20706 |  CHEMBL1912074 CHEMBL1912074 | C13H19N3O2S3 | 345.494 | 7 / 2 | 2.8 | Yes |
| 21094 |  CHEMBL137638 CHEMBL137638 | C16H24N4 | 272.396 | 3 / 1 | 3.9 | Yes |
| 21345 | 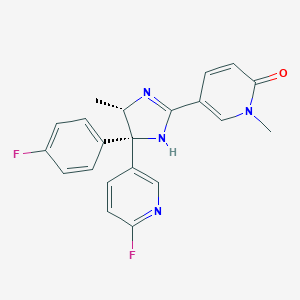 CHEMBL1782077 CHEMBL1782077 | C21H18F2N4O | 380.399 | 5 / 1 | 1.9 | Yes |
| 21525 | 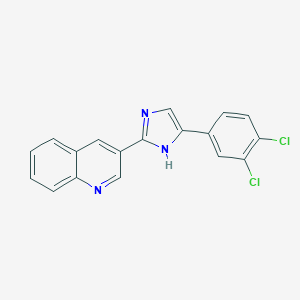 CHEMBL420541 CHEMBL420541 | C18H11Cl2N3 | 340.207 | 2 / 1 | 4.9 | Yes |
| 22025 | 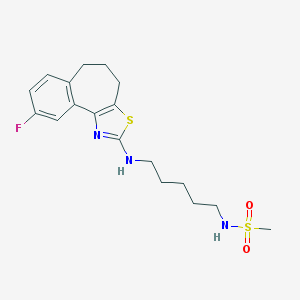 CHEMBL1836319 CHEMBL1836319 | C18H24FN3O2S2 | 397.527 | 7 / 2 | 3.9 | Yes |
| 22580 | 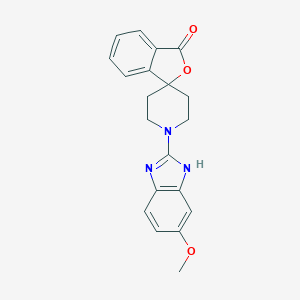 CHEMBL525543 CHEMBL525543 | C20H19N3O3 | 349.39 | 5 / 1 | 3.4 | Yes |
| 22652 |  CHEMBL312169 CHEMBL312169 | C21H26FN3O3S | 419.515 | 7 / 1 | 3.1 | Yes |
| 23373 |  CHEMBL311159 CHEMBL311159 | C23H23N3O5 | 421.453 | 5 / 2 | 3.7 | Yes |
| 23749 |  CHEMBL585711 CHEMBL585711 | C22H26FN5OS2 | 459.602 | 9 / 1 | 3.8 | Yes |
| 23796 |  CHEMBL159134 CHEMBL159134 | C16H12ClN3O | 297.742 | 2 / 2 | 3.7 | Yes |
| 24067 |  CHEMBL119726 CHEMBL119726 | C22H27N3O2 | 365.477 | 2 / 1 | 3.6 | Yes |
| 24337 |  CHEMBL495169 CHEMBL495169 | C24H25N5O3S | 463.556 | 6 / 1 | 2.5 | Yes |
| 24895 |  CHEMBL1912087 CHEMBL1912087 | C18H23N5O2S3 | 437.595 | 9 / 2 | 3.8 | Yes |
| 25679 |  CHEMBL425012 CHEMBL425012 | C23H24BrF3N4O3S | 573.429 | 10 / 2 | 6.1 | No |
| 26657 | 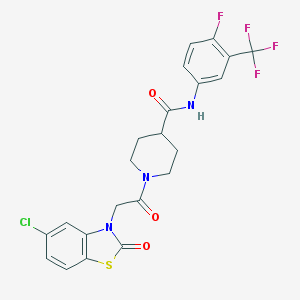 CHEMBL16986 CHEMBL16986 | C22H18ClF4N3O3S | 515.908 | 8 / 1 | 4.3 | No |
| 26690 | 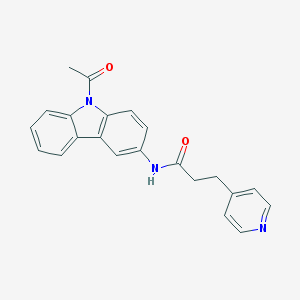 CHEMBL116210 CHEMBL116210 | C22H19N3O2 | 357.413 | 3 / 1 | 3.2 | Yes |
