

You can:
| Name | Corticotropin-releasing factor receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CRHR2 |
| Synonym | CRH-R2 CRH-R-2 CRFR2beta CRFR2alpha CRFR2 [ Show all ] |
| Disease | Generalized anxiety disorder Anxiety disorder Congestive heart failure Eating disorders stimulate food consumption anxiety |
| Length | 411 |
| Amino acid sequence | MDAALLHSLLEANCSLALAEELLLDGWGPPLDPEGPYSYCNTTLDQIGTCWPRSAAGALVERPCPEYFNGVKYNTTRNAYRECLENGTWASKINYSQCEPILDDKQRKYDLHYRIALVVNYLGHCVSVAALVAAFLLFLALRSIRCLRNVIHWNLITTFILRNVMWFLLQLVDHEVHESNEVWCRCITTIFNYFVVTNFFWMFVEGCYLHTAIVMTYSTERLRKCLFLFIGWCIPFPIIVAWAIGKLYYENEQCWFGKEPGDLVDYIYQGPIILVLLINFVFLFNIVRILMTKLRASTTSETIQYRKAVKATLVLLPLLGITYMLFFVNPGEDDLSQIMFIYFNSFLQSFQGFFVSVFYCFFNGEVRSAVRKRWHRWQDHHSLRVPMARAMSIPTSPTRISFHSIKQTAAV |
| UniProt | Q13324 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q13324 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q13324. |
| BioLiP | N/A |
| Therapeutic Target Database | T11011 |
| ChEMBL | CHEMBL4069 |
| IUPHAR | 213 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 464062 | 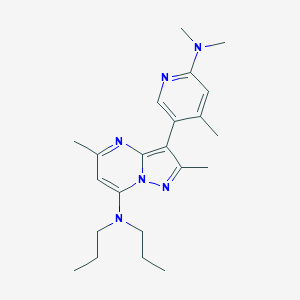 195055-03-9 195055-03-9 | C22H32N6 | 380.54 | 5 / 0 | 4.7 | Yes |
| 11473 | 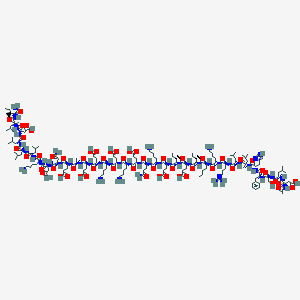 CHEMBL2370927 CHEMBL2370927 | C183H307N47O56 | 4061.74 | 63 / 55 | -17.2 | No |
| 14543 | 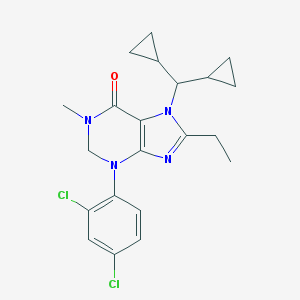 CHEMBL426607 CHEMBL426607 | C21H24Cl2N4O | 419.35 | 3 / 0 | 5.2 | No |
| 19631 | 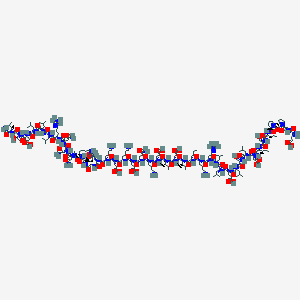 CHEMBL2370916 CHEMBL2370916 | C211H362N58O66 | 4767.56 | 73 / 64 | -23.3 | No |
| 22937 |  CHEMBL2370940 CHEMBL2370940 | C183H305N47O55 | 4043.73 | 61 / 54 | -14.5 | No |
| 24481 |  CHEMBL427807 CHEMBL427807 | C204H337N63O64 | 4696.32 | 71 / 69 | -18.8 | No |
| 467058 |  CHEMBL186203 CHEMBL186203 | C20H24Cl2N4O | 407.339 | 4 / 1 | 5.2 | No |
