

You can:
| Name | Type-1B angiotensin II receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Agtr1b |
| Synonym | Angiotensin II type-1B receptor AT1B AT3 |
| Disease | N/A for non-human GPCRs |
| Length | 359 |
| Amino acid sequence | MTLNSSTEDGIKRIQDDCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNHLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLMAGLASLPAVIYRNVYFIENTNITVCAFHYESQNSTLPIGLGLTKNILGFVFPFLIILTSYTLIWKALKKAYKIQKNTPRNDDIFRIIMAIVLFFFFSWVPHQIFTFLDVLIQLGIIRDCEIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKKYFLQLLKYIPPTAKSHAGLSTKMSTLSYRPSDNMSSSAKKSASFFEVE |
| UniProt | P29089 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL263 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 460 | 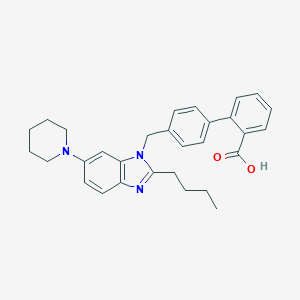 CHEMBL434815 CHEMBL434815 | C30H33N3O2 | 467.613 | 4 / 1 | 6.9 | No |
| 1149 | 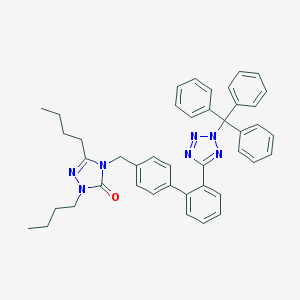 CHEMBL89246 CHEMBL89246 | C43H43N7O | 673.865 | 5 / 0 | 9.9 | No |
| 1292 | 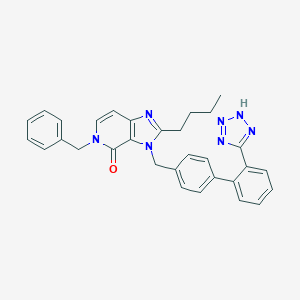 CHEMBL42397 CHEMBL42397 | C31H29N7O | 515.621 | 5 / 1 | 5.6 | No |
| 1987 | 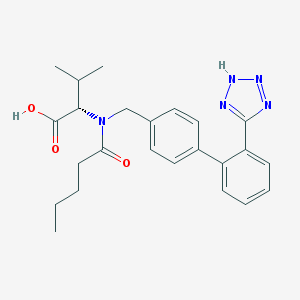 valsartan valsartan | C24H29N5O3 | 435.528 | 6 / 2 | 4.4 | Yes |
| 3705 | 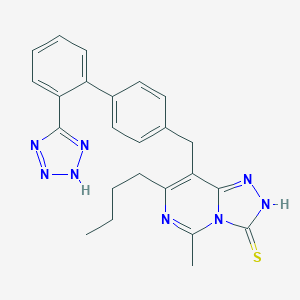 CHEMBL78532 CHEMBL78532 | C24H24N8S | 456.572 | 6 / 2 | 4.2 | Yes |
| 5903 | 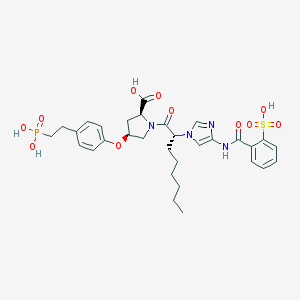 CHEMBL343723 CHEMBL343723 | C31H39N4O11PS | 706.704 | 12 / 5 | 2.4 | No |
| 6032 | 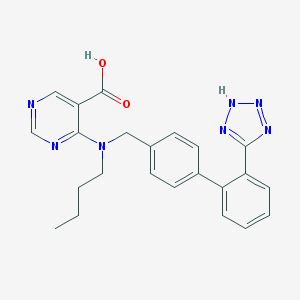 141872-46-0 141872-46-0 | C23H23N7O2 | 429.484 | 8 / 2 | 3.8 | Yes |
