

You can:
| Name | Metabotropic glutamate receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GRM1 |
| Synonym | SCAR13 MGLUR1 mGlu1 receptor metabotropic glutamate receptor 1 GPRC1A [ Show all ] |
| Disease | Alzheimer disease; Huntington's disease Neurological disease Neuropathic pain Schizophrenia Pain |
| Length | 1194 |
| Amino acid sequence | MVGLLLFFFPAIFLEVSLLPRSPGRKVLLAGASSQRSVARMDGDVIIGALFSVHHQPPAEKVPERKCGEIREQYGIQRVEAMFHTLDKINADPVLLPNITLGSEIRDSCWHSSVALEQSIEFIRDSLISIRDEKDGINRCLPDGQSLPPGRTKKPIAGVIGPGSSSVAIQVQNLLQLFDIPQIAYSATSIDLSDKTLYKYFLRVVPSDTLQARAMLDIVKRYNWTYVSAVHTEGNYGESGMDAFKELAAQEGLCIAHSDKIYSNAGEKSFDRLLRKLRERLPKARVVVCFCEGMTVRGLLSAMRRLGVVGEFSLIGSDGWADRDEVIEGYEVEANGGITIKLQSPEVRSFDDYFLKLRLDTNTRNPWFPEFWQHRFQCRLPGHLLENPNFKRICTGNESLEENYVQDSKMGFVINAIYAMAHGLQNMHHALCPGHVGLCDAMKPIDGSKLLDFLIKSSFIGVSGEEVWFDEKGDAPGRYDIMNLQYTEANRYDYVHVGTWHEGVLNIDDYKIQMNKSGVVRSVCSEPCLKGQIKVIRKGEVSCCWICTACKENEYVQDEFTCKACDLGWWPNADLTGCEPIPVRYLEWSNIESIIAIAFSCLGILVTLFVTLIFVLYRDTPVVKSSSRELCYIILAGIFLGYVCPFTLIAKPTTTSCYLQRLLVGLSSAMCYSALVTKTNRIARILAGSKKKICTRKPRFMSAWAQVIIASILISVQLTLVVTLIIMEPPMPILSYPSIKEVYLICNTSNLGVVAPLGYNGLLIMSCTYYAFKTRNVPANFNEAKYIAFTMYTTCIIWLAFVPIYFGSNYKIITTCFAVSLSVTVALGCMFTPKMYIIIAKPERNVRSAFTTSDVVRMHVGDGKLPCRSNTFLNIFRRKKAGAGNANSNGKSVSWSEPGGGQVPKGQHMWHRLSVHVKTNETACNQTAVIKPLTKSYQGSGKSLTFSDTSTKTLYNVEEEEDAQPIRFSPPGSPSMVVHRRVPSAATTPPLPSHLTAEETPLFLAEPALPKGLPPPLQQQQQPPPQQKSLMDQLQGVVSNFSTAIPDFHAVLAGPGGPGNGLRSLYPPPPPPQHLQMLPLQLSTFGEELVSPPADDDDDSERFKLLQEYVYEHEREGNTEEDELEEEEEDLQAASKLTPDDSPALTPPSPFRDSVASGSSVPSSPVSESVLCTPPNVSYASVILRDYKQSSSTL |
| UniProt | Q13255 |
| Protein Data Bank | 4or2, 3ks9 |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4or2. |
| BioLiP | BL0271871,BL0271876, BL0271872,BL0271873,BL0271874,, BL0174211,BL0174213, BL0174210,BL0174212 |
