

You can:
| Name | C-C chemokine receptor type 3 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Ccr3 |
| Synonym | CKR3 chemokine (C-C motif) receptor 3 CD193 CCR3 CCR-3 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 359 |
| Amino acid sequence | MASNEEELKTVVETFETTPYEYEWAPPCEKVSIRELGSWLLPPLYSLVFIVGLLGNMMVVLILIKYRKLQIMTNIYLLNLAISDLLFLFTVPFWIHYVLWNEWGFGHCMCKMLSGLYYLALYSEIFFIILLTIDRYLAIVHAVLALRARTVTFATITSIITWGFAVLAALPEFIFHESQDNFGDLSCSPRYPEGEEDSWKRFHALRMNIFGLALPLLIMVICYSGIIKTLLRCPNKKKHKAIQLIFVVMIVFFIFWTPYNLVLLLSAFHSTFLETSCQQSIHLDLAMQVTEVITHTHCCINPIIYAFVGERFRKHLRLFFHRNVAIYLRKYISFLPGEKLERTSSVSPSTGEQEISVVF |
| UniProt | O54814 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3928 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 26129 | 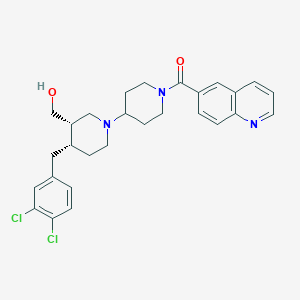 CHEMBL178447 CHEMBL178447 | C28H31Cl2N3O2 | 512.475 | 4 / 1 | 5.4 | No |
| 443624 | 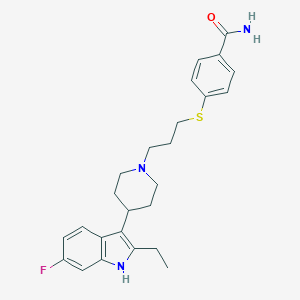 CHEMBL3355937 CHEMBL3355937 | C25H30FN3OS | 439.593 | 4 / 2 | 5.0 | Yes |
| 443639 | 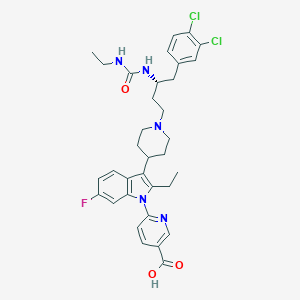 CHEMBL3355965 CHEMBL3355965 | C34H38Cl2FN5O3 | 654.608 | 6 / 3 | 4.8 | No |
| 444379 | 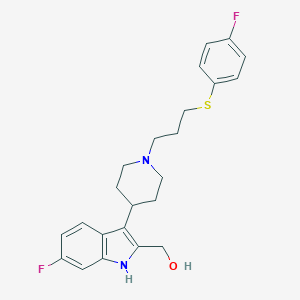 CHEMBL3355939 CHEMBL3355939 | C23H26F2N2OS | 416.531 | 5 / 2 | 4.5 | Yes |