| 7337 | 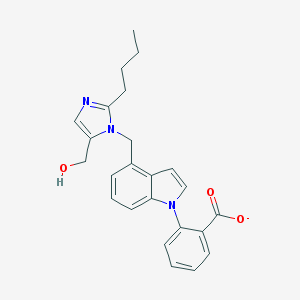 CHEMBL288691 CHEMBL288691 | C24H24N3O3- | 402.474 | 4 / 1 | 4.4 | Yes |
| 7776 | 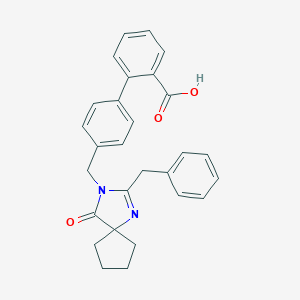 CHEMBL115771 CHEMBL115771 | C28H26N2O3 | 438.527 | 4 / 1 | 4.7 | Yes |
| 8123 | 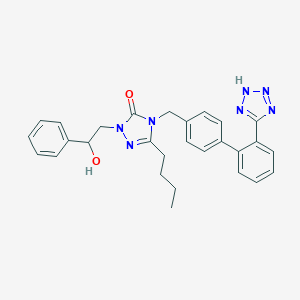 CHEMBL304329 CHEMBL304329 | C28H29N7O2 | 495.587 | 6 / 2 | 4.1 | Yes |
| 9655 | 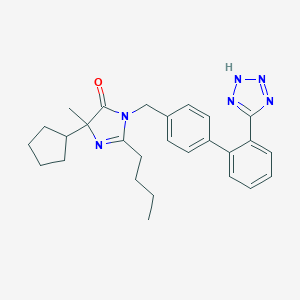 CHEMBL47985 CHEMBL47985 | C27H32N6O | 456.594 | 5 / 1 | 5.1 | No |
| 9851 | 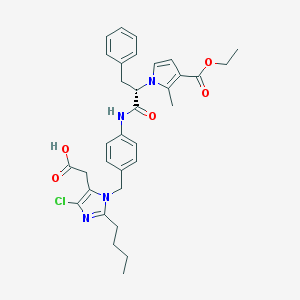 CHEMBL45659 CHEMBL45659 | C33H37ClN4O5 | 605.132 | 6 / 2 | 6.2 | No |
| 13488 | 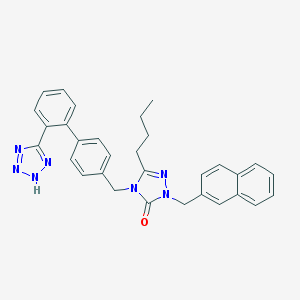 CHEMBL86986 CHEMBL86986 | C31H29N7O | 515.621 | 5 / 1 | 6.0 | No |
| 15788 | 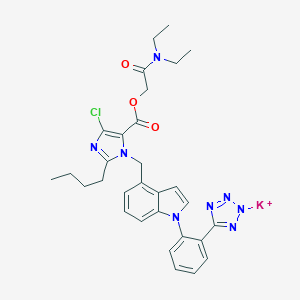 CHEMBL2021415 CHEMBL2021415 | C30H32ClKN8O3 | 627.187 | 8 / 0 | N/A | No |
| 16632 | 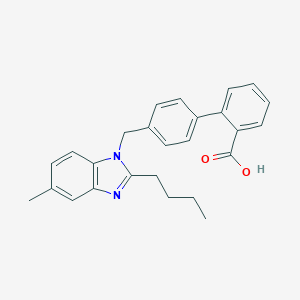 CHEMBL131722 CHEMBL131722 | C26H26N2O2 | 398.506 | 3 / 1 | 6.3 | No |
| 17487 | 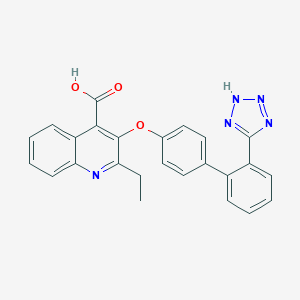 CHEMBL57907 CHEMBL57907 | C25H19N5O3 | 437.459 | 7 / 2 | 4.9 | Yes |
| 19527 |  CHEMBL86437 CHEMBL86437 | C27H24N4O2 | 436.515 | 5 / 1 | 5.3 | No |
| 19809 |  CHEMBL275437 CHEMBL275437 | C29H36N2O3 | 460.618 | 4 / 1 | 6.5 | No |
| 19827 |  CHEMBL113330 CHEMBL113330 | C27H28F3N3O4 | 515.533 | 8 / 1 | 4.4 | No |
| 20699 |  CHEMBL325201 CHEMBL325201 | C27H32N2O3 | 432.564 | 4 / 1 | 5.5 | No |
| 21107 | 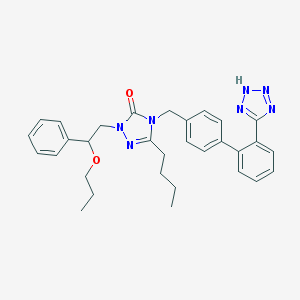 CHEMBL308239 CHEMBL308239 | C31H35N7O2 | 537.668 | 6 / 1 | 5.5 | No |