| Therapeutic Target Database | T27137 |
| ChEMBL | CHEMBL3772 |
| IUPHAR | 289 |
| DrugBank | BE0000824 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 463128 | 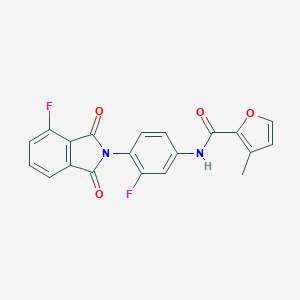 CHEMBL3634421 CHEMBL3634421 | C20H12F2N2O4 | 382.323 | 6 / 1 | 3.3 | Yes |
| 2929 | 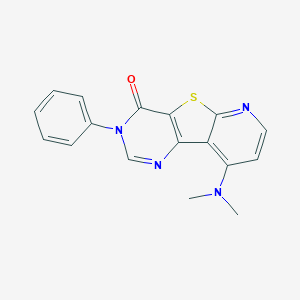 CHEMBL224615 CHEMBL224615 | C17H14N4OS | 322.386 | 5 / 0 | 3.2 | Yes |
| 4136 | 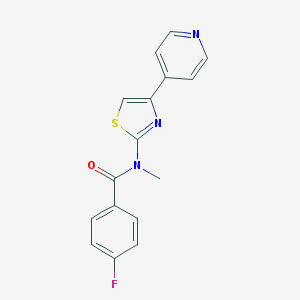 CHEMBL578580 CHEMBL578580 | C16H12FN3OS | 313.35 | 5 / 0 | 3.0 | Yes |
| 4247 | 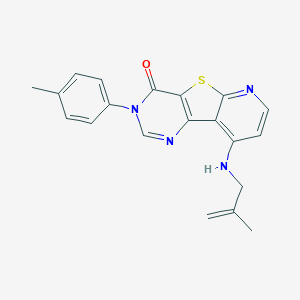 CHEMBL1783868 CHEMBL1783868 | C20H18N4OS | 362.451 | 5 / 1 | 4.7 | Yes |
| 4944 | 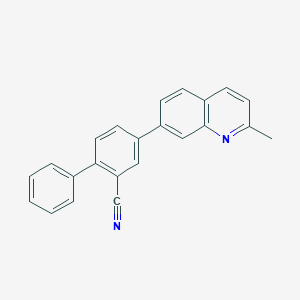 CHEMBL1099113 CHEMBL1099113 | C23H16N2 | 320.395 | 2 / 0 | 5.6 | No |
| 5563 | 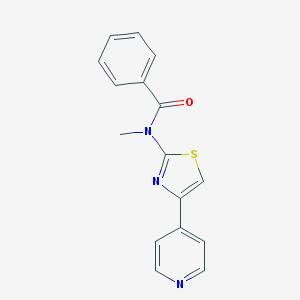 CHEMBL576121 CHEMBL576121 | C16H13N3OS | 295.36 | 4 / 0 | 2.9 | Yes |
| 521625 | 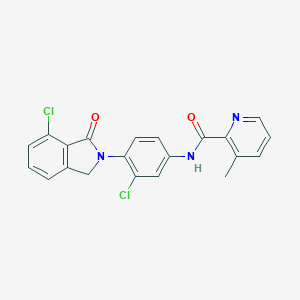 CHEMBL3787159 CHEMBL3787159 | C21H15Cl2N3O2 | 412.27 | 3 / 1 | 4.2 | Yes |
| 6310 | 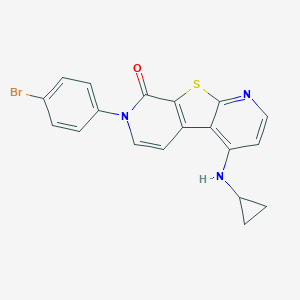 CHEMBL1951882 CHEMBL1951882 | C19H14BrN3OS | 412.305 | 4 / 1 | 4.9 | Yes |
| 441956 | 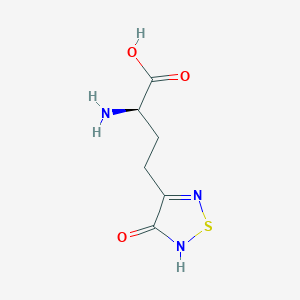 CHEMBL334842 CHEMBL334842 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 441962 | 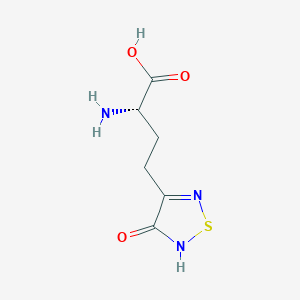 CHEMBL131922 CHEMBL131922 | C6H9N3O3S | 203.216 | 6 / 3 | -3.4 | Yes |
| 536135 | 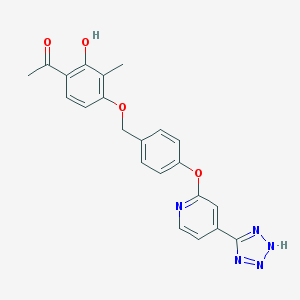 CHEMBL3899832 CHEMBL3899832 | C22H19N5O4 | 417.425 | 8 / 2 | 3.6 | Yes |
| 7201 | 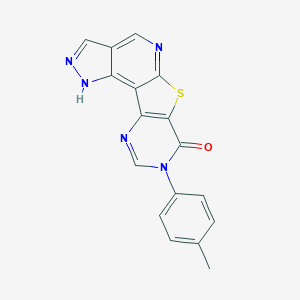 CHEMBL238262 CHEMBL238262 | C17H11N5OS | 333.369 | 5 / 1 | 3.1 | Yes |
| 7790 | 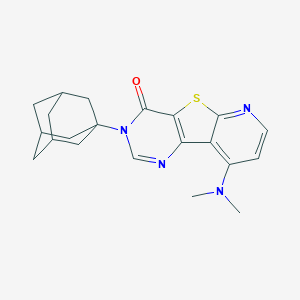 CHEMBL224322 CHEMBL224322 | C21H24N4OS | 380.51 | 5 / 0 | 4.3 | Yes |
