

You can:
| Name | Type-1 angiotensin II receptor |
|---|---|
| Species | Oryctolagus cuniculus (Rabbit) |
| Gene | AGTR1 |
| Synonym | Angiotensin II type-1 receptor AT1 |
| Disease | N/A for non-human GPCRs |
| Length | 359 |
| Amino acid sequence | MMLNSSTEDGIKRIQDDCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLAVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFFIENTNITVCAFHYESQNSTLPIGLGLTKNILGFLFPFLIILTSYTLIWKALKKAYEIQKNKPRNDDIFKIIMAIVLFFFFSWVPHQIFTFLDVLIQLGVIHDCRIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKKYFLQLLKYIPPKAKSHSNLSTKMSTLSYRPSDNVSSSSKKPVPCFEVE |
| UniProt | P34976 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3948 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 101 | 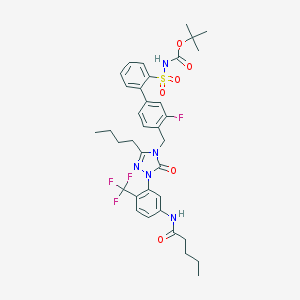 CHEMBL338434 CHEMBL338434 | C36H41F4N5O6S | 747.807 | 11 / 2 | 7.3 | No |
| 116 | 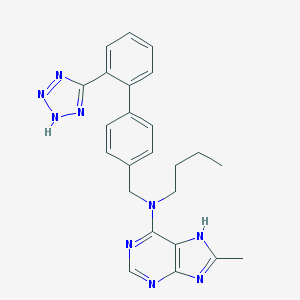 CHEMBL52036 CHEMBL52036 | C24H25N9 | 439.527 | 7 / 2 | 4.5 | Yes |
| 139 | 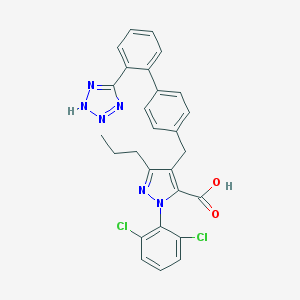 CHEMBL338740 CHEMBL338740 | C27H22Cl2N6O2 | 533.413 | 6 / 2 | 7.0 | No |
| 385 | 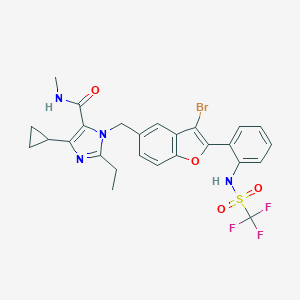 CHEMBL63072 CHEMBL63072 | C26H24BrF3N4O4S | 625.461 | 9 / 2 | 5.6 | No |
| 600 | 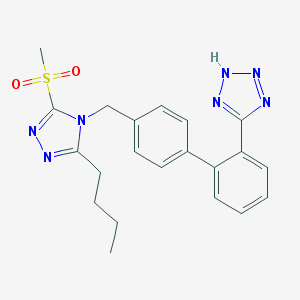 CHEMBL156307 CHEMBL156307 | C21H23N7O2S | 437.522 | 7 / 1 | 3.2 | Yes |
| 753 | 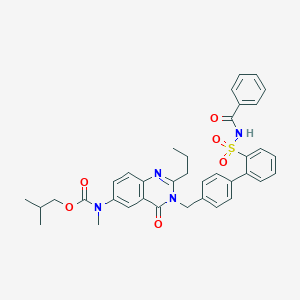 CHEMBL350554 CHEMBL350554 | C37H38N4O6S | 666.793 | 7 / 1 | 6.3 | No |
| 1133 | 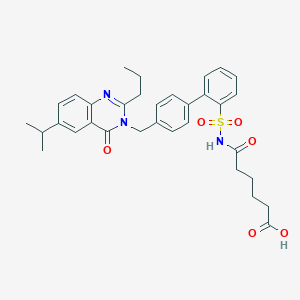 CHEMBL166428 CHEMBL166428 | C33H37N3O6S | 603.734 | 7 / 2 | 4.8 | No |