| 113228 | 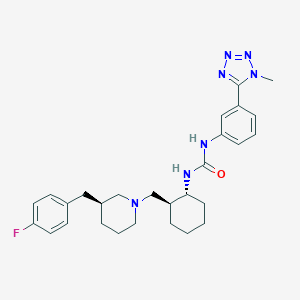 CHEMBL363840 CHEMBL363840 | C28H36FN7O | 505.642 | 6 / 2 | 4.8 | No |
| 446571 | 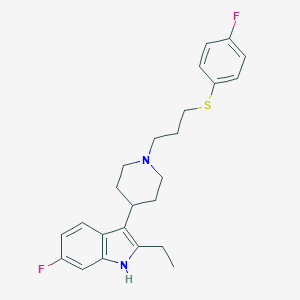 CHEMBL3355932 CHEMBL3355932 | C24H28F2N2S | 414.559 | 4 / 1 | 6.2 | No |
| 447204 | 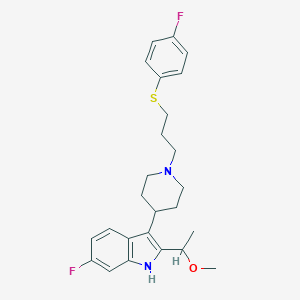 CHEMBL3355940 CHEMBL3355940 | C25H30F2N2OS | 444.585 | 5 / 1 | 5.5 | No |
| 447312 | 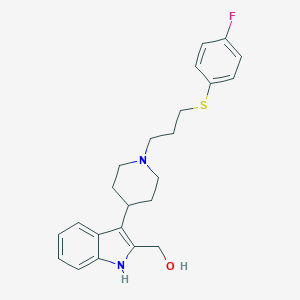 CHEMBL3355943 CHEMBL3355943 | C23H27FN2OS | 398.54 | 4 / 2 | 4.4 | Yes |
| 447512 |  CHEMBL3355933 CHEMBL3355933 | C24H29FN2S | 396.568 | 3 / 1 | 6.1 | No |
| 447917 |  CHEMBL3355958 CHEMBL3355958 | C33H36F2N2O2S | 562.72 | 6 / 0 | 8.1 | No |
| 448757 |  CHEMBL3355964 CHEMBL3355964 | C28H35Cl2FN4O | 533.513 | 3 / 3 | 6.7 | No |
| 448831 |  CHEMBL3355944 CHEMBL3355944 | C25H31FN2OS | 426.594 | 4 / 1 | 5.4 | No |
| 449031 | 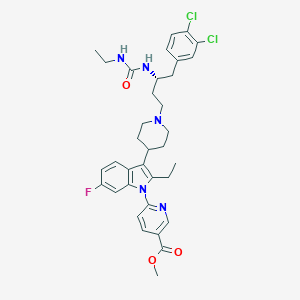 CHEMBL3355966 CHEMBL3355966 | C35H40Cl2FN5O3 | 668.635 | 6 / 2 | 7.4 | No |
| 449317 | 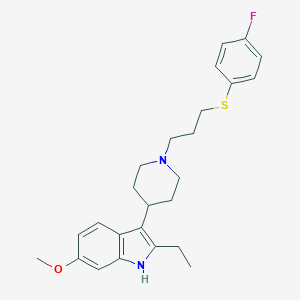 CHEMBL3355947 CHEMBL3355947 | C25H31FN2OS | 426.594 | 4 / 1 | 6.1 | No |
| 449725 | 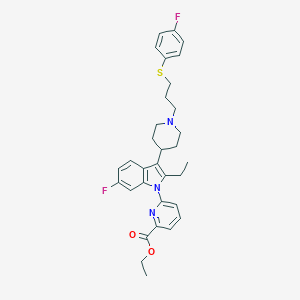 CHEMBL3355961 CHEMBL3355961 | C32H35F2N3O2S | 563.708 | 7 / 0 | 7.7 | No |