| 7994 | 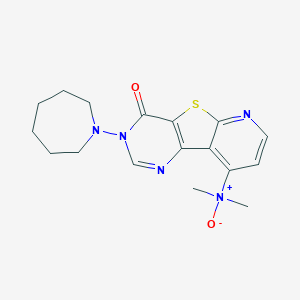 CHEMBL386408 CHEMBL386408 | C17H21N5O2S | 359.448 | 6 / 0 | 2.3 | Yes |
| 8269 | 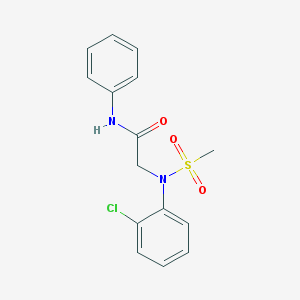 CHEMBL2208408 CHEMBL2208408 | C15H15ClN2O3S | 338.806 | 4 / 1 | 2.6 | Yes |
| 8693 | 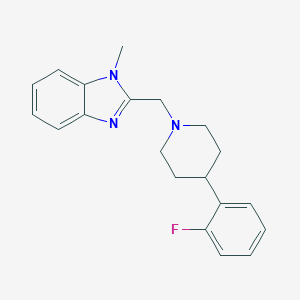 CHEMBL607689 CHEMBL607689 | C20H22FN3 | 323.415 | 3 / 0 | 3.6 | Yes |
| 463944 | 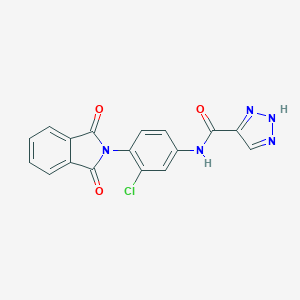 CHEMBL3628092 CHEMBL3628092 | C17H10ClN5O3 | 367.749 | 5 / 2 | 1.9 | Yes |
| 521719 | 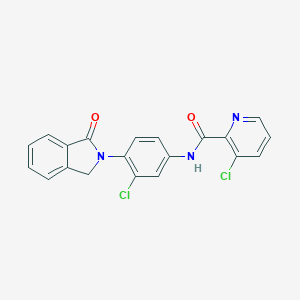 CHEMBL3786967 CHEMBL3786967 | C20H13Cl2N3O2 | 398.243 | 3 / 1 | 3.9 | Yes |
| 464128 | 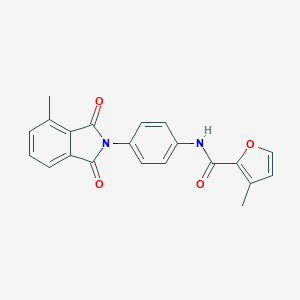 CHEMBL3634415 CHEMBL3634415 | C21H16N2O4 | 360.369 | 4 / 1 | 3.4 | Yes |
| 10406 | 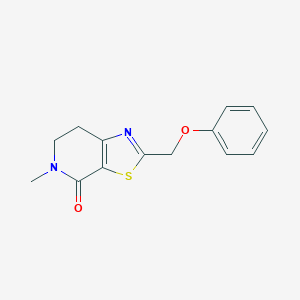 CHEMBL2426616 CHEMBL2426616 | C14H14N2O2S | 274.338 | 4 / 0 | 2.3 | Yes |
| 10815 | 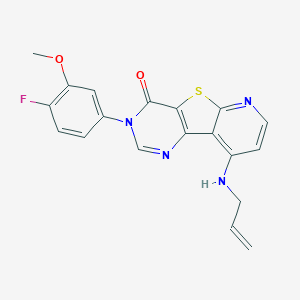 CHEMBL1783862 CHEMBL1783862 | C19H15FN4O2S | 382.413 | 7 / 1 | 3.8 | Yes |
| 11775 | 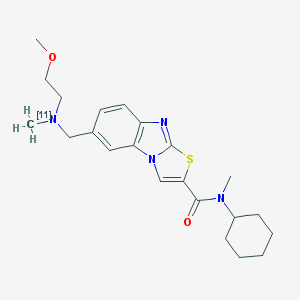 CHEMBL1645351 CHEMBL1645351 | C22H30N4O2S | 413.568 | 5 / 0 | 4.6 | Yes |
| 11776 | 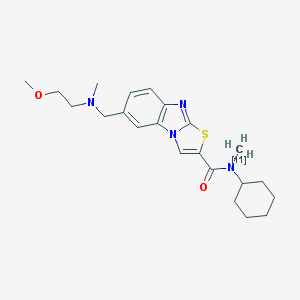 CHEMBL1771388 CHEMBL1771388 | C22H30N4O2S | 413.568 | 5 / 0 | 4.6 | Yes |
| 459323 | 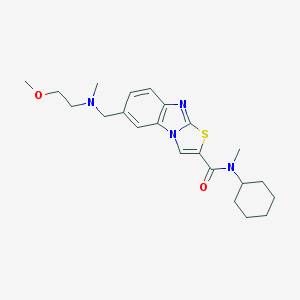 CHEMBL1645351 CHEMBL1645351 | C22H30N4O2S | 414.568 | 5 / 0 | 4.6 | Yes |
| 442162 | 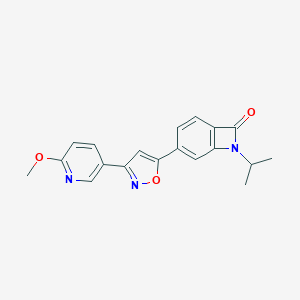 CHEMBL3398289 CHEMBL3398289 | C19H17N3O3 | 335.363 | 5 / 0 | 2.3 | Yes |
| 13315 | 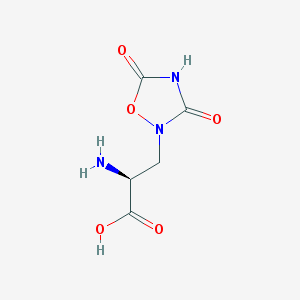 QUISQUALIC ACID QUISQUALIC ACID | C5H7N3O5 | 189.127 | 6 / 3 | -3.9 | Yes |
| 13572 | 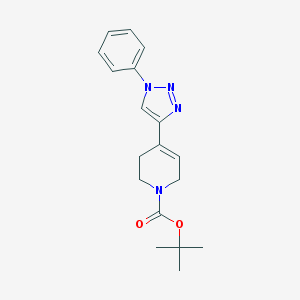 CHEMBL511355 CHEMBL511355 | C18H22N4O2 | 326.4 | 4 / 0 | 2.4 | Yes |