| 28766 | 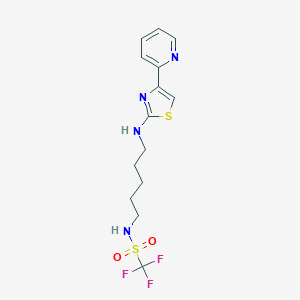 CHEMBL1912067 CHEMBL1912067 | C14H17F3N4O2S2 | 394.431 | 10 / 2 | 3.3 | Yes |
| 29409 | 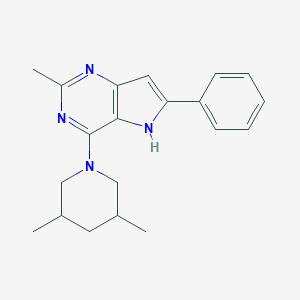 CHEMBL137360 CHEMBL137360 | C20H24N4 | 320.44 | 3 / 1 | 4.7 | Yes |
| 30527 |  CHEMBL278677 CHEMBL278677 | C26H23N5O2S | 469.563 | 7 / 3 | 4.7 | Yes |
| 30651 |  CHEMBL312739 CHEMBL312739 | C24H26N2O2 | 374.484 | 2 / 2 | 4.3 | Yes |
| 30804 |  CHEMBL160722 CHEMBL160722 | C22H23N3O2 | 361.445 | 3 / 2 | 4.1 | Yes |
| 30939 |  CHEMBL526084 CHEMBL526084 | C22H18ClN3O4 | 423.853 | 5 / 1 | 3.6 | Yes |
| 31200 |  CHEMBL16965 CHEMBL16965 | C26H23N5O2S | 469.563 | 7 / 3 | 4.7 | Yes |
| 31254 |  CHEMBL165506 CHEMBL165506 | C29H37N3O3S | 507.693 | 6 / 2 | 5.0 | No |
| 31342 |  CHEMBL418546 CHEMBL418546 | C23H25ClN4O4S | 488.987 | 6 / 2 | 2.0 | Yes |
| 31494 |  CID 9930634 CID 9930634 | C24H28O5 | 396.483 | 5 / 1 | 3.6 | Yes |
| 31579 | 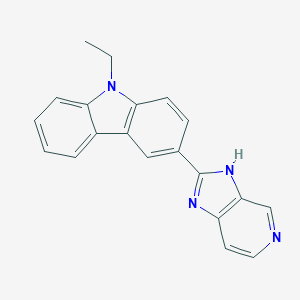 CHEMBL417225 CHEMBL417225 | C20H16N4 | 312.376 | 2 / 1 | 3.8 | Yes |
| 31604 | 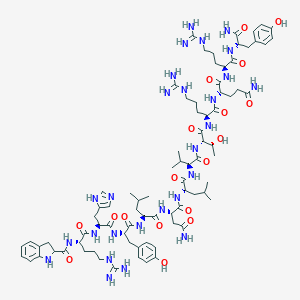 CID 44439571 CID 44439571 | C81H123N27O18 | 1763.05 | 23 / 29 | -2.0 | No |
| 32150 | 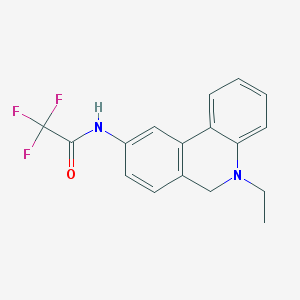 CHEMBL304245 CHEMBL304245 | C17H15F3N2O | 320.315 | 5 / 1 | 3.9 | Yes |
| 32967 | 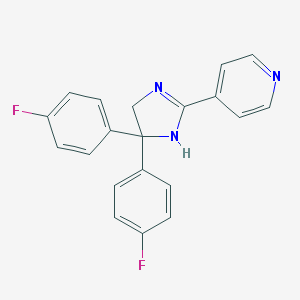 CHEMBL521862 CHEMBL521862 | C20H15F2N3 | 335.358 | 4 / 1 | 3.0 | Yes |
| 33819 |  BDBM50261898 BDBM50261898 | C23H25FO4 | 384.447 | 5 / 0 | 3.5 | Yes |
| 33838 |  CHEMBL77589 CHEMBL77589 | C24H26N2O2 | 374.484 | 2 / 1 | 5.6 | No |
| 33849 |  CHEMBL292729 CHEMBL292729 | C21H25N3O | 335.451 | 2 / 1 | 3.3 | Yes |