| 21647 | 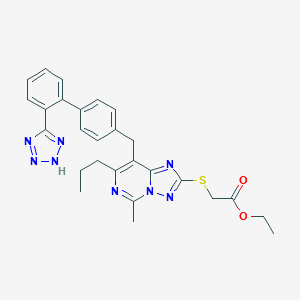 CHEMBL80161 CHEMBL80161 | C27H28N8O2S | 528.635 | 9 / 1 | 5.6 | No |
| 22758 | 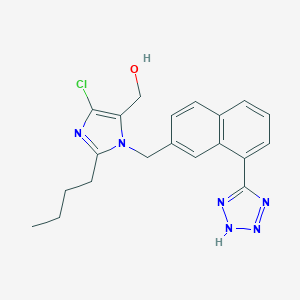 CHEMBL323179 CHEMBL323179 | C20H21ClN6O | 396.879 | 5 / 2 | 3.9 | Yes |
| 25192 | 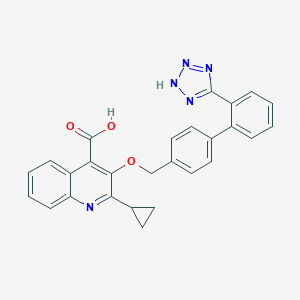 BDBM50049201 BDBM50049201 | C27H21N5O3 | 463.497 | 7 / 2 | 4.6 | Yes |
| 26751 | 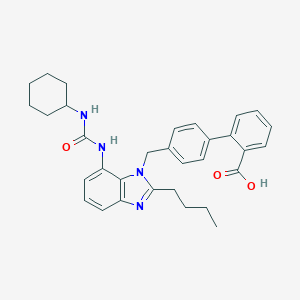 CHEMBL131209 CHEMBL131209 | C32H36N4O3 | 524.665 | 4 / 3 | 6.7 | No |
| 27724 | 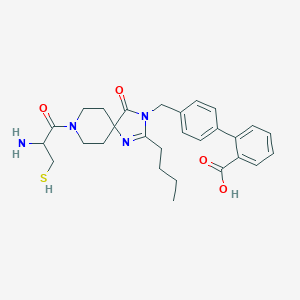 CHEMBL114368 CHEMBL114368 | C28H34N4O4S | 522.664 | 7 / 3 | 0.4 | No |
| 29817 | 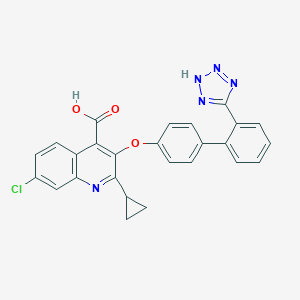 CHEMBL64567 CHEMBL64567 | C26H18ClN5O3 | 483.912 | 7 / 2 | 5.3 | No |
| 30655 | 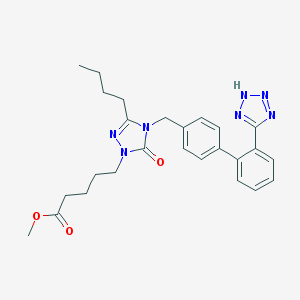 CHEMBL66173 CHEMBL66173 | C26H31N7O3 | 489.58 | 7 / 1 | 3.8 | Yes |
| 31132 | 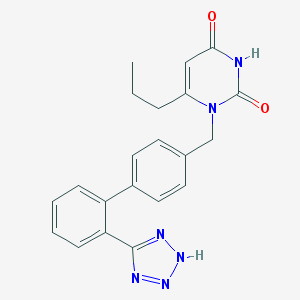 CHEMBL304032 CHEMBL304032 | C21H20N6O2 | 388.431 | 5 / 2 | 2.7 | Yes |
| 31328 | 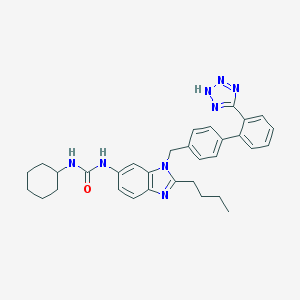 CHEMBL337025 CHEMBL337025 | C32H36N8O | 548.695 | 5 / 3 | 6.4 | No |
| 32787 | 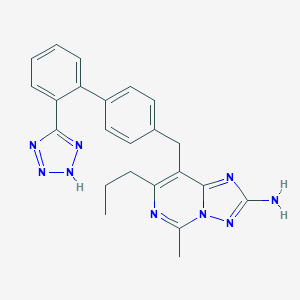 CHEMBL77959 CHEMBL77959 | C23H23N9 | 425.5 | 7 / 2 | 4.1 | Yes |
| 35043 | 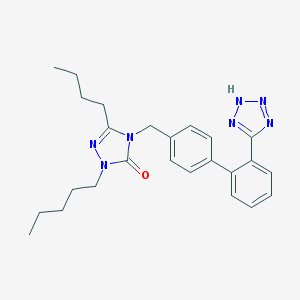 CHEMBL70029 CHEMBL70029 | C25H31N7O | 445.571 | 5 / 1 | 5.1 | No |
| 36323 | 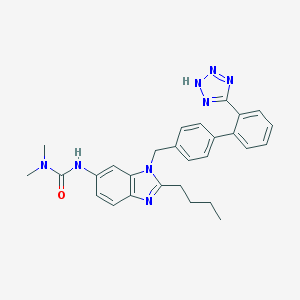 CHEMBL130746 CHEMBL130746 | C28H30N8O | 494.603 | 5 / 2 | 4.8 | Yes |
| 37444 | 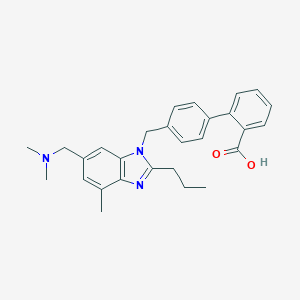 CHEMBL130322 CHEMBL130322 | C28H31N3O2 | 441.575 | 4 / 1 | 3.3 | Yes |