| 14197 | 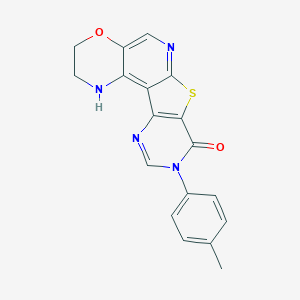 CHEMBL235975 CHEMBL235975 | C18H14N4O2S | 350.396 | 6 / 1 | 3.2 | Yes |
| 521895 | 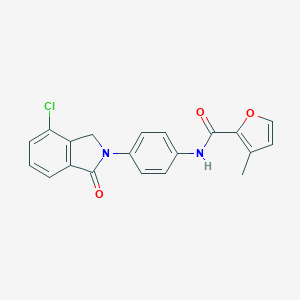 CHEMBL3786739 CHEMBL3786739 | C20H15ClN2O3 | 366.801 | 3 / 1 | 3.7 | Yes |
| 555542 | 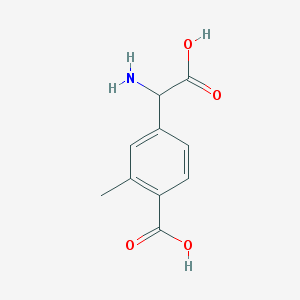 2Me4CPG 2Me4CPG | C10H11NO4 | 209.201 | 5 / 3 | -1.8 | Yes |
| 521901 | 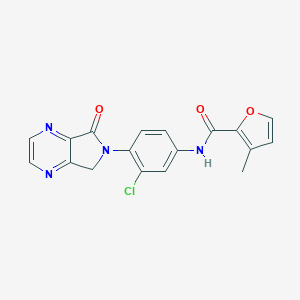 CHEMBL3786738 CHEMBL3786738 | C18H13ClN4O3 | 368.777 | 5 / 1 | 2.0 | Yes |
| 15217 | 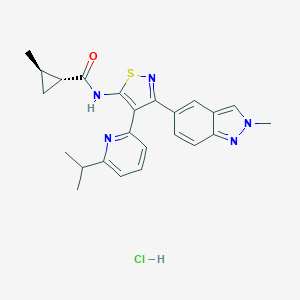 CHEMBL2334981 CHEMBL2334981 | C24H26ClN5OS | 468.016 | 5 / 2 | N/A | N/A |
| 15348 | 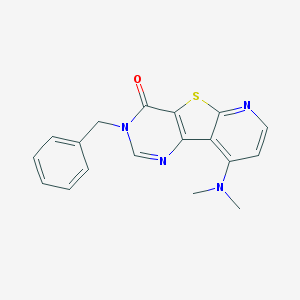 CHEMBL388087 CHEMBL388087 | C18H16N4OS | 336.413 | 5 / 0 | 3.1 | Yes |
| 442254 | 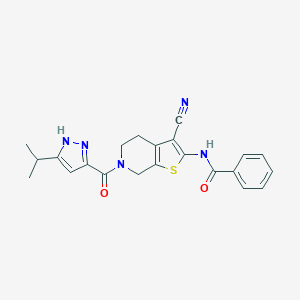 CHEMBL3422865 CHEMBL3422865 | C22H21N5O2S | 419.503 | 5 / 2 | 3.9 | Yes |
| 15575 | 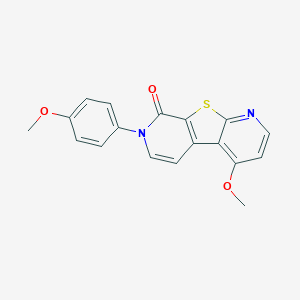 CHEMBL1951670 CHEMBL1951670 | C18H14N2O3S | 338.381 | 5 / 0 | 3.6 | Yes |
| 555543 | 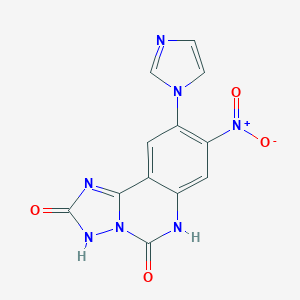 SCHEMBL6909945 SCHEMBL6909945 | C12H7N7O4 | 313.233 | 5 / 2 | -0.1 | Yes |
| 464842 | 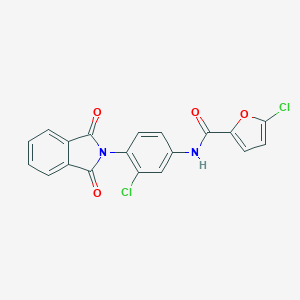 CHEMBL3628098 CHEMBL3628098 | C19H10Cl2N2O4 | 401.199 | 4 / 1 | 4.3 | Yes |
| 16006 | 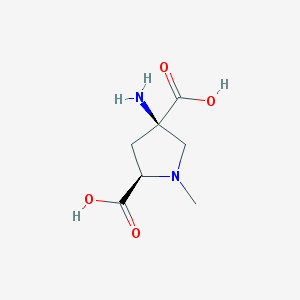 CHEMBL300330 CHEMBL300330 | C7H12N2O4 | 188.183 | 6 / 3 | -6.0 | Yes |
| 16228 | 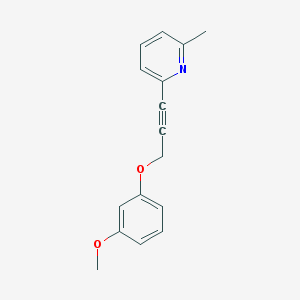 CHEMBL213102 CHEMBL213102 | C16H15NO2 | 253.301 | 3 / 0 | 3.1 | Yes |
| 16274 | 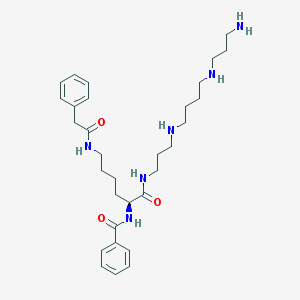 CHEMBL1269131 CHEMBL1269131 | C31H48N6O3 | 552.764 | 6 / 6 | 2.2 | No |
| 517410 | 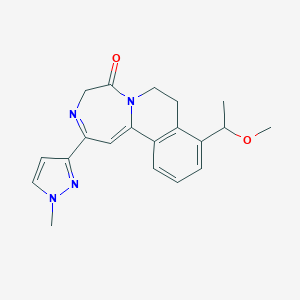 CHEMBL3644417 CHEMBL3644417 | C20H22N4O2 | 350.422 | 4 / 0 | 1.5 | Yes |
