

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MDAALLHSLLEANCSLALAEELLLDGWGPPLDPEGPYSYCNTTLDQIGTCWPRSAAGALVERPCPEYFNGVKYNTTRNAYRECLENGTWASKINYSQCEPILDDKQRKYDLHYRIALVVNYLGHCVSVAALVAAFLLFLALRSIRCLRNVIHWNLITTFILRNVMWFLLQLVDHEVHESNEVWCRCITTIFNYFVVTNFFWMFVEGCYLHTAIVMTYSTERLRKCLFLFIGWCIPFPIIVAWAIGKLYYENEQCWFGKEPGDLVDYIYQGPIILVLLINFVFLFNIVRILMTKLRASTTSETIQYRKAVKATLVLLPLLGITYMLFFVNPGEDDLSQIMFIYFNSFLQSFQGFFVSVFYCFFNGEVRSAVRKRWHRWQDHHSLRVPMARAMSIPTSPTRISFHSIKQTAAV | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCHHHCCCCCCCCCCCSSCCCCCCSSCCCCCCCCHHHHCCCCCCCCCHHHCCCCSSSSSHHHHHHHHHHHHHHHHHSSHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHCCCCCSSSSSSSSCCCCCCCCCCSSSCCCCCCSSSSSSCHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 962267787558899999999999862477788899999988008984207989999889805922527786788884531128996156547865133310112233210001442345311178799899999875022031255379999999999999999999987502201478653279999999999999999999998760666741578522566644210465330578887203333566504514898505899954267787778899998542145422554321157888899899999999999999886322457526999999999999999999888622189999999999987236687778877788988989987578887778889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MDAALLHSLLEANCSLALAEELLLDGWGPPLDPEGPYSYCNTTLDQIGTCWPRSAAGALVERPCPEYFNGVKYNTTRNAYRECLENGTWASKINYSQCEPILDDKQRKYDLHYRIALVVNYLGHCVSVAALVAAFLLFLALRSIRCLRNVIHWNLITTFILRNVMWFLLQLVDHEVHESNEVWCRCITTIFNYFVVTNFFWMFVEGCYLHTAIVMTYSTERLRKCLFLFIGWCIPFPIIVAWAIGKLYYENEQCWFGKEPGDLVDYIYQGPIILVLLINFVFLFNIVRILMTKLRASTTSETIQYRKAVKATLVLLPLLGITYMLFFVNPGEDDLSQIMFIYFNSFLQSFQGFFVSVFYCFFNGEVRSAVRKRWHRWQDHHSLRVPMARAMSIPTSPTRISFHSIKQTAAV | |
| 753621332133413234324302641454354755331011210111000130321330312002002002133432010202320233321100201213343343341122001000120110001002201000000030324101000000000002100100000102223441201000000000001000000000002100200010132732100000000011000000000000001334100022442200000130110000200320123001000210335635324200100100000001122011000000246210100100010000031001001000011440051023103413463324353344334323334334433443647 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSSCCCCHHHCCCCCCCCCCCSSCCCCCCSSCCCCCCCCHHHHCCCCCCCCCHHHCCCCSSSSSHHHHHHHHHHHHHHHHHSSHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHCCCCCSSSSSSSSCCCCCCCCCCSSSCCCCCCSSSSSSCHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDAALLHSLLEANCSLALAEELLLDGWGPPLDPEGPYSYCNTTLDQIGTCWPRSAAGALVERPCPEYFNGVKYNTTRNAYRECLENGTWASKINYSQCEPILDDKQRKYDLHYRIALVVNYLGHCVSVAALVAAFLLFLALRSIRCLRNVIHWNLITTFILRNVMWFLLQLVDHEVHESNEVWCRCITTIFNYFVVTNFFWMFVEGCYLHTAIVMTYSTERLRKCLFLFIGWCIPFPIIVAWAIGKLYYENEQCWFGKEPGDLVDYIYQGPIILVLLINFVFLFNIVRILMTKLRASTTSETIQYRKAVKATLVLLPLLGITYMLFFVNPGEDDLSQIMFIYFNSFLQSFQGFFVSVFYCFFNGEVRSAVRKRWHRWQDHHSLRVPMARAMSIPTSPTRISFHSIKQTAAV | |||||||||||||||||||||||||