| 1406 | 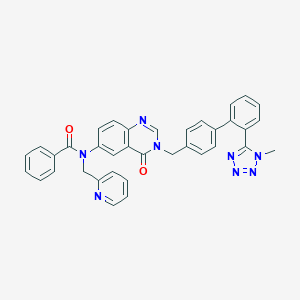 CHEMBL30086 CHEMBL30086 | C36H28N8O2 | 604.674 | 7 / 0 | 4.8 | No |
| 1609 | 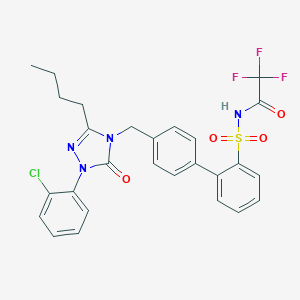 CHEMBL94582 CHEMBL94582 | C27H24ClF3N4O4S | 593.018 | 8 / 1 | 6.1 | No |
| 1860 | 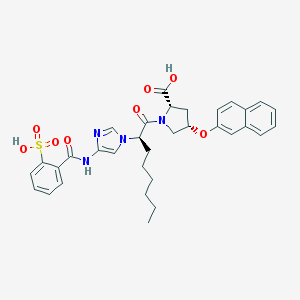 CHEMBL139457 CHEMBL139457 | C33H36N4O8S | 648.731 | 9 / 3 | 5.2 | No |
| 1937 | 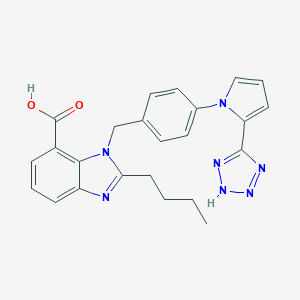 CHEMBL400424 CHEMBL400424 | C24H23N7O2 | 441.495 | 6 / 2 | 3.9 | Yes |
| 2230 | 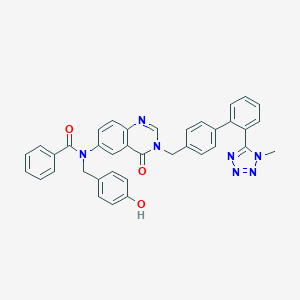 CHEMBL30015 CHEMBL30015 | C37H29N7O3 | 619.685 | 7 / 1 | 5.5 | No |
| 2430 | 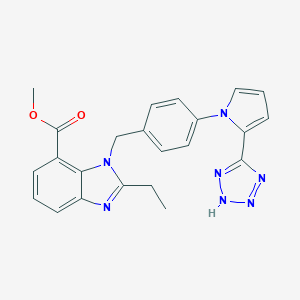 CHEMBL241684 CHEMBL241684 | C23H21N7O2 | 427.468 | 6 / 1 | 3.4 | Yes |
| 3537 | 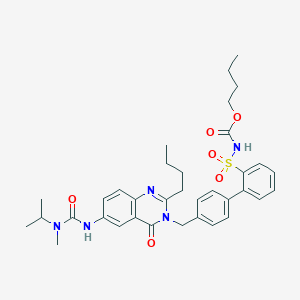 CHEMBL74476 CHEMBL74476 | C35H43N5O6S | 661.818 | 7 / 2 | 5.8 | No |
| 3681 | 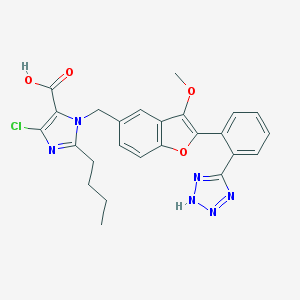 CHEMBL157552 CHEMBL157552 | C25H23ClN6O4 | 506.947 | 8 / 2 | 5.4 | No |
| 3734 | 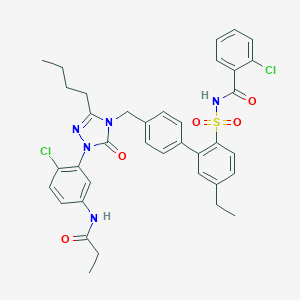 CHEMBL319730 CHEMBL319730 | C37H37Cl2N5O5S | 734.693 | 6 / 2 | 7.7 | No |
| 3822 |  CHEMBL62754 CHEMBL62754 | C30H42N4O5 | 538.689 | 5 / 1 | 5.3 | No |
| 4288 |  CHEMBL155877 CHEMBL155877 | C28H27ClN8OS | 559.089 | 7 / 2 | 5.8 | No |
| 4361 |  CHEMBL132408 CHEMBL132408 | C27H24N8O | 476.544 | 6 / 1 | 4.7 | Yes |
| 4375 |  CHEMBL350697 CHEMBL350697 | C27H26N4O3S2 | 518.65 | 7 / 2 | 5.2 | No |
| 4694 | 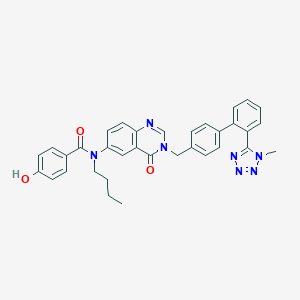 CHEMBL282168 CHEMBL282168 | C34H31N7O3 | 585.668 | 7 / 1 | 5.2 | No |