| 35444 |  CHEMBL2370939 CHEMBL2370939 | C186H312N52O55 | 4156.85 | 62 / 57 | -16.7 | No |
| 36171 |  CHEMBL1794007 CHEMBL1794007 | C160H266N50O44 | 3594.19 | 51 / 50 | -12.6 | No |
| 43216 |  CHEMBL1794010 CHEMBL1794010 | C157H263N45O44 | 3485.1 | 49 / 47 | -9.3 | No |
| 44192 |  Cipargamin Cipargamin | C19H14Cl2FN3O | 390.239 | 3 / 3 | 3.9 | Yes |
| 45857 |  CHEMBL2369741 CHEMBL2369741 | C158H267N47O43 | 3513.16 | 49 / 48 | -9.9 | No |
| 470249 | 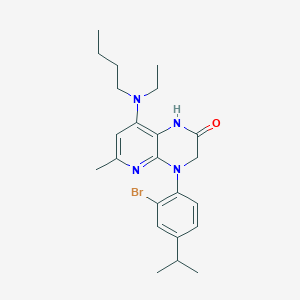 CHEMBL186252 CHEMBL186252 | C23H31BrN4O | 459.432 | 4 / 1 | 5.8 | No |
| 62021 | 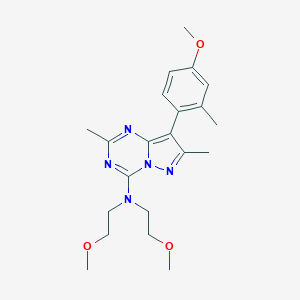 UNII-7FC996ZZL5 UNII-7FC996ZZL5 | C21H29N5O3 | 399.495 | 7 / 0 | 3.0 | Yes |
| 63474 |  CHEMBL2370921 CHEMBL2370921 | C182H305N49O54 | 4043.73 | 61 / 55 | -15.5 | No |
| 68461 | 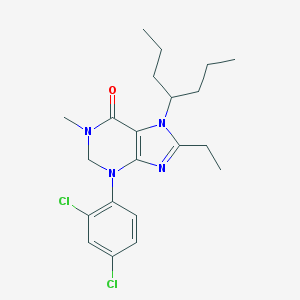 CHEMBL184938 CHEMBL184938 | C21H28Cl2N4O | 423.382 | 3 / 0 | 6.3 | No |
| 87398 |  CID 44290454 CID 44290454 | C202H349N57O63S | 4616.41 | 71 / 63 | -24.0 | No |
| 92459 |  CHEMBL2372650 CHEMBL2372650 | C208H344N60O63S2 | 4757.52 | 72 / 64 | -15.4 | No |
| 460042 |  CHEMBL265635 CHEMBL265635 | C208H344N60O63S2 | 4757.52 | 72 / 64 | -15.4 | No |
| 110328 |  CHEMBL2370931 CHEMBL2370931 | C163H272N50O44 | 3636.27 | 51 / 50 | -11.3 | No |
| 553889 |  Astressin Astressin | C161H269N49O42 | 3563.22 | 49 / 51 | -8.0 | No |
| 121132 |  CHEMBL1794009 CHEMBL1794009 | C161H269N49O42 | 3563.22 | 49 / 48 | -9.1 | No |
| 128216 |  CHEMBL2370914 CHEMBL2370914 | C170H286N44O50 | 3746.42 | 57 / 50 | -15.8 | No |
| 479813 |  CHEMBL3628739 CHEMBL3628739 | C161H274N48O46S | 3650.31 | 55 / 53 | -20.2 | No |
| 139344 |  CHEMBL2370917 CHEMBL2370917 | C183H307N51O53 | 4069.77 | 59 / 55 | -14.4 | No |
| 165497 |  CHEMBL2370938 CHEMBL2370938 | C185H314N50O53 | 4086.84 | 60 / 55 | -14.5 | No |
| 483424 |  CHEMBL3628728 CHEMBL3628728 | C20H24Br2N4O | 496.247 | 4 / 1 | 5.4 | No |