| 37635 | 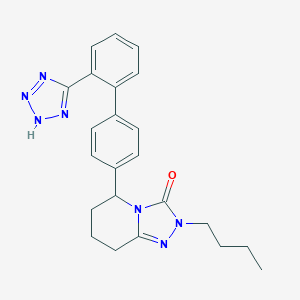 CHEMBL314799 CHEMBL314799 | C23H25N7O | 415.501 | 5 / 1 | 3.6 | Yes |
| 38200 | 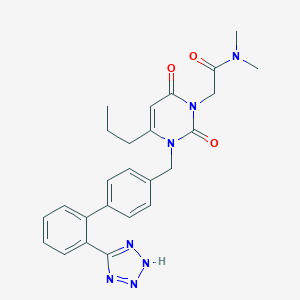 CHEMBL62269 CHEMBL62269 | C25H27N7O3 | 473.537 | 6 / 1 | 2.4 | Yes |
| 39484 | 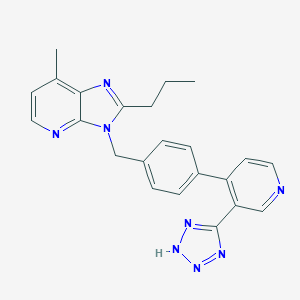 CHEMBL47382 CHEMBL47382 | C23H22N8 | 410.485 | 6 / 1 | 3.6 | Yes |
| 40708 | 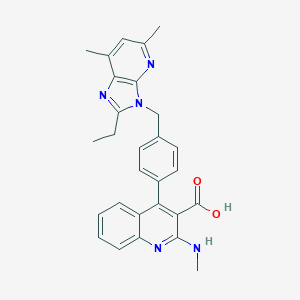 CHEMBL86562 CHEMBL86562 | C28H27N5O2 | 465.557 | 6 / 2 | 6.2 | No |
| 40782 | 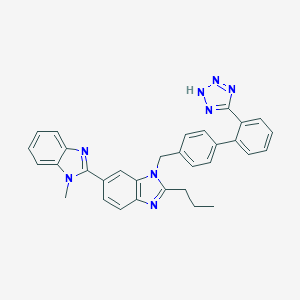 CHEMBL337232 CHEMBL337232 | C32H28N8 | 524.632 | 5 / 1 | 6.2 | No |
| 41905 | 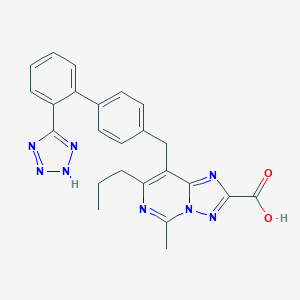 CHEMBL419835 CHEMBL419835 | C24H22N8O2 | 454.494 | 8 / 2 | 4.3 | Yes |
| 43207 | 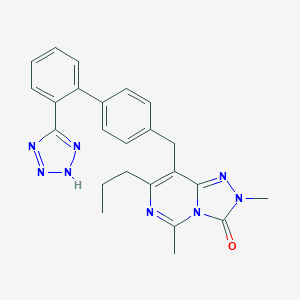 CHEMBL76421 CHEMBL76421 | C24H24N8O | 440.511 | 6 / 1 | 3.2 | Yes |
| 43476 | 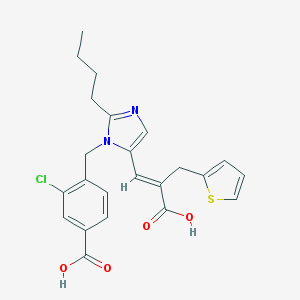 CHEMBL37679 CHEMBL37679 | C23H23ClN2O4S | 458.957 | 6 / 2 | 5.1 | No |
| 43605 | 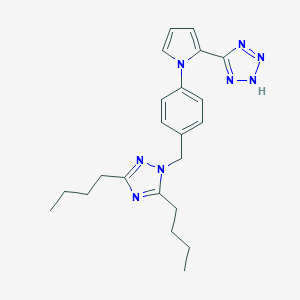 CHEMBL12810 CHEMBL12810 | C22H28N8 | 404.522 | 5 / 1 | 4.7 | Yes |
| 44745 | 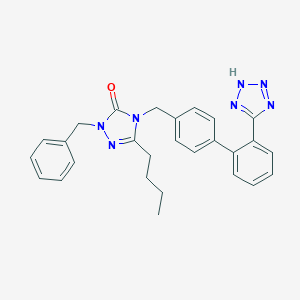 CHEMBL422593 CHEMBL422593 | C27H27N7O | 465.561 | 5 / 1 | 4.8 | Yes |
| 45167 | 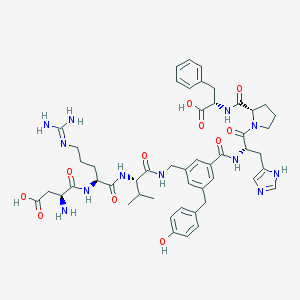 CHEMBL428986 CHEMBL428986 | C50H64N12O11 | 1009.14 | 14 / 12 | -1.4 | No |
| 46102 | 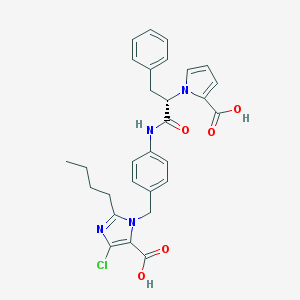 CHEMBL48195 CHEMBL48195 | C29H29ClN4O5 | 549.024 | 6 / 3 | 5.8 | No |
| 47567 | 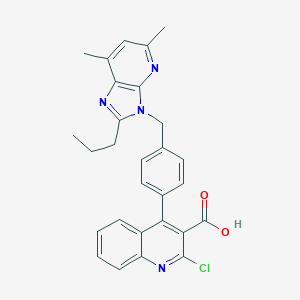 CHEMBL314315 CHEMBL314315 | C28H25ClN4O2 | 484.984 | 5 / 1 | 6.6 | No |