| 521974 | 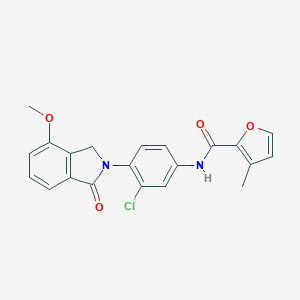 CHEMBL3787140 CHEMBL3787140 | C21H17ClN2O4 | 396.827 | 4 / 1 | 3.7 | Yes |
| 17048 | 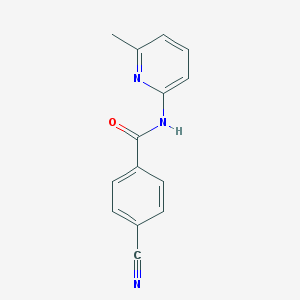 CHEMBL493571 CHEMBL493571 | C14H11N3O | 237.262 | 3 / 1 | 2.1 | Yes |
| 17226 | 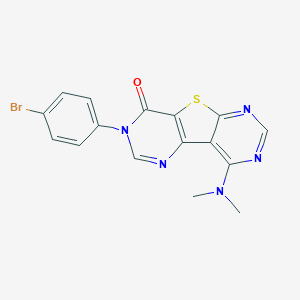 CHEMBL2205923 CHEMBL2205923 | C16H12BrN5OS | 402.27 | 6 / 0 | 3.6 | Yes |
| 442371 | 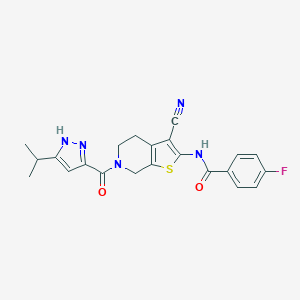 CHEMBL3422867 CHEMBL3422867 | C22H20FN5O2S | 437.493 | 6 / 2 | 4.0 | Yes |
| 522033 | 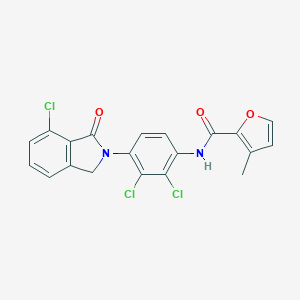 CHEMBL3787654 CHEMBL3787654 | C20H13Cl3N2O3 | 435.685 | 3 / 1 | 5.0 | Yes |
| 19478 | 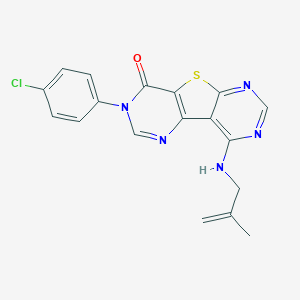 CHEMBL1784054 CHEMBL1784054 | C18H14ClN5OS | 383.854 | 6 / 1 | 4.6 | Yes |
| 442467 | 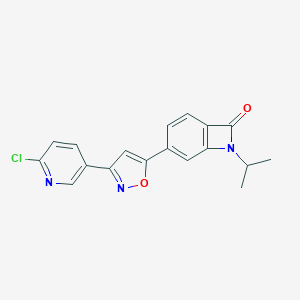 CHEMBL3398288 CHEMBL3398288 | C18H14ClN3O2 | 339.779 | 4 / 0 | 2.9 | Yes |
| 20309 | 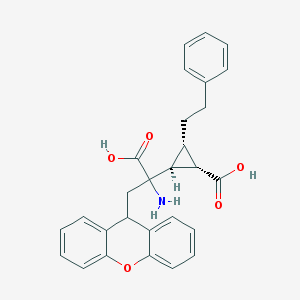 CHEMBL319279 CHEMBL319279 | C28H27NO5 | 457.526 | 6 / 3 | 2.1 | Yes |
| 465378 | 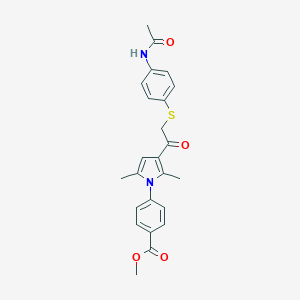 AC1N2AWG AC1N2AWG | C24H24N2O4S | 436.526 | 5 / 1 | 4.1 | Yes |
| 20859 | 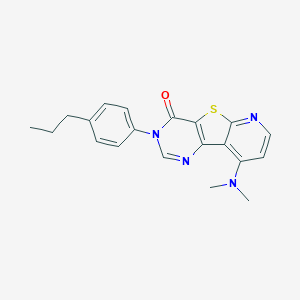 CHEMBL426018 CHEMBL426018 | C20H20N4OS | 364.467 | 5 / 0 | 4.5 | Yes |
| 20878 | 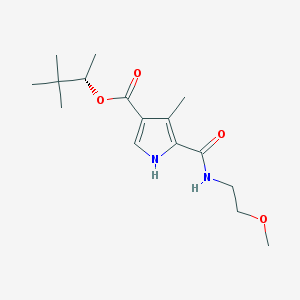 CHEMBL233708 CHEMBL233708 | C16H26N2O4 | 310.394 | 4 / 2 | 2.6 | Yes |
| 442500 | 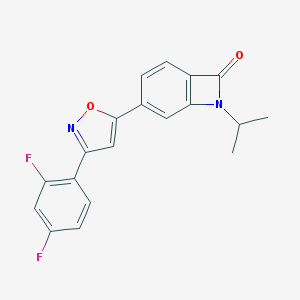 CHEMBL3398283 CHEMBL3398283 | C19H14F2N2O2 | 340.33 | 5 / 0 | 3.2 | Yes |
| 442518 | 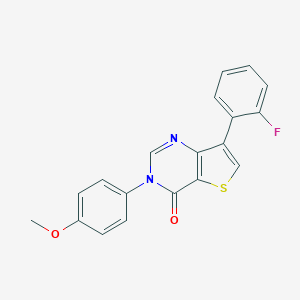 CHEMBL3330817 CHEMBL3330817 | C19H13FN2O2S | 352.383 | 5 / 0 | 4.2 | Yes |
| 23675 | 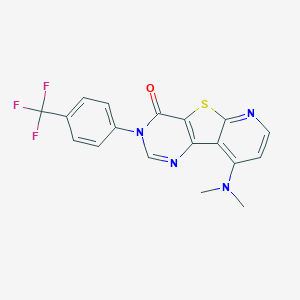 CHEMBL224375 CHEMBL224375 | C18H13F3N4OS | 390.384 | 8 / 0 | 4.1 | Yes |