| 5902 | 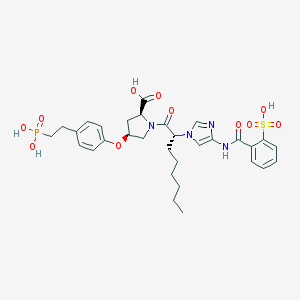 CHEMBL343723 CHEMBL343723 | C31H39N4O11PS | 706.704 | 12 / 5 | 2.4 | No |
| 6030 | 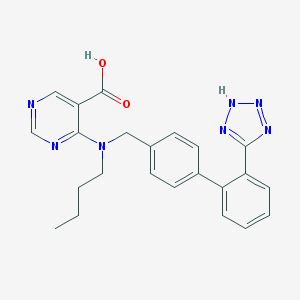 141872-46-0 141872-46-0 | C23H23N7O2 | 429.484 | 8 / 2 | 3.8 | Yes |
| 6127 | 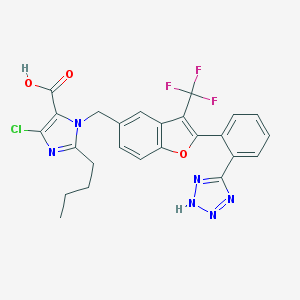 CHEMBL348312 CHEMBL348312 | C25H20ClF3N6O3 | 544.919 | 10 / 2 | 6.3 | No |
| 6698 |  CHEMBL160031 CHEMBL160031 | C23H21BrN2O3 | 453.336 | 4 / 1 | 5.6 | No |
| 6780 |  CHEMBL2079788 CHEMBL2079788 | C32H32KN9O2 | 613.767 | 8 / 0 | N/A | No |
| 6782 |  CHEMBL50174 CHEMBL50174 | C25H23N7O | 437.507 | 7 / 2 | 5.0 | Yes |
| 6887 |  CHEMBL37566 CHEMBL37566 | C25H23Cl2N3O3 | 484.377 | 5 / 1 | 6.4 | No |
| 7147 | 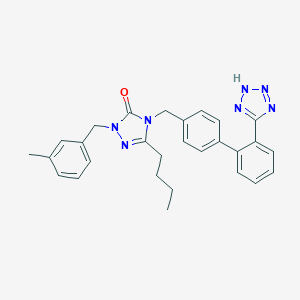 CHEMBL87129 CHEMBL87129 | C28H29N7O | 479.588 | 5 / 1 | 5.1 | No |
| 7382 | 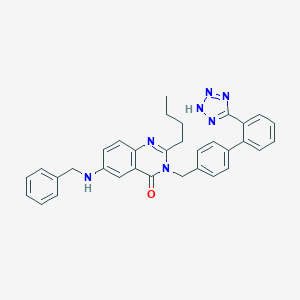 CHEMBL26217 CHEMBL26217 | C33H31N7O | 541.659 | 6 / 2 | 6.1 | No |
| 7502 | 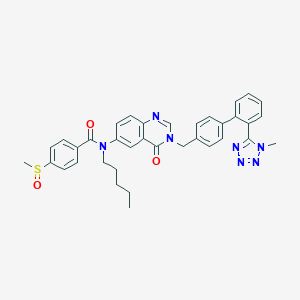 CHEMBL29730 CHEMBL29730 | C36H35N7O3S | 645.782 | 8 / 0 | 5.2 | No |
| 8685 | 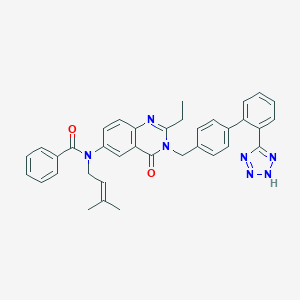 CHEMBL276853 CHEMBL276853 | C36H33N7O2 | 595.707 | 6 / 1 | 6.2 | No |
| 8813 | 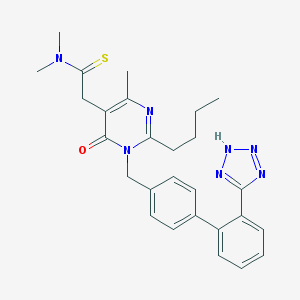 FIMASARTAN FIMASARTAN | C27H31N7OS | 501.653 | 6 / 1 | 3.5 | No |
| 9364 | 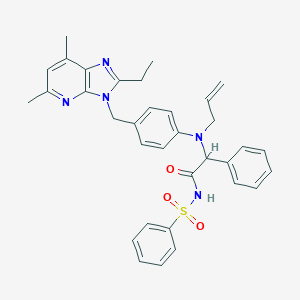 CHEMBL337006 CHEMBL337006 | C34H35N5O3S | 593.746 | 6 / 1 | 6.8 | No |
| 9466 | 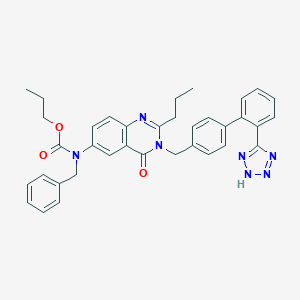 CHEMBL322471 CHEMBL322471 | C36H35N7O3 | 613.722 | 7 / 1 | 6.2 | No |