| 48664 | 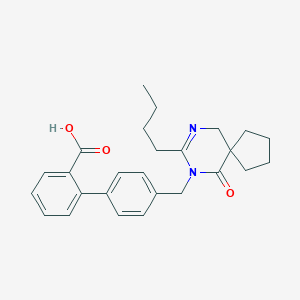 CHEMBL295646 CHEMBL295646 | C26H30N2O3 | 418.537 | 4 / 1 | 4.6 | Yes |
| 48859 | 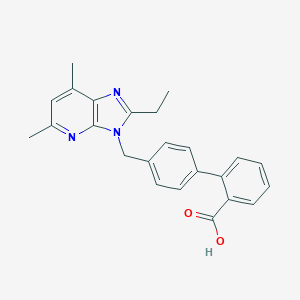 CHEMBL87842 CHEMBL87842 | C24H23N3O2 | 385.467 | 4 / 1 | 5.0 | Yes |
| 49025 | 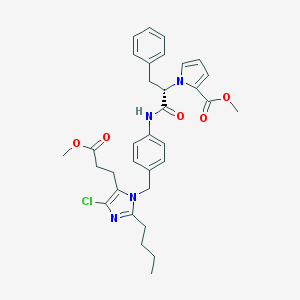 CHEMBL295311 CHEMBL295311 | C33H37ClN4O5 | 605.132 | 6 / 1 | 6.3 | No |
| 49658 | 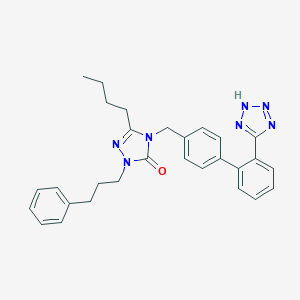 CHEMBL313222 CHEMBL313222 | C29H31N7O | 493.615 | 5 / 1 | 5.6 | No |
| 50672 | 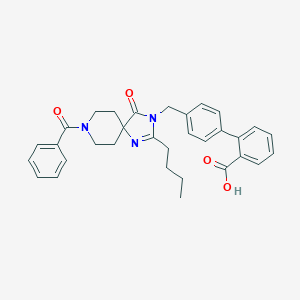 CHEMBL114720 CHEMBL114720 | C32H33N3O4 | 523.633 | 5 / 1 | 4.9 | No |
| 51040 | 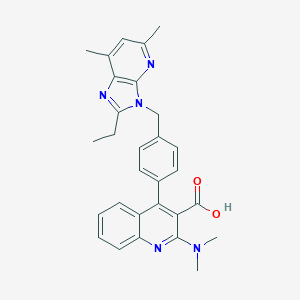 CHEMBL86436 CHEMBL86436 | C29H29N5O2 | 479.584 | 6 / 1 | 5.8 | No |
| 52180 | 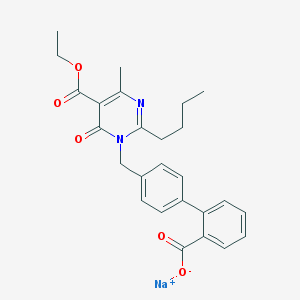 CHEMBL145276 CHEMBL145276 | C26H27N2NaO5 | 470.501 | 6 / 0 | N/A | N/A |
| 52642 | 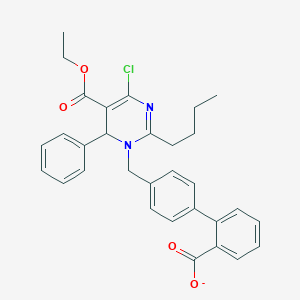 CHEMBL343622 CHEMBL343622 | C31H30ClN2O4- | 530.041 | 5 / 0 | 6.9 | No |
| 53950 | 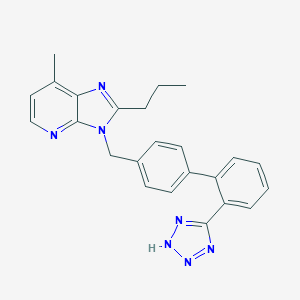 L-158338 L-158338 | C24H23N7 | 409.497 | 5 / 1 | 4.7 | Yes |
| 54281 | 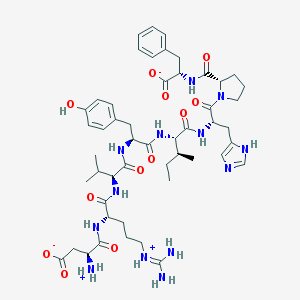 CHEBI:58506 CHEBI:58506 | C50H71N13O12 | 1046.2 | 13 / 12 | -0.4 | No |
| 56984 | 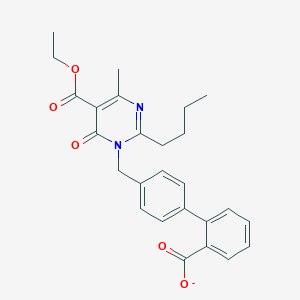 CHEMBL145276 CHEMBL145276 | C26H27N2O5- | 447.511 | 6 / 0 | 4.9 | Yes |
| 57026 | 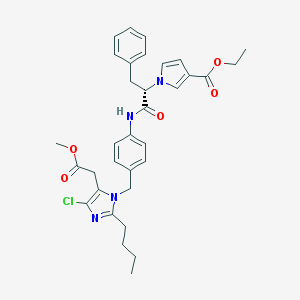 CHEMBL301147 CHEMBL301147 | C33H37ClN4O5 | 605.132 | 6 / 1 | 6.1 | No |
| 58003 | 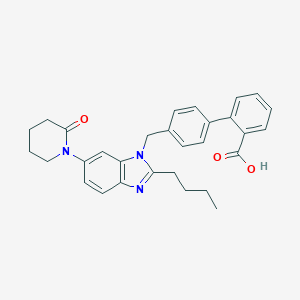 CHEMBL434640 CHEMBL434640 | C30H31N3O3 | 481.596 | 4 / 1 | 5.7 | No |