| 23925 | 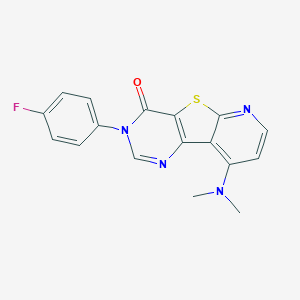 CHEMBL225124 CHEMBL225124 | C17H13FN4OS | 340.376 | 6 / 0 | 3.3 | Yes |
| 459427 |  CHEMBL3702371 CHEMBL3702371 | C22H22N2O3 | 362.429 | 4 / 0 | 3.2 | Yes |
| 442647 |  CHEMBL3422887 CHEMBL3422887 | C22H18FN3OS | 391.464 | 5 / 1 | 4.8 | Yes |
| 25091 |  CHEMBL523693 CHEMBL523693 | C20H12N2 | 280.33 | 2 / 0 | 4.3 | Yes |
| 25354 |  CHEMBL1093561 CHEMBL1093561 | C16H14N6OS | 338.389 | 7 / 2 | 2.7 | Yes |
| 459441 |  CHEMBL3702413 CHEMBL3702413 | C17H15IN4O2 | 434.237 | 4 / 1 | 0.8 | Yes |
| 26249 |  CHEMBL2205916 CHEMBL2205916 | C19H17N5O2S | 379.438 | 7 / 1 | 3.4 | Yes |
| 26270 |  CHEMBL1269126 CHEMBL1269126 | C31H49N5O2 | 523.766 | 5 / 5 | 4.5 | No |
| 522386 |  CHEMBL3785813 CHEMBL3785813 | C20H15ClN2O3 | 366.801 | 3 / 1 | 3.7 | Yes |
| 28317 | 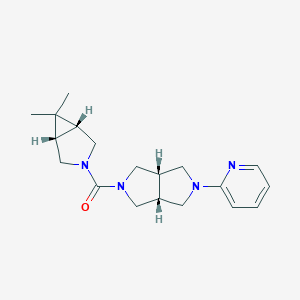 CHEMBL2418357 CHEMBL2418357 | C19H26N4O | 326.444 | 3 / 0 | 2.1 | Yes |
| 442839 | 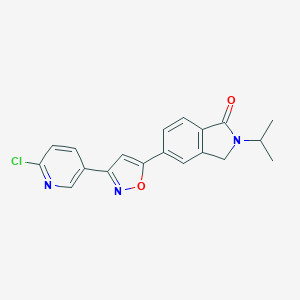 CHEMBL3398278 CHEMBL3398278 | C19H16ClN3O2 | 353.806 | 4 / 0 | 3.4 | Yes |
| 522460 | 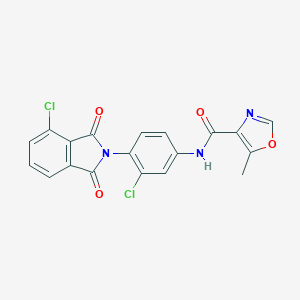 CHEMBL3759867 CHEMBL3759867 | C19H11Cl2N3O4 | 416.214 | 5 / 1 | 3.7 | Yes |
| 459483 | 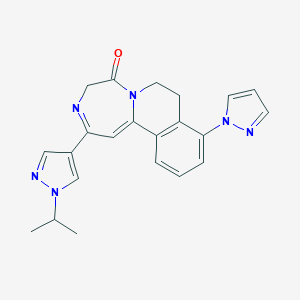 CHEMBL3702393 CHEMBL3702393 | C22H22N6O | 386.459 | 4 / 0 | 1.8 | Yes |
| 459489 | 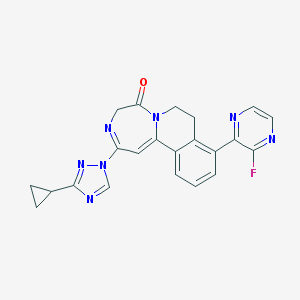 CHEMBL3702438 CHEMBL3702438 | C22H18FN7O | 415.432 | 7 / 0 | 1.6 | Yes |
| 31039 | 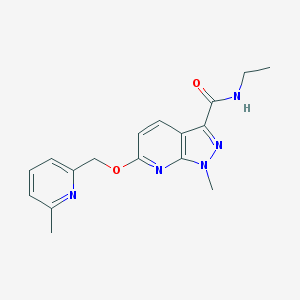 CHEMBL3122222 CHEMBL3122222 | C17H19N5O2 | 325.372 | 5 / 1 | 2.0 | Yes |
| 466734 | 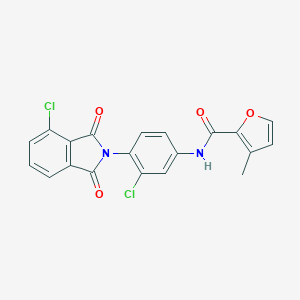 CHEMBL3628111 CHEMBL3628111 | C20H12Cl2N2O4 | 415.226 | 4 / 1 | 4.3 | Yes |
| 31623 | 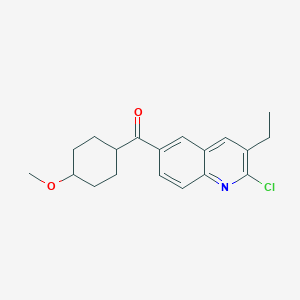 CHEMBL178592 CHEMBL178592 | C19H22ClNO2 | 331.84 | 3 / 0 | 4.7 | Yes |