| 450005 | 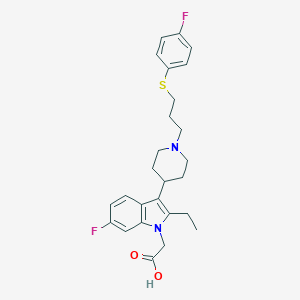 CHEMBL3355952 CHEMBL3355952 | C26H30F2N2O2S | 472.595 | 6 / 1 | 3.6 | Yes |
| 221020 | 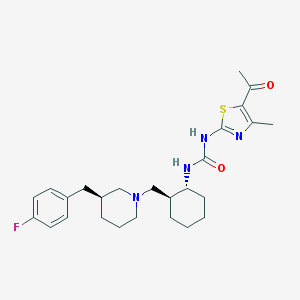 CHEMBL195433 CHEMBL195433 | C26H35FN4O2S | 486.65 | 6 / 2 | 5.3 | No |
| 451613 | 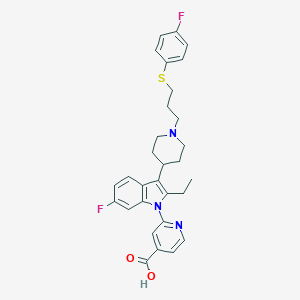 CHEMBL3355962 CHEMBL3355962 | C30H31F2N3O2S | 535.654 | 7 / 1 | 4.3 | No |
| 451623 | 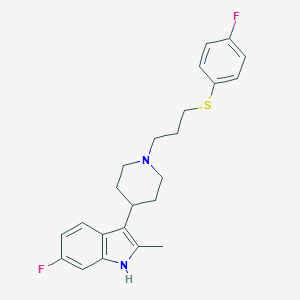 CHEMBL3355938 CHEMBL3355938 | C23H26F2N2S | 400.532 | 4 / 1 | 5.8 | No |
| 451845 | 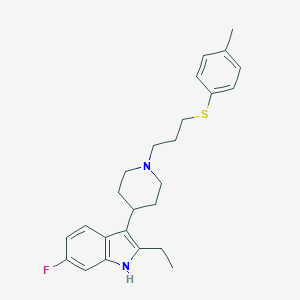 CHEMBL3355934 CHEMBL3355934 | C25H31FN2S | 410.595 | 3 / 1 | 6.5 | No |
| 452975 | 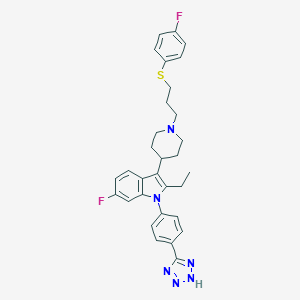 CHEMBL3355954 CHEMBL3355954 | C31H32F2N6S | 558.696 | 7 / 1 | 7.0 | No |
| 453030 | 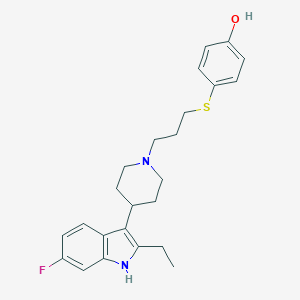 CHEMBL3355935 CHEMBL3355935 | C24H29FN2OS | 412.567 | 4 / 2 | 5.8 | No |
| 453097 | 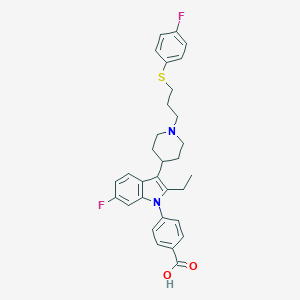 CHEMBL3355953 CHEMBL3355953 | C31H32F2N2O2S | 534.666 | 6 / 1 | 5.1 | No |
| 453534 | 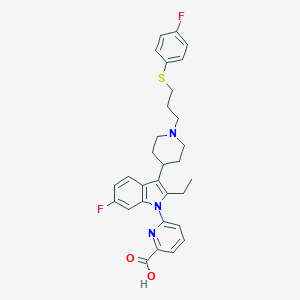 CHEMBL3355960 CHEMBL3355960 | C30H31F2N3O2S | 535.654 | 7 / 1 | 4.7 | No |
| 453780 | 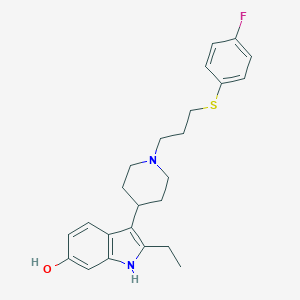 CHEMBL3355946 CHEMBL3355946 | C24H29FN2OS | 412.567 | 4 / 2 | 5.8 | No |