| 59338 | 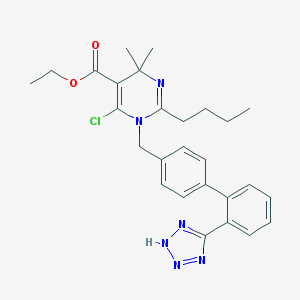 CHEMBL145564 CHEMBL145564 | C27H31ClN6O2 | 507.035 | 6 / 1 | 4.9 | No |
| 60813 | 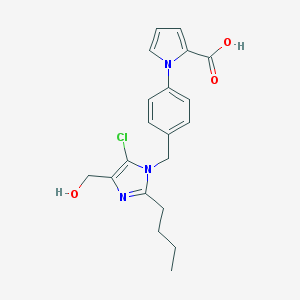 CHEMBL267547 CHEMBL267547 | C20H22ClN3O3 | 387.864 | 4 / 2 | 3.7 | Yes |
| 61055 | 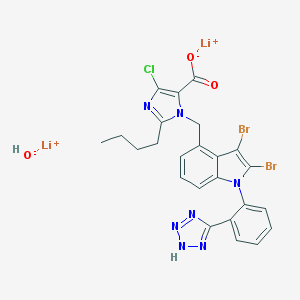 CHEMBL2021418 CHEMBL2021418 | C24H20Br2ClLi2N7O3 | 663.608 | 7 / 2 | N/A | No |
| 61740 | 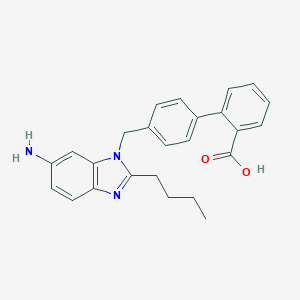 CHEMBL130828 CHEMBL130828 | C25H25N3O2 | 399.494 | 4 / 2 | 5.2 | No |
| 63112 | 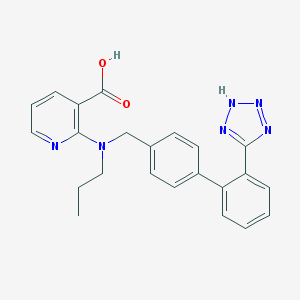 Abbott-81988 Abbott-81988 | C23H22N6O2 | 414.469 | 7 / 2 | 4.1 | Yes |
| 63140 | 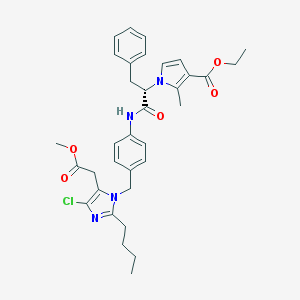 CHEMBL297265 CHEMBL297265 | C34H39ClN4O5 | 619.159 | 6 / 1 | 6.5 | No |
| 67892 | 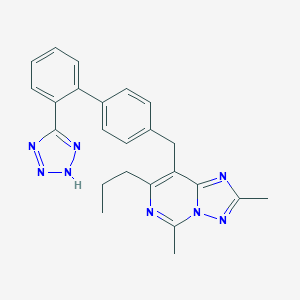 CHEMBL79171 CHEMBL79171 | C24H24N8 | 424.512 | 6 / 1 | 4.8 | Yes |
| 69379 | 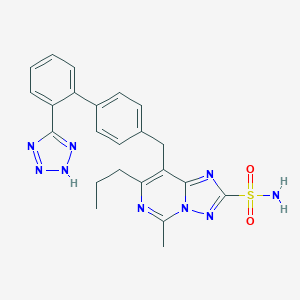 CHEMBL79879 CHEMBL79879 | C23H23N9O2S | 489.558 | 9 / 2 | 3.3 | Yes |
| 70046 |  CHEMBL49748 CHEMBL49748 | C27H35ClN4O4 | 515.051 | 5 / 2 | 5.2 | No |
| 70072 |  CHEMBL105525 CHEMBL105525 | C20H25ClN6O | 400.911 | 5 / 2 | 3.4 | Yes |
| 70153 |  CHEMBL71613 CHEMBL71613 | C29H39N7O | 501.679 | 5 / 1 | 6.7 | No |
| 72302 |  CHEMBL326049 CHEMBL326049 | C29H38N2O | 430.636 | 2 / 0 | 6.0 | No |
| 73948 | 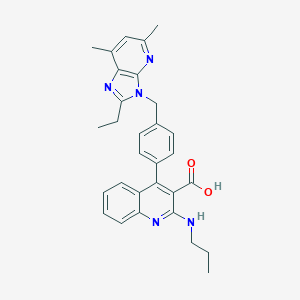 CHEMBL312889 CHEMBL312889 | C30H31N5O2 | 493.611 | 6 / 2 | 7.1 | No |
| 75476 | 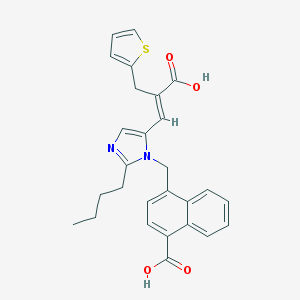 UNII-A9T9WVAH03 UNII-A9T9WVAH03 | C27H26N2O4S | 474.575 | 6 / 2 | 5.8 | No |
| 76054 | 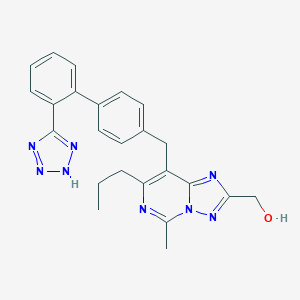 CHEMBL80773 CHEMBL80773 | C24H24N8O | 440.511 | 7 / 2 | 3.6 | Yes |