| 31632 | 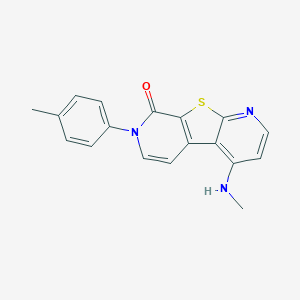 CHEMBL1951688 CHEMBL1951688 | C18H15N3OS | 321.398 | 4 / 1 | 4.0 | Yes |
| 32067 | 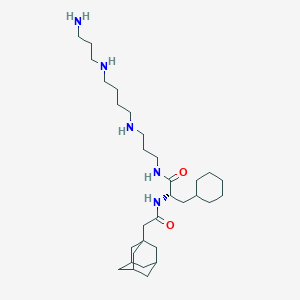 CHEMBL1269127 CHEMBL1269127 | C31H57N5O2 | 531.83 | 5 / 5 | 5.0 | No |
| 466897 | 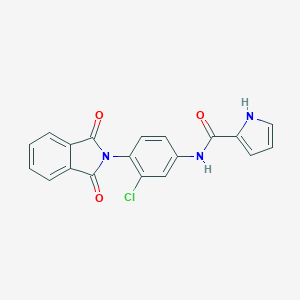 CHEMBL3628083 CHEMBL3628083 | C19H12ClN3O3 | 365.773 | 3 / 2 | 3.0 | Yes |
| 32526 | 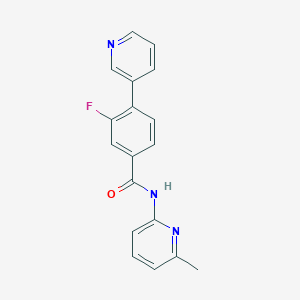 CHEMBL469121 CHEMBL469121 | C18H14FN3O | 307.328 | 4 / 1 | 3.1 | Yes |
| 32612 | 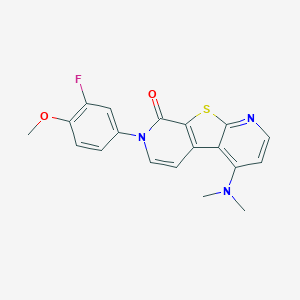 CHEMBL1951666 CHEMBL1951666 | C19H16FN3O2S | 369.414 | 6 / 0 | 3.9 | Yes |
| 522564 | 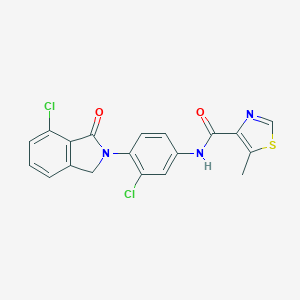 CHEMBL3787296 CHEMBL3787296 | C19H13Cl2N3O2S | 418.292 | 4 / 1 | 4.4 | Yes |
| 34615 | 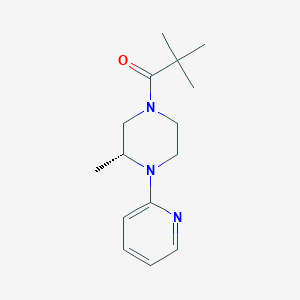 CHEMBL2397355 CHEMBL2397355 | C15H23N3O | 261.369 | 3 / 0 | 2.4 | Yes |
| 34832 | 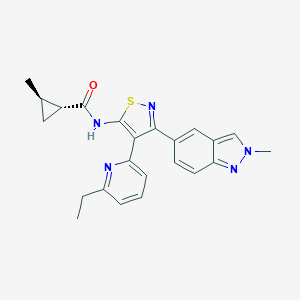 CHEMBL2334976 CHEMBL2334976 | C23H23N5OS | 417.531 | 5 / 1 | 4.1 | Yes |
| 522586 | 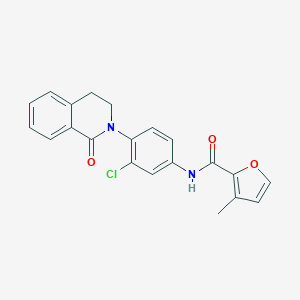 CHEMBL3785252 CHEMBL3785252 | C21H17ClN2O3 | 380.828 | 3 / 1 | 4.2 | Yes |
| 35425 | 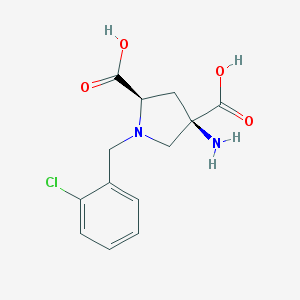 CHEMBL418462 CHEMBL418462 | C13H15ClN2O4 | 298.723 | 6 / 3 | -3.9 | Yes |
| 35693 | 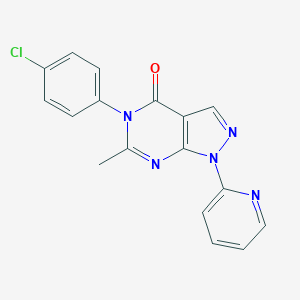 CHEMBL245991 CHEMBL245991 | C17H12ClN5O | 337.767 | 4 / 0 | 2.6 | Yes |
| 35748 | 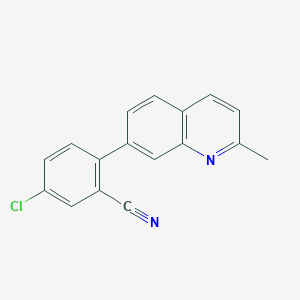 CHEMBL1099112 CHEMBL1099112 | C17H11ClN2 | 278.739 | 2 / 0 | 4.6 | Yes |
| 35792 | 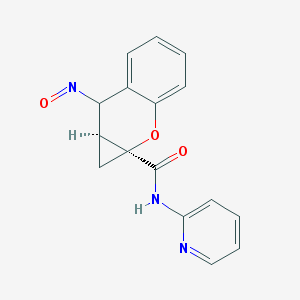 BDBM50388941 BDBM50388941 | C16H13N3O3 | 295.298 | 5 / 1 | 1.4 | Yes |
| 459540 | 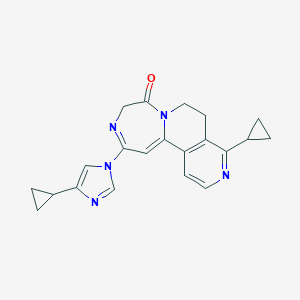 CHEMBL3644383 CHEMBL3644383 | C21H21N5O | 359.433 | 4 / 0 | 1.1 | Yes |