| 168112 |  BDBM50158983 BDBM50158983 | C202H345N55O64S | 4600.36 | 71 / 64 | -19.2 | No |
| 460722 | 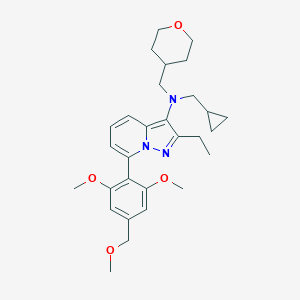 UNII-O90RQ1XQ79 UNII-O90RQ1XQ79 | C29H39N3O4 | 493.648 | 6 / 0 | 4.6 | Yes |
| 183663 | 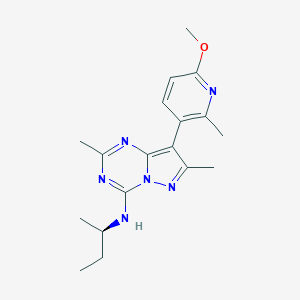 PEXACERFONT PEXACERFONT | C18H24N6O | 340.431 | 6 / 1 | 3.8 | Yes |
| 487424 | 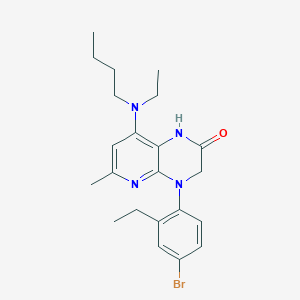 CHEMBL3628729 CHEMBL3628729 | C22H29BrN4O | 445.405 | 4 / 1 | 5.5 | No |
| 487620 | 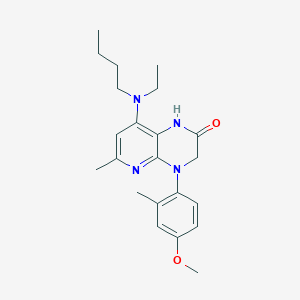 CHEMBL189089 CHEMBL189089 | C22H30N4O2 | 382.508 | 5 / 1 | 4.3 | Yes |
| 205740 | 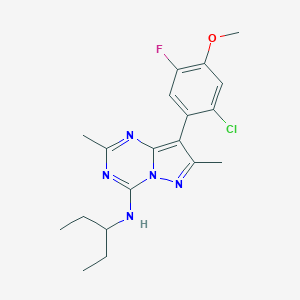 CHEMBL484158 CHEMBL484158 | C19H23ClFN5O | 391.875 | 6 / 1 | 5.4 | No |
| 488594 | 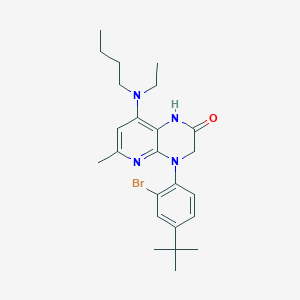 CHEMBL3628734 CHEMBL3628734 | C24H33BrN4O | 473.459 | 4 / 1 | 6.4 | No |
| 207704 | 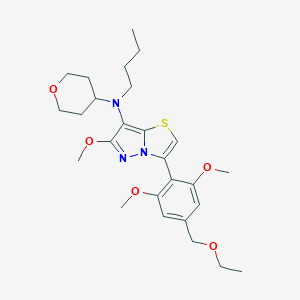 CHEMBL2179195 CHEMBL2179195 | C26H37N3O5S | 503.658 | 8 / 0 | 4.8 | No |
| 228647 | 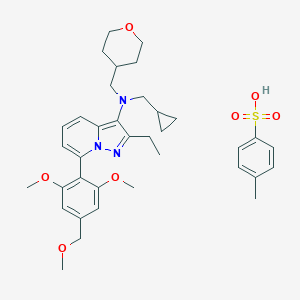 UNII-YP6748BALI UNII-YP6748BALI | C36H47N3O7S | 665.846 | 9 / 1 | N/A | No |
| 240926 | 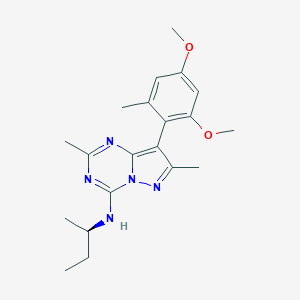 CHEMBL506214 CHEMBL506214 | C20H27N5O2 | 369.469 | 6 / 1 | 4.5 | Yes |
| 241140 | 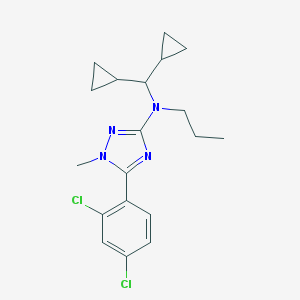 CHEMBL110609 CHEMBL110609 | C19H24Cl2N4 | 379.329 | 3 / 0 | 5.8 | No |
| 264146 | 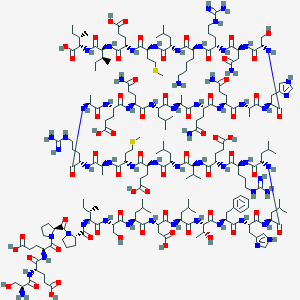 UNII-D8690V3244 UNII-D8690V3244 | C208H343N59O64S2 | 4758.5 | 73 / 67 | -13.6 | No |
| 265184 |  CHEMBL1794006 CHEMBL1794006 | C161H271N49O43 | 3581.24 | 51 / 49 | -11.8 | No |
| 270028 | 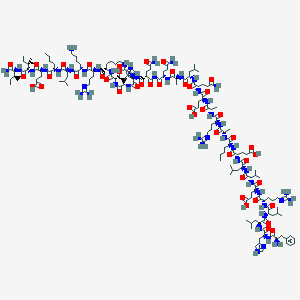 CHEMBL2369740 CHEMBL2369740 | C163H271N49O44 | 3621.26 | 51 / 49 | -8.8 | No |
| 274172 |  CHEMBL2369693 CHEMBL2369693 | C160H265N49O45 | 3595.18 | 52 / 50 | -12.0 | No |
| 274572 |  CHEMBL2368116 CHEMBL2368116 | C181H305N49O54 | 4031.72 | 61 / 55 | -15.4 | No |