| 454751 | 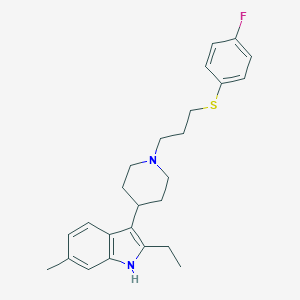 CHEMBL3355945 CHEMBL3355945 | C25H31FN2S | 410.595 | 3 / 1 | 6.5 | No |
| 455802 | 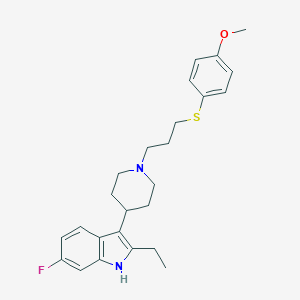 CHEMBL3355936 CHEMBL3355936 | C25H31FN2OS | 426.594 | 4 / 1 | 6.1 | No |
| 455867 | 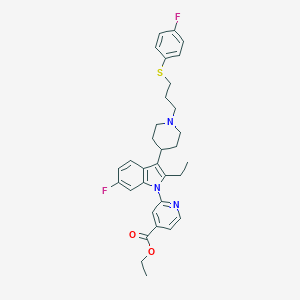 CHEMBL3355963 CHEMBL3355963 | C32H35F2N3O2S | 563.708 | 7 / 0 | 7.3 | No |
| 456388 | 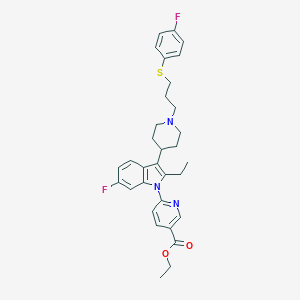 CHEMBL3355959 CHEMBL3355959 | C32H35F2N3O2S | 563.708 | 7 / 0 | 7.3 | No |
| 458103 | 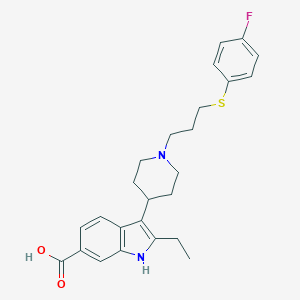 CHEMBL3355949 CHEMBL3355949 | C25H29FN2O2S | 440.577 | 5 / 2 | 3.4 | Yes |
| 458324 | 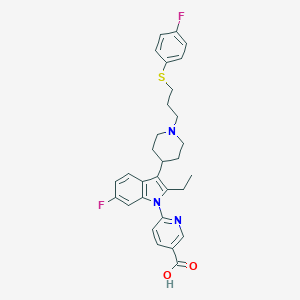 CHEMBL3355956 CHEMBL3355956 | C30H31F2N3O2S | 535.654 | 7 / 1 | 4.3 | No |
| 458388 | 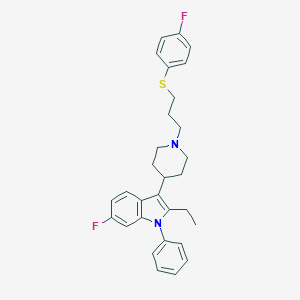 CHEMBL3355951 CHEMBL3355951 | C30H32F2N2S | 490.657 | 4 / 0 | 7.8 | No |
| 458570 | 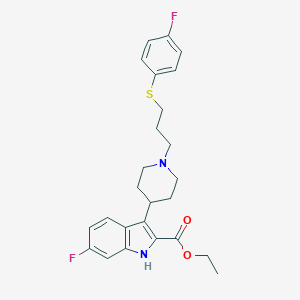 CHEMBL3355941 CHEMBL3355941 | C25H28F2N2O2S | 458.568 | 6 / 1 | 5.9 | No |
| 458955 | 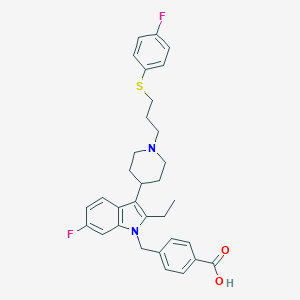 CHEMBL3355955 CHEMBL3355955 | C32H34F2N2O2S | 548.693 | 6 / 1 | 5.0 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417