| 9517 | 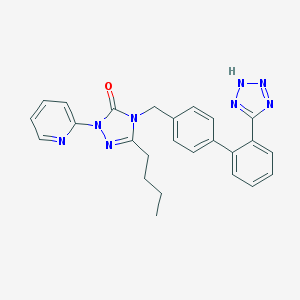 CHEMBL86741 CHEMBL86741 | C25H24N8O | 452.522 | 6 / 1 | 4.1 | Yes |
| 9668 | 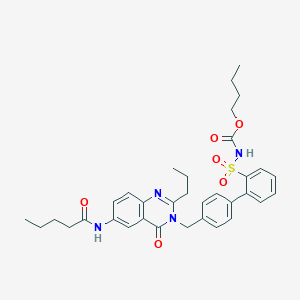 CHEMBL352503 CHEMBL352503 | C34H40N4O6S | 632.776 | 7 / 2 | 5.8 | No |
| 9686 | 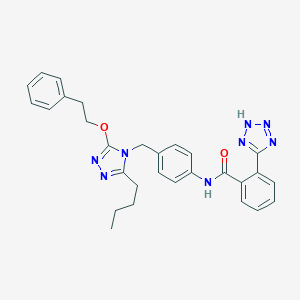 CHEMBL2237609 CHEMBL2237609 | C29H30N8O2 | 522.613 | 7 / 2 | 5.1 | No |
| 10033 | 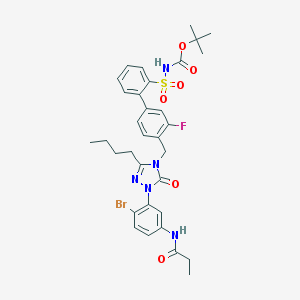 CHEMBL124771 CHEMBL124771 | C33H37BrFN5O6S | 730.65 | 8 / 2 | 6.2 | No |
| 10037 | 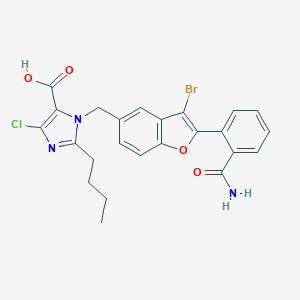 CHEMBL155599 CHEMBL155599 | C24H21BrClN3O4 | 530.803 | 5 / 2 | 5.8 | No |
| 10455 | 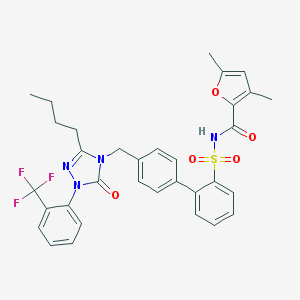 CHEMBL329693 CHEMBL329693 | C33H31F3N4O5S | 652.689 | 9 / 1 | 7.1 | No |
| 10513 | 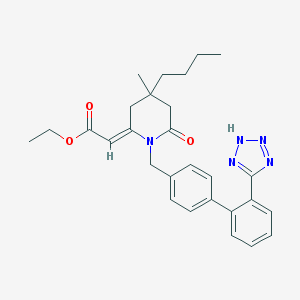 CHEMBL299151 CHEMBL299151 | C28H33N5O3 | 487.604 | 6 / 1 | 5.2 | No |
| 10865 | 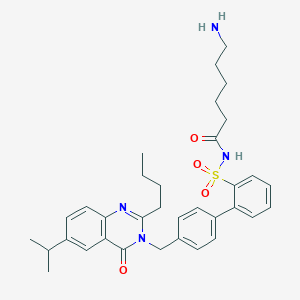 CHEMBL165541 CHEMBL165541 | C34H42N4O4S | 602.794 | 6 / 2 | 5.3 | No |
| 10942 | 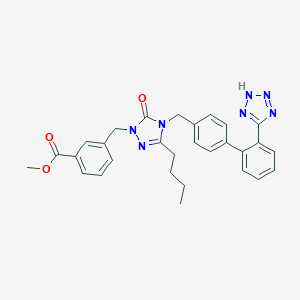 CHEMBL83182 CHEMBL83182 | C29H29N7O3 | 523.597 | 7 / 1 | 4.6 | No |
| 11331 | 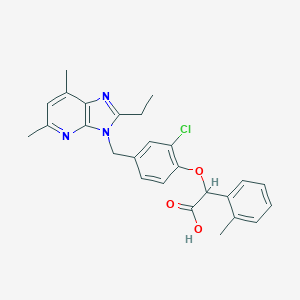 CHEMBL15517 CHEMBL15517 | C26H26ClN3O3 | 463.962 | 5 / 1 | 6.1 | No |
| 11379 | 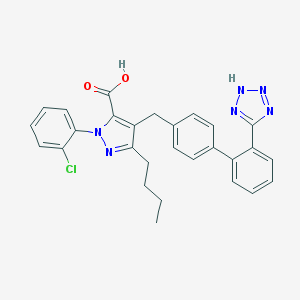 CHEMBL93226 CHEMBL93226 | C28H25ClN6O2 | 512.998 | 6 / 2 | 6.9 | No |
| 11929 | 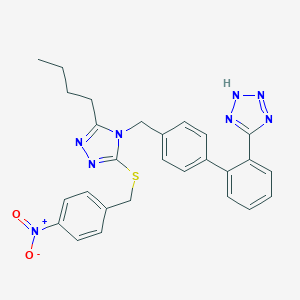 CHEMBL346542 CHEMBL346542 | C27H26N8O2S | 526.619 | 8 / 1 | 5.8 | No |