| 276069 |  BDBM50313853 BDBM50313853 | C205H339N59O63S | 4670.38 | 71 / 64 | -19.4 | No |
| 461605 |  CHEMBL1790241 CHEMBL1790241 | C205H339N59O63S | 4670.38 | 71 / 64 | -19.4 | No |
| 301918 | 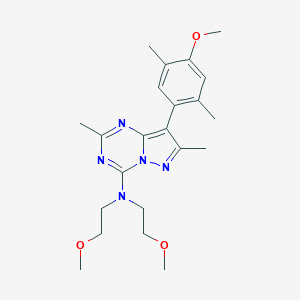 CHEMBL483993 CHEMBL483993 | C22H31N5O3 | 413.522 | 7 / 0 | 3.4 | Yes |
| 501555 | 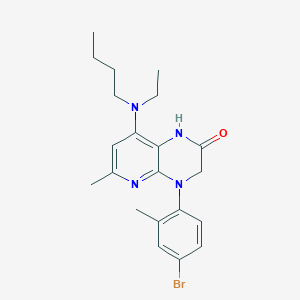 CHEMBL3628735 CHEMBL3628735 | C21H27BrN4O | 431.378 | 4 / 1 | 5.0 | Yes |
| 321168 | 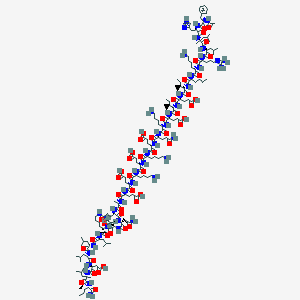 CHEMBL2370915 CHEMBL2370915 | C170H284N44O49 | 3728.4 | 55 / 49 | -13.1 | No |
| 504659 | 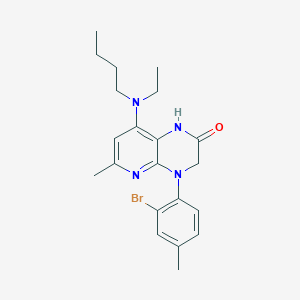 CHEMBL3628736 CHEMBL3628736 | C21H27BrN4O | 431.378 | 4 / 1 | 5.0 | Yes |
| 339697 |  CHEMBL2370941 CHEMBL2370941 | C187H315N49O56 | 4145.86 | 63 / 56 | -14.3 | No |
| 342574 |  CHEMBL2370929 CHEMBL2370929 | C184H310N50O55 | 4102.8 | 62 / 56 | -16.3 | No |
| 356310 |  CP-376395 CP-376395 | C21H30N2O | 326.484 | 3 / 1 | 6.4 | No |
| 507585 |  CHEMBL3628733 CHEMBL3628733 | C22H30N4O | 366.509 | 4 / 1 | 4.7 | Yes |
| 369375 |  CHEMBL2370930 CHEMBL2370930 | C178H302N56O46 | 3962.72 | 55 / 55 | -15.5 | No |
| 391589 |  CHEMBL2370920 CHEMBL2370920 | C180H299N47O55 | 4001.65 | 61 / 54 | -15.8 | No |
| 399233 |  CHEMBL128100 CHEMBL128100 | C18H19Cl3F3N3S | 472.776 | 7 / 1 | 7.1 | No |
| 403778 |  CHEMBL2370924 CHEMBL2370924 | C185H312N50O53 | 4084.83 | 60 / 55 | -14.8 | No |
| 409487 |  CHEMBL295563 CHEMBL295563 | C22H30N4O2 | 382.508 | 5 / 0 | 4.9 | Yes |
| 409638 |  CHEMBL2370926 CHEMBL2370926 | C185H310N48O56 | 4102.79 | 62 / 55 | -15.3 | No |
| 410853 |  CHEMBL2373995 CHEMBL2373995 | C210H340N62O67S2 | 4869.52 | 76 / 67 | -18.9 | No |
| 422902 |  CHEMBL2369694 CHEMBL2369694 | C159H266N48O42 | 3522.17 | 49 / 48 | -9.0 | No |
| 440028 |  CHEMBL2370933 CHEMBL2370933 | C182H305N49O54 | 4043.73 | 61 / 55 | -15.5 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417