| 33892 |  CHEMBL167860 CHEMBL167860 | C27H39N3O4S | 501.686 | 7 / 3 | 3.0 | No |
| 35088 | 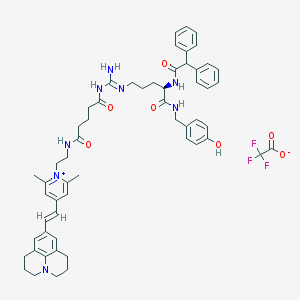 CHEMBL1774211 CHEMBL1774211 | C57H65F3N8O7 | 1031.19 | 12 / 6 | N/A | No |
| 36648 | 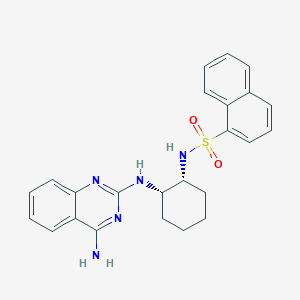 CHEMBL277551 CHEMBL277551 | C24H25N5O2S | 447.557 | 7 / 3 | 4.8 | Yes |
| 37036 | 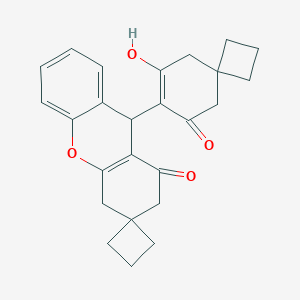 CID 9886706 CID 9886706 | C25H26O4 | 390.479 | 4 / 1 | 4.3 | Yes |
| 37424 | 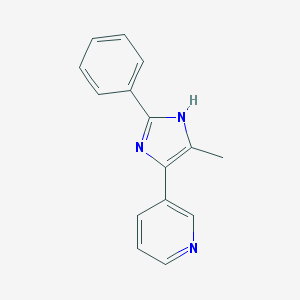 CHEMBL6893 CHEMBL6893 | C15H13N3 | 235.29 | 2 / 1 | 2.7 | Yes |
| 37602 |  CHEMBL418459 CHEMBL418459 | C20H23N3O2 | 337.423 | 3 / 1 | 2.7 | Yes |
| 37703 |  CHEMBL3040658 CHEMBL3040658 | C84H116N18O16 | 1633.96 | 18 / 17 | 2.5 | No |
| 38167 |  CHEMBL489726 CHEMBL489726 | C20H16F2N4O | 366.372 | 5 / 2 | 2.3 | Yes |
| 38215 |  CHEMBL123410 CHEMBL123410 | C22H24Cl2N2O | 403.347 | 2 / 0 | 5.4 | No |
| 38354 |  CHEMBL294560 CHEMBL294560 | C20H16N4 | 312.376 | 2 / 1 | 4.1 | Yes |
| 38634 |  SCHEMBL14994383 SCHEMBL14994383 | C23H26O4 | 366.457 | 4 / 1 | 3.6 | Yes |
| 38635 |  AC1OF52R AC1OF52R | C23H26O4 | 366.457 | 4 / 1 | 3.6 | Yes |
| 38640 |  CHEMBL491762 CHEMBL491762 | C23H26O4 | 366.457 | 4 / 1 | 3.6 | Yes |
| 38961 |  CHEMBL117684 CHEMBL117684 | C22H21N3O | 343.43 | 2 / 1 | 3.6 | Yes |
| 39441 |  CHEMBL138172 CHEMBL138172 | C20H22N4O | 334.423 | 4 / 1 | 3.5 | Yes |
| 40340 |  CHEMBL521844 CHEMBL521844 | C21H15ClF2N2 | 368.812 | 3 / 1 | 4.7 | Yes |
| 40438 |  CHEMBL137102 CHEMBL137102 | C22H30N4 | 350.51 | 3 / 2 | 6.5 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417