| 36254 | 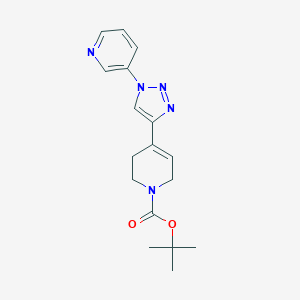 CHEMBL453249 CHEMBL453249 | C17H21N5O2 | 327.388 | 5 / 0 | 1.4 | Yes |
| 36477 | 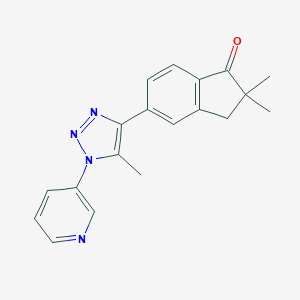 CHEMBL568451 CHEMBL568451 | C19H18N4O | 318.38 | 4 / 0 | 3.0 | Yes |
| 467496 | 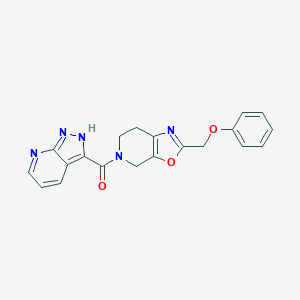 CHEMBL3605296 CHEMBL3605296 | C20H17N5O3 | 375.388 | 6 / 1 | 2.1 | Yes |
| 459554 | 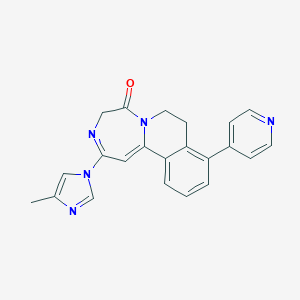 CHEMBL3702428 CHEMBL3702428 | C22H19N5O | 369.428 | 4 / 0 | 2.0 | Yes |
| 467723 | 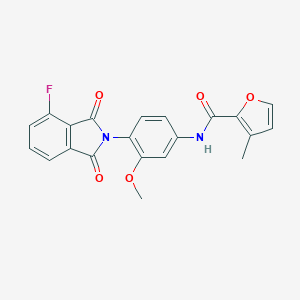 CHEMBL3634432 CHEMBL3634432 | C21H15FN2O5 | 394.358 | 6 / 1 | 3.1 | Yes |
| 39424 | 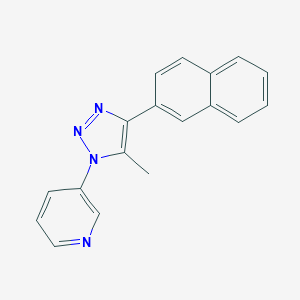 CHEMBL567584 CHEMBL567584 | C18H14N4 | 286.338 | 3 / 0 | 3.6 | Yes |
| 39962 | 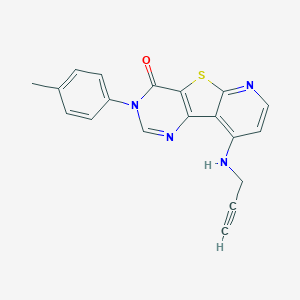 CHEMBL1783875 CHEMBL1783875 | C19H14N4OS | 346.408 | 5 / 1 | 3.5 | Yes |
| 467839 | 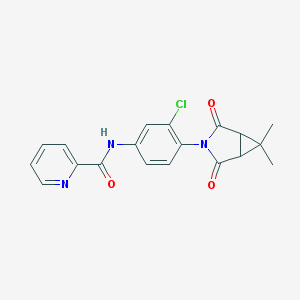 CHEMBL3628281 CHEMBL3628281 | C19H16ClN3O3 | 369.805 | 4 / 1 | 2.5 | Yes |
| 40177 | 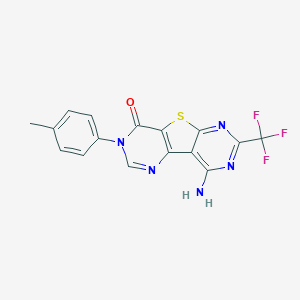 CHEMBL1093902 CHEMBL1093902 | C16H10F3N5OS | 377.345 | 9 / 1 | 3.3 | Yes |
| 40638 | 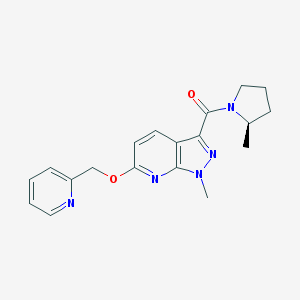 CHEMBL3122225 CHEMBL3122225 | C19H21N5O2 | 351.41 | 5 / 0 | 2.3 | Yes |
| 40653 | 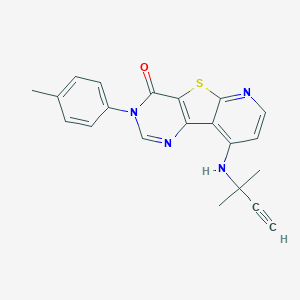 CHEMBL1784080 CHEMBL1784080 | C21H18N4OS | 374.462 | 5 / 1 | 4.1 | Yes |
| 40734 | 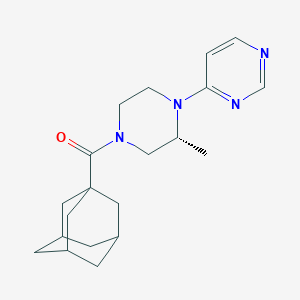 CHEMBL2397377 CHEMBL2397377 | C20H28N4O | 340.471 | 4 / 0 | 2.9 | Yes |
| 40955 | 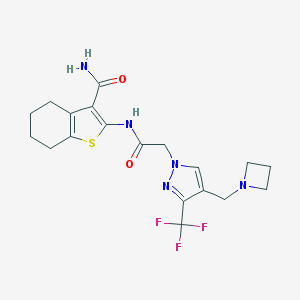 CHEMBL1257182 CHEMBL1257182 | C19H22F3N5O2S | 441.473 | 8 / 2 | 2.8 | Yes |
| 41085 | 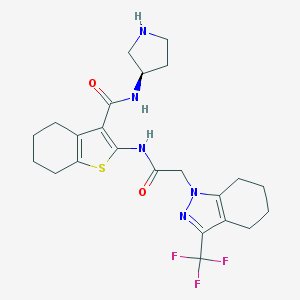 CHEMBL1234889 CHEMBL1234889 | C23H28F3N5O2S | 495.565 | 8 / 3 | 4.2 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417