| 1 | 5vaiR | 0.29 | 0.29 | 0.90 | 2.07 | Download | -----TVSLSETVQKWREYRRQCQHFLTEAPPLA-TGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCW-TRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHATLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 2 | 5vaiA | 0.29 | 0.29 | 0.90 | 4.09 | Download | -------SLSETVQKWREYRRQCQHFLTEAPPLA-TGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCW-TRNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARGRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 3 | 5vaiR | 0.29 | 0.29 | 0.90 | 3.34 | Download | -----TVSLSETVQKWREYRRQCQHFLTEAPPLATG-LFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNY-WLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARGRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 4 | 5vai | 0.31 | 0.29 | 0.88 | 2.43 | Download | VSLSETWREYRRQCQHFLTEA-------P---PLATGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKRDLSECEESSPE------ERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMN-YWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHATLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 5 | 5vai | 0.29 | 0.29 | 0.90 | 3.14 | Download | -----TVSLSETVQKWREYRRQCQHFLTEAPP-LATGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESSPERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMN-YWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHATLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 6 | 5vaiR | 0.29 | 0.29 | 0.90 | 3.41 | Download | ------VSLSETVQKWREYRRQCQHFLTEAPPLA-TGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWT-RNSNMNYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 7 | 5vai | 0.29 | 0.29 | 0.88 | 4.52 | Download | --------------KWREYRRQCQHFLTEAPPL-ATGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSECEESSPERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNM-NYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHGTLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 8 | 4k5yA | 0.73 | 0.44 | 0.60 | 3.80 | Download | ---------------------------------------------------------------------------------------------------------------HYHVAAIINYLGHCISLVALLVAFVLFLRARSIRCLRNIIHANLIAAFILRNATWFVVQLTMSEVHQSNVGWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIVL---TDRLRAWMFICIGWGVPFPIIVAWAIGKLYYDNEKCWAGKRPGVYTDYIYQGPMALVLLINFIFLFNIVRILMTKLRASTTSETIQARKAVKATLVLLPLLGITYMLAFV----NEVSRVVFIYFNAFLESFQGFFVSVFACFLNS----------------------------------------------- | |||||||||||||||||||
| 9 | 5vaiR | 0.29 | 0.29 | 0.90 | 2.80 | Download | -----TVSLSETVQKWREYRRQCQHFLTEA-PPLATGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLSEEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMN-YWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHARLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| 10 | 5vaiR | 0.29 | 0.29 | 0.90 | 2.03 | Download | -----TVSLSETVQKWREYRRQCQHFLTE-APPLATGLFCNRTFDDY-ACWPDGAPGSFVNVSCPWYLPWASNVLQGHVYRFCTAEGHWLPKDNSSLPWRDLECEESSPEERLLSLYIIYTVGYALSFSALVIASAILLGFRHLHCTRNYIHLNLFASFILRALSVFIKDAALKWMYYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFAVFSEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNMNYW-LIIRLPILFAIGVNFLIFIRVICIVVSKLKANLMCKTDIKCRLAKSTLTLIPLLGTHEVIFAFVMDEHGTLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
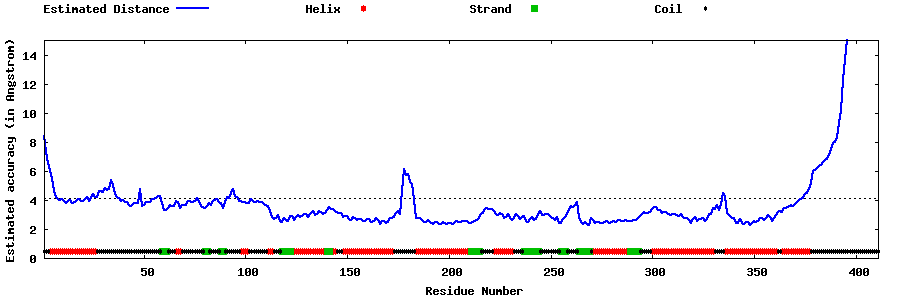 | |||
|
|
|||
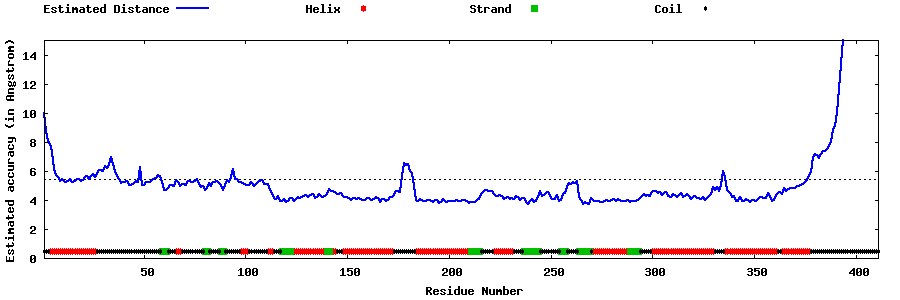 | |||
|
| |||
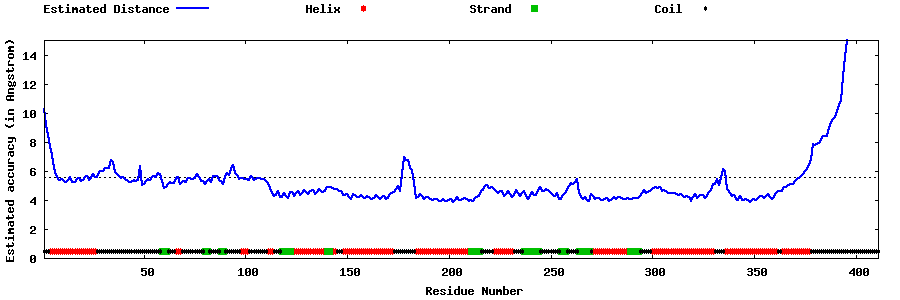 | |||
|
| |||
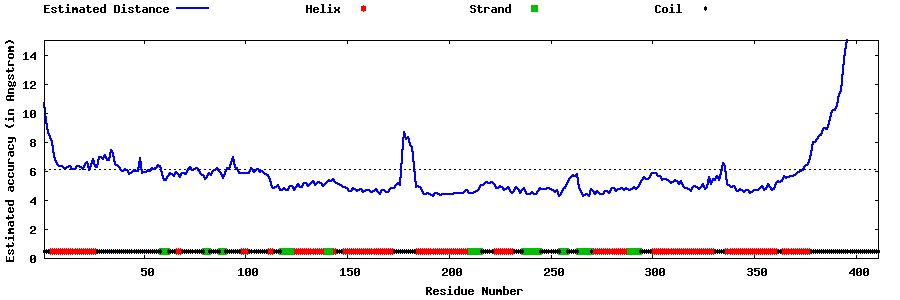 | |||
|
| |||
 | |||
|
| |||