| 12180 | 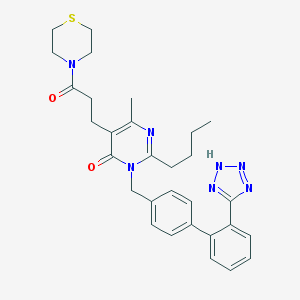 CHEMBL1951138 CHEMBL1951138 | C30H35N7O2S | 557.717 | 7 / 1 | 3.7 | No |
| 12347 | 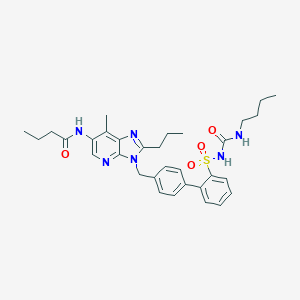 CHEMBL306641 CHEMBL306641 | C32H40N6O4S | 604.77 | 6 / 3 | 5.3 | No |
| 12480 | 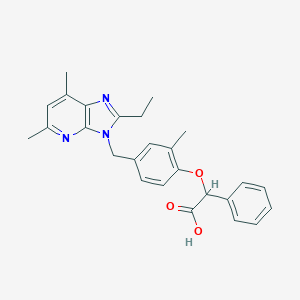 CHEMBL15730 CHEMBL15730 | C26H27N3O3 | 429.52 | 5 / 1 | 5.5 | No |
| 12483 | 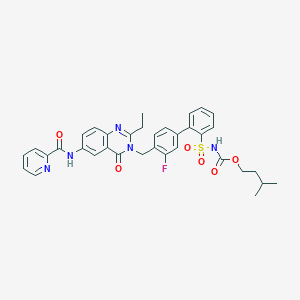 CHEMBL74144 CHEMBL74144 | C35H34FN5O6S | 671.744 | 9 / 2 | 5.5 | No |
| 12590 | 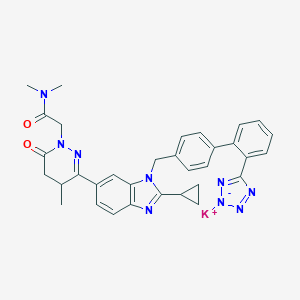 CHEMBL2079790 CHEMBL2079790 | C33H32KN9O2 | 625.778 | 8 / 0 | N/A | No |
| 12812 | 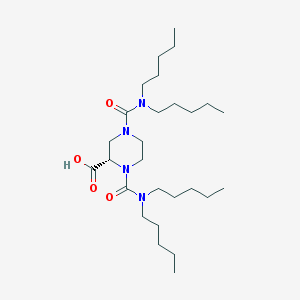 CHEMBL62157 CHEMBL62157 | C27H52N4O4 | 496.737 | 4 / 1 | 5.8 | No |
| 12861 | 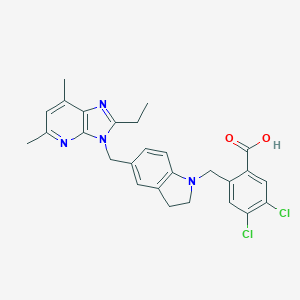 CHEMBL134567 CHEMBL134567 | C27H26Cl2N4O2 | 509.431 | 5 / 1 | 6.3 | No |
| 12932 | 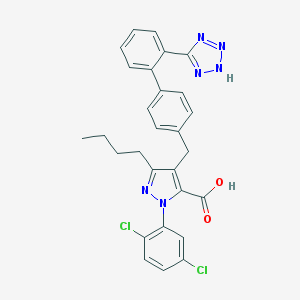 CHEMBL315878 CHEMBL315878 | C28H24Cl2N6O2 | 547.44 | 6 / 2 | 7.5 | No |
| 12944 | 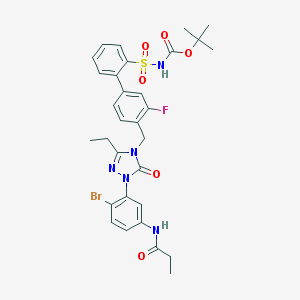 CHEMBL122157 CHEMBL122157 | C31H33BrFN5O6S | 702.596 | 8 / 2 | 5.3 | No |
| 13311 | 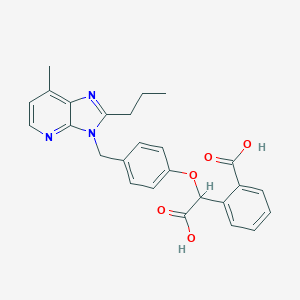 CHEMBL290465 CHEMBL290465 | C26H25N3O5 | 459.502 | 7 / 2 | 4.6 | Yes |
| 13344 | 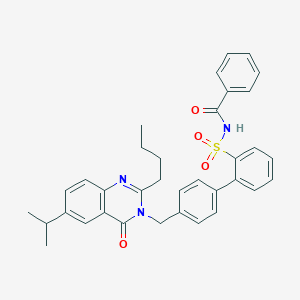 CHEMBL167662 CHEMBL167662 | C35H35N3O4S | 593.742 | 5 / 1 | 6.9 | No |
| 13487 | 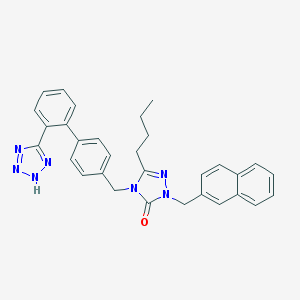 CHEMBL86986 CHEMBL86986 | C31H29N7O | 515.621 | 5 / 1 | 6.0 | No |
| 13954 | 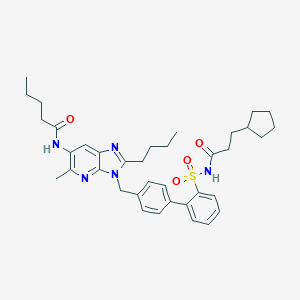 CHEMBL304947 CHEMBL304947 | C37H47N5O4S | 657.874 | 6 / 2 | 7.8 | No |