| 77060 | 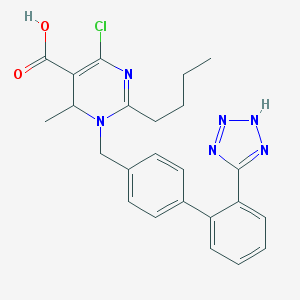 CHEMBL358335 CHEMBL358335 | C24H25ClN6O2 | 464.954 | 6 / 2 | 4.0 | Yes |
| 77944 | 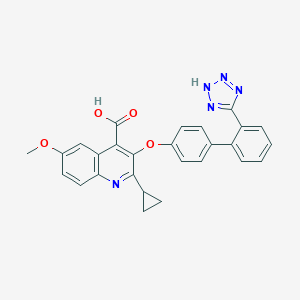 CHEMBL64220 CHEMBL64220 | C27H21N5O4 | 479.496 | 8 / 2 | 4.6 | Yes |
| 79118 | 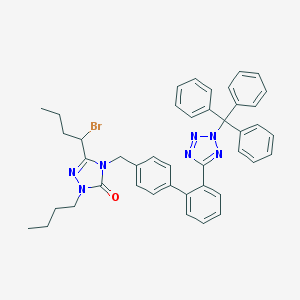 CHEMBL87356 CHEMBL87356 | C43H42BrN7O | 752.761 | 5 / 0 | 10.6 | No |
| 80347 | 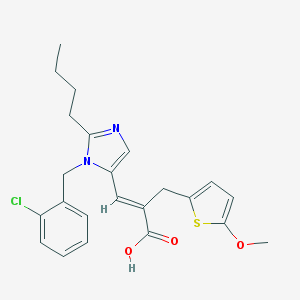 CHEMBL127194 CHEMBL127194 | C23H25ClN2O3S | 444.974 | 5 / 1 | 5.9 | No |
| 81679 | 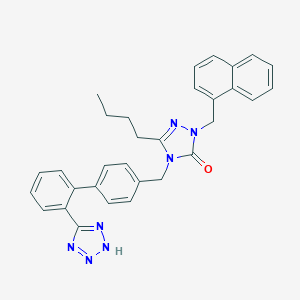 CHEMBL87184 CHEMBL87184 | C31H29N7O | 515.621 | 5 / 1 | 6.0 | No |
| 82705 | 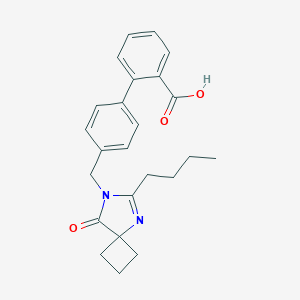 CHEMBL113674 CHEMBL113674 | C24H26N2O3 | 390.483 | 4 / 1 | 4.1 | Yes |
| 82725 | 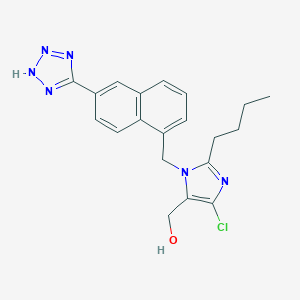 CHEMBL320951 CHEMBL320951 | C20H21ClN6O | 396.879 | 5 / 2 | 3.9 | Yes |
| 83073 | 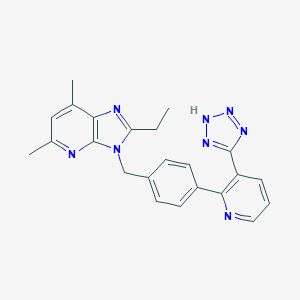 CHEMBL86346 CHEMBL86346 | C23H22N8 | 410.485 | 6 / 1 | 3.7 | Yes |
| 85133 | 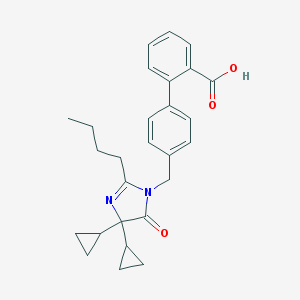 CHEMBL44268 CHEMBL44268 | C27H30N2O3 | 430.548 | 4 / 1 | 4.7 | Yes |
| 87500 | 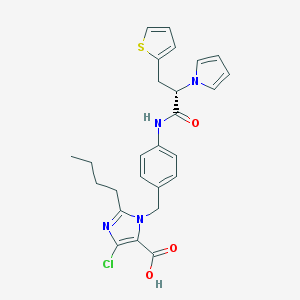 CHEMBL49954 CHEMBL49954 | C26H27ClN4O3S | 511.037 | 5 / 2 | 5.6 | No |
| 87532 | 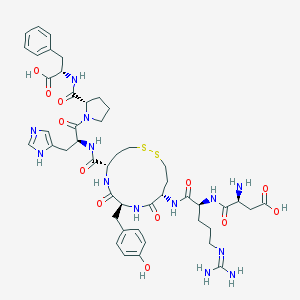 CHEMBL411997 CHEMBL411997 | C47H63N13O12S2 | 1066.22 | 17 / 13 | -3.5 | No |
| 89341 | 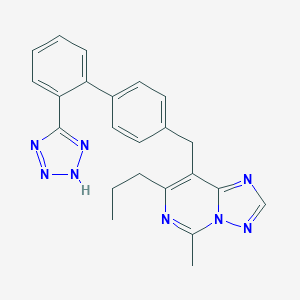 CHEMBL309384 CHEMBL309384 | C23H22N8 | 410.485 | 6 / 1 | 4.4 | Yes |
| 89652 | 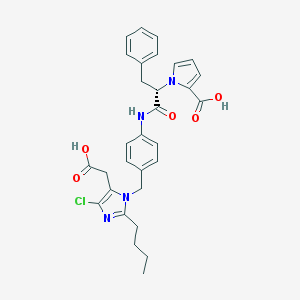 CHEMBL297653 CHEMBL297653 | C30H31ClN4O5 | 563.051 | 6 / 3 | 5.4 | No |
| 90788 | 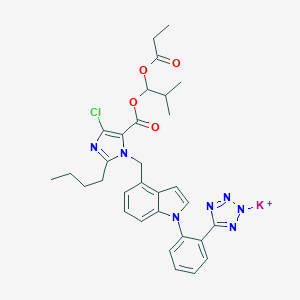 CHEMBL2079768 CHEMBL2079768 | C31H33ClKN7O4 | 642.198 | 9 / 0 | N/A | No |
| 95888 | 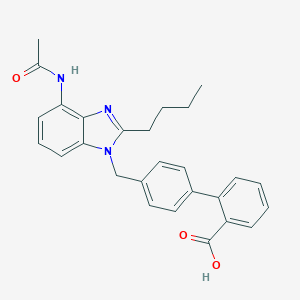 CHEMBL334741 CHEMBL334741 | C27H27N3O3 | 441.531 | 4 / 2 | 5.1 | No |
| 97769 | 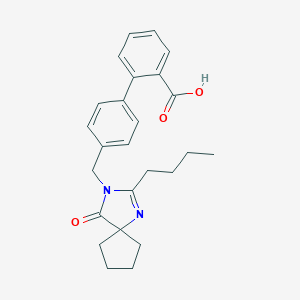 CHEMBL439968 CHEMBL439968 | C25H28N2O3 | 404.51 | 4 / 1 | 4.5 | Yes |
| 97901 | 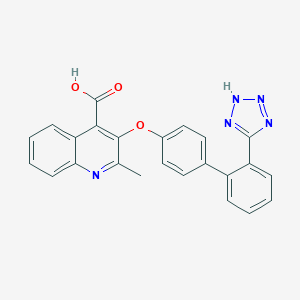 CHEMBL418122 CHEMBL418122 | C24H17N5O3 | 423.432 | 7 / 2 | 4.5 | Yes |
| 98943 | 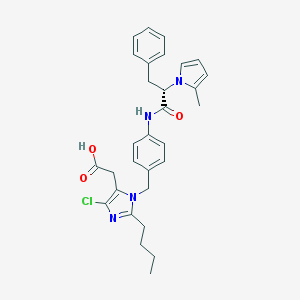 CHEMBL48168 CHEMBL48168 | C30H33ClN4O3 | 533.069 | 4 / 2 | 5.9 | No |
| 99463 | 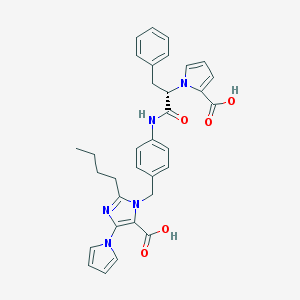 CHEMBL300487 CHEMBL300487 | C33H33N5O5 | 579.657 | 6 / 3 | 5.5 | No |
| 101392 | 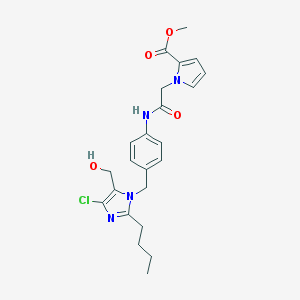 CHEMBL48526 CHEMBL48526 | C23H27ClN4O4 | 458.943 | 5 / 2 | 3.4 | Yes |
| 101831 | 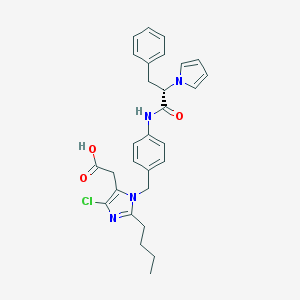 CHEMBL297202 CHEMBL297202 | C29H31ClN4O3 | 519.042 | 4 / 2 | 5.5 | No |
| 101959 | 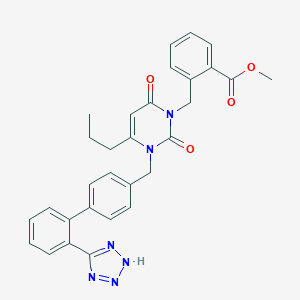 CHEMBL65503 CHEMBL65503 | C30H28N6O4 | 536.592 | 7 / 1 | 4.2 | No |
| 103236 | 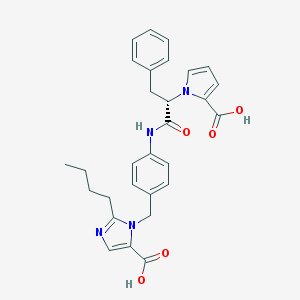 CHEMBL301705 CHEMBL301705 | C29H30N4O5 | 514.582 | 6 / 3 | 4.8 | No |
| 104294 | 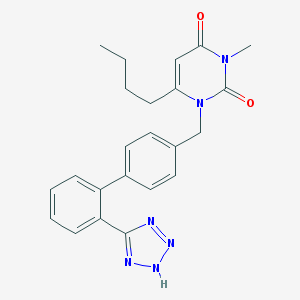 CHEMBL292316 CHEMBL292316 | C23H24N6O2 | 416.485 | 5 / 1 | 3.4 | Yes |
| 106239 | 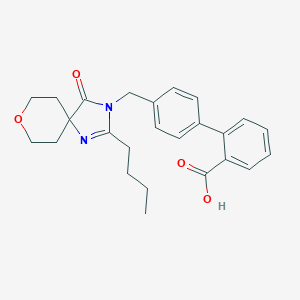 CHEMBL325445 CHEMBL325445 | C25H28N2O4 | 420.509 | 5 / 1 | 3.6 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417