| 14080 | 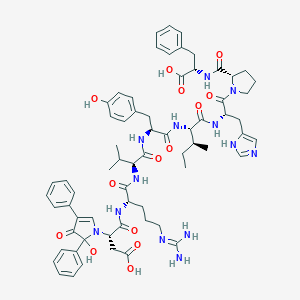 CHEMBL3275931 CHEMBL3275931 | C66H81N13O14 | 1280.45 | 17 / 13 | 4.1 | No |
| 14363 | 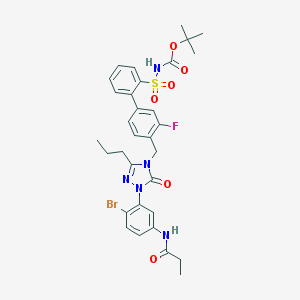 CHEMBL436396 CHEMBL436396 | C32H35BrFN5O6S | 716.623 | 8 / 2 | 5.6 | No |
| 14856 | 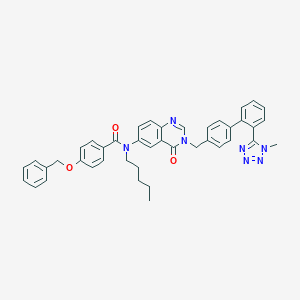 CHEMBL263038 CHEMBL263038 | C42H39N7O3 | 689.82 | 7 / 0 | 7.6 | No |
| 15538 | 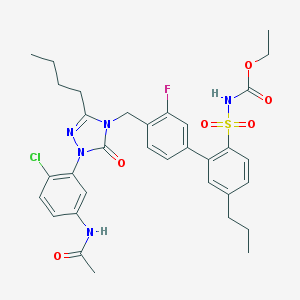 CHEMBL321313 CHEMBL321313 | C33H37ClFN5O6S | 686.196 | 8 / 2 | 6.4 | No |
| 16477 | 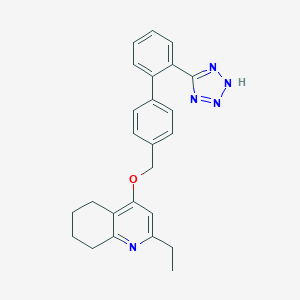 UNII-V85UF0B9WX UNII-V85UF0B9WX | C25H25N5O | 411.509 | 5 / 1 | 5.2 | No |
| 16516 | 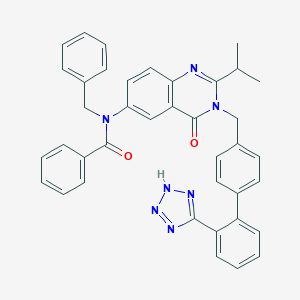 CHEMBL290669 CHEMBL290669 | C39H33N7O2 | 631.74 | 6 / 1 | 6.8 | No |
| 16614 | 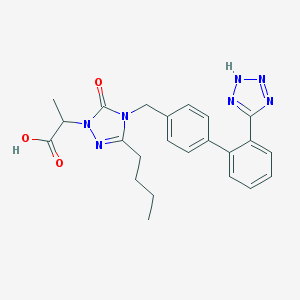 CHEMBL315469 CHEMBL315469 | C23H25N7O3 | 447.499 | 7 / 2 | 3.3 | Yes |
| 16740 | 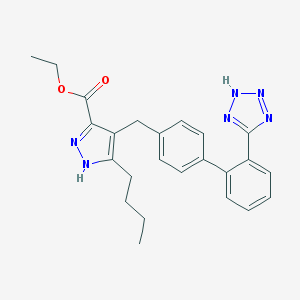 CHEMBL120716 CHEMBL120716 | C24H26N6O2 | 430.512 | 6 / 2 | 5.3 | No |
| 17190 | 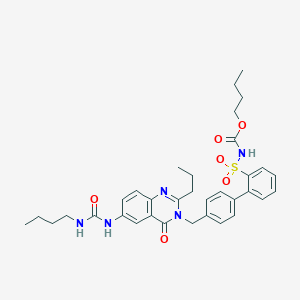 CHEMBL353615 CHEMBL353615 | C34H41N5O6S | 647.791 | 7 / 3 | 5.5 | No |
| 17260 | 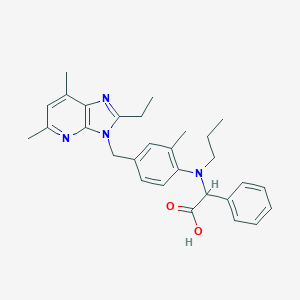 CHEMBL335066 CHEMBL335066 | C29H34N4O2 | 470.617 | 5 / 1 | 6.6 | No |
| 17787 | 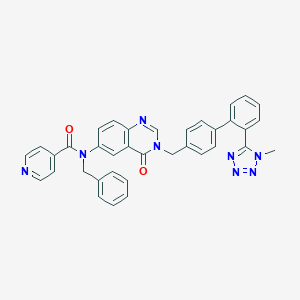 CHEMBL29063 CHEMBL29063 | C36H28N8O2 | 604.674 | 7 / 0 | 4.8 | No |
| 18112 | 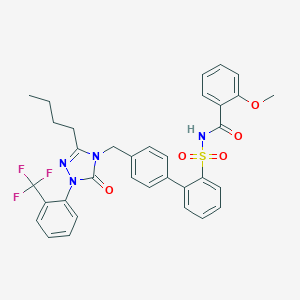 CHEMBL316904 CHEMBL316904 | C34H31F3N4O5S | 664.7 | 9 / 1 | 6.9 | No |
| 18443 |  CHEMBL26277 CHEMBL26277 | C30H26N2O3 | 462.549 | 4 / 1 | 6.1 | No |
| 18765 |  CHEMBL311386 CHEMBL311386 | C34H40FN5O6S | 665.781 | 8 / 2 | 4.5 | No |
| 19638 |  CHEMBL139325 CHEMBL139325 | C39H33Cl2N5O5S | 754.683 | 6 / 2 | 8.1 | No |
| 20477 |  CHEMBL316482 CHEMBL316482 | C31H33F3N4O5S | 630.683 | 9 / 1 | 6.9 | No |
| 23045 | 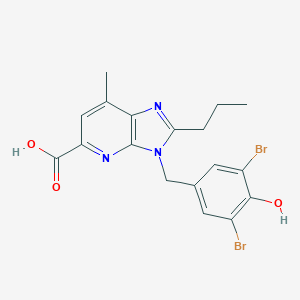 CHEMBL293926 CHEMBL293926 | C18H17Br2N3O3 | 483.16 | 5 / 2 | 4.7 | Yes |
| 23471 | 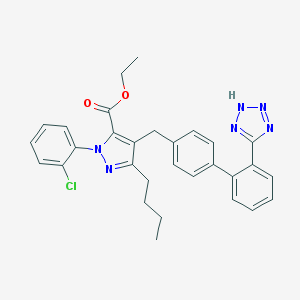 CHEMBL92821 CHEMBL92821 | C30H29ClN6O2 | 541.052 | 6 / 1 | 7.6 | No |
| 24994 | 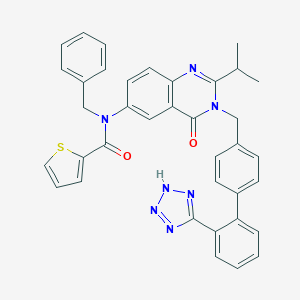 CHEMBL290768 CHEMBL290768 | C37H31N7O2S | 637.762 | 7 / 1 | 6.8 | No |
| 25002 | 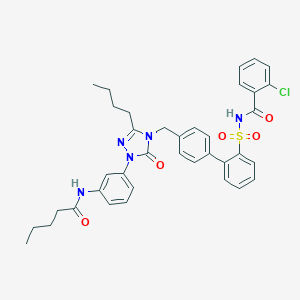 CHEMBL140619 CHEMBL140619 | C37H38ClN5O5S | 700.251 | 6 / 2 | 7.2 | No |
| 26250 |  CHEMBL299897 CHEMBL299897 | C31H34N4O3S2Si | 602.843 | 6 / 1 | N/A | No |
| 26328 |  CHEMBL345894 CHEMBL345894 | C26H25N5O3 | 455.518 | 6 / 2 | 3.7 | Yes |
| 26721 |  CHEMBL108981 CHEMBL108981 | C37H37N7O3 | 627.749 | 7 / 1 | 6.6 | No |
| 26807 |  CHEMBL24802 CHEMBL24802 | C27H23F3N6O | 504.517 | 8 / 1 | 5.5 | No |
| 26816 | 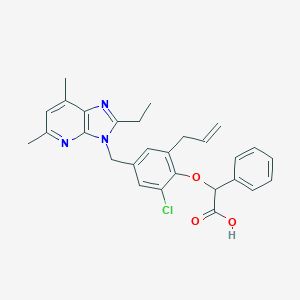 CHEMBL15729 CHEMBL15729 | C28H28ClN3O3 | 490.0 | 5 / 1 | 6.9 | No |
| 27391 | 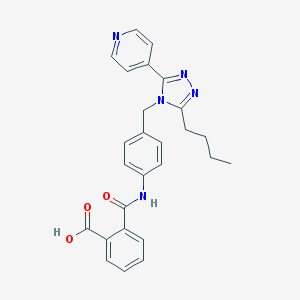 CHEMBL347050 CHEMBL347050 | C26H25N5O3 | 455.518 | 6 / 2 | 3.7 | Yes |
| 27703 | 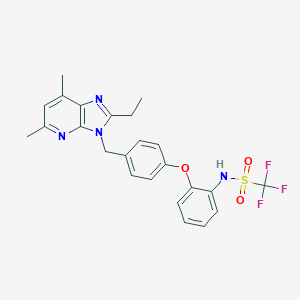 CHEMBL63280 CHEMBL63280 | C24H23F3N4O3S | 504.528 | 9 / 1 | 5.6 | No |
| 28047 | 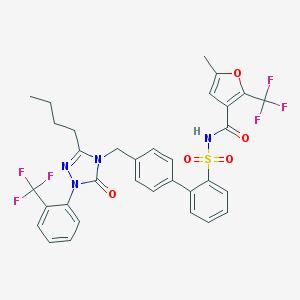 CHEMBL97075 CHEMBL97075 | C33H28F6N4O5S | 706.66 | 12 / 1 | 7.3 | No |
| 28173 | 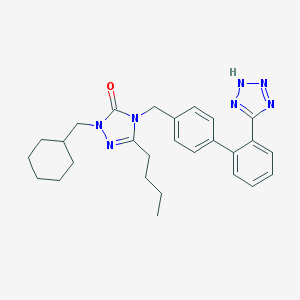 CHEMBL87239 CHEMBL87239 | C27H33N7O | 471.609 | 5 / 1 | 5.6 | No |
| 29135 | 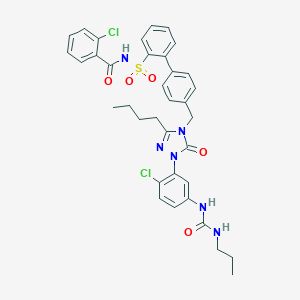 CHEMBL139452 CHEMBL139452 | C36H36Cl2N6O5S | 735.681 | 6 / 3 | 7.2 | No |
| 29386 | 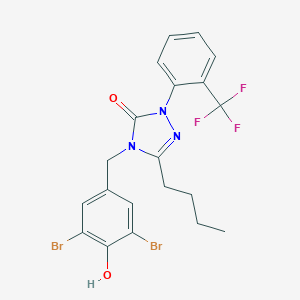 CHEMBL304655 CHEMBL304655 | C20H18Br2F3N3O2 | 549.186 | 6 / 1 | 5.9 | No |
| 29480 | 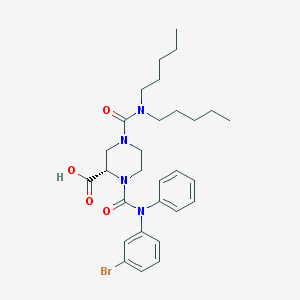 CHEMBL64861 CHEMBL64861 | C29H39BrN4O4 | 587.559 | 4 / 1 | 6.0 | No |
| 29560 | 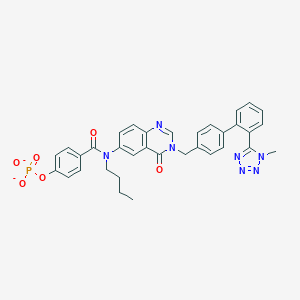 CHEMBL281890 CHEMBL281890 | C34H30N7O6P-2 | 663.631 | 10 / 0 | 3.8 | No |
| 29641 | 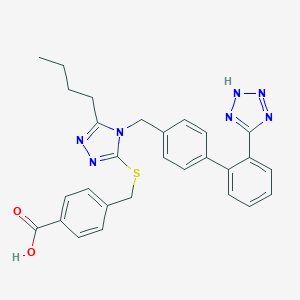 CHEMBL155008 CHEMBL155008 | C28H27N7O2S | 525.631 | 8 / 2 | 5.5 | No |
| 29686 | 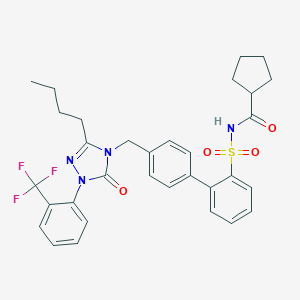 CHEMBL316273 CHEMBL316273 | C32H33F3N4O4S | 626.695 | 8 / 1 | 6.8 | No |
| 29758 | 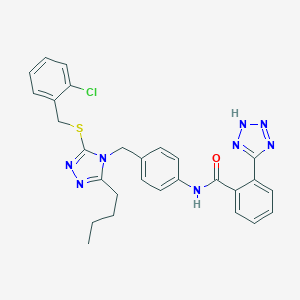 CHEMBL2237591 CHEMBL2237591 | C28H27ClN8OS | 559.089 | 7 / 2 | 5.8 | No |
| 29959 | 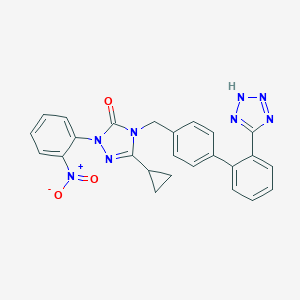 CHEMBL86853 CHEMBL86853 | C25H20N8O3 | 480.488 | 7 / 1 | 3.7 | Yes |
| 30176 | 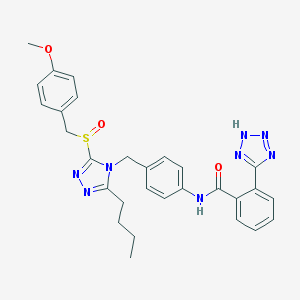 CHEMBL155554 CHEMBL155554 | C29H30N8O3S | 570.672 | 9 / 2 | 3.8 | No |
| 30293 | 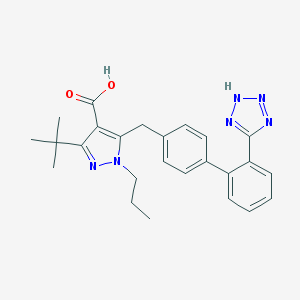 UR-7280 UR-7280 | C25H28N6O2 | 444.539 | 6 / 2 | 5.1 | No |
| 30841 | 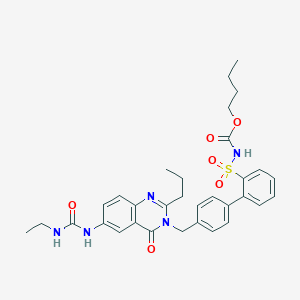 CHEMBL169221 CHEMBL169221 | C32H37N5O6S | 619.737 | 7 / 3 | 4.6 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417