

You can:
| Name | Platelet-activating factor receptor |
|---|---|
| Species | Cavia porcellus (Guinea pig) |
| Gene | PTAFR |
| Synonym | PAF-R PAFr |
| Disease | N/A for non-human GPCRs |
| Length | 342 |
| Amino acid sequence | MELNSSSRVDSEFRYTLFPIVYSIIFVLGIIANGYVLWVFARLYPSKKLNEIKIFMVNLTVADLLFLITLPLWIVYYSNQGNWFLPKFLCNLAGCLFFINTYCSVAFLGVITYNRFQAVKYPIKTAQATTRKRGIALSLVIWVAIVAAASYFLVMDSTNVVSNKAGSGNITRCFEHYEKGSKPVLIIHICIVLGFFIVFLLILFCNLVIIHTLLRQPVKQQRNAEVRRRALWMVCTVLAVFVICFVPHHMVQLPWTLAELGMWPSSNHQAINDAHQVTLCLLSTNCVLDPVIYCFLTKKFRKHLSEKLNIMRSSQKCSRVTTDTGTEMAIPINHTPVNPIKN |
| UniProt | P21556 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5136 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 200 | 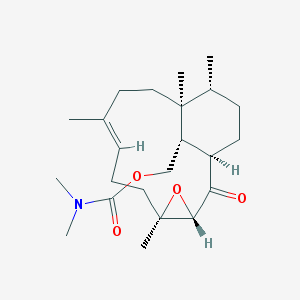 CHEMBL151310 CHEMBL151310 | C23H37NO4 | 391.552 | 4 / 0 | 4.2 | Yes |
| 916 | 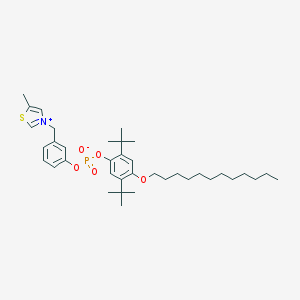 CHEMBL46754 CHEMBL46754 | C37H56NO5PS | 657.891 | 6 / 0 | 12.6 | No |
| 1683 | 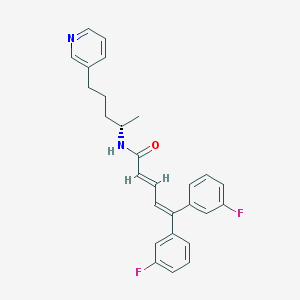 CHEMBL52933 CHEMBL52933 | C27H26F2N2O | 432.515 | 4 / 1 | 6.4 | No |
| 1685 | 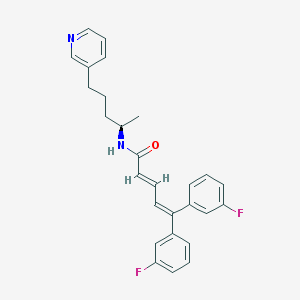 CHEMBL54935 CHEMBL54935 | C27H26F2N2O | 432.515 | 4 / 1 | 6.4 | No |
| 519709 | 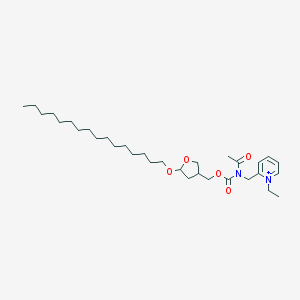 CHEMBL351410 CHEMBL351410 | C32H55N2O5+ | 547.801 | 5 / 0 | 8.8 | No |
| 519710 | 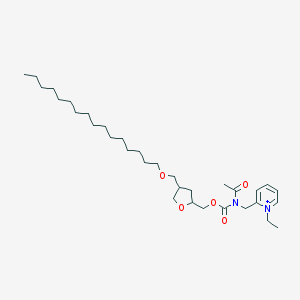 CHEMBL126981 CHEMBL126981 | C33H57N2O5+ | 561.828 | 5 / 0 | 8.7 | No |
| 519711 | 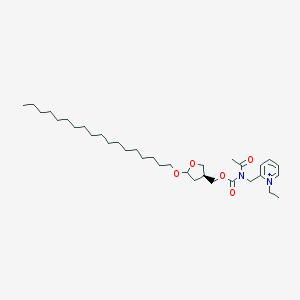 CHEMBL157989 CHEMBL157989 | C34H59N2O5+ | 575.855 | 5 / 0 | 9.9 | No |
| 519712 | 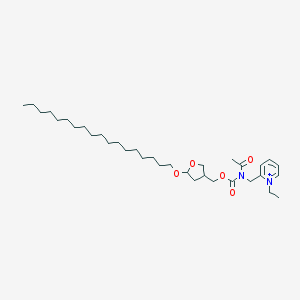 CHEMBL290840 CHEMBL290840 | C34H59N2O5+ | 575.855 | 5 / 0 | 9.9 | No |
| 519713 |  CHEMBL160994 CHEMBL160994 | C34H59N2O5+ | 575.855 | 5 / 0 | 9.9 | No |
| 3370 |  CID 15005336 CID 15005336 | C32H30ClN5O3 | 568.074 | 6 / 2 | 5.6 | No |
| 4126 |  CHEMBL158974 CHEMBL158974 | C33H57IN2O6 | 704.731 | 7 / 0 | N/A | No |
| 4252 |  2-[4-(2-Methyl-1H-imidazo[4,5-c]pyridin-1-yl)phenyl]-1,4-dihydro-5H-1-benzazepin-5-one 2-[4-(2-Methyl-1H-imidazo[4,5-c]pyridin-1-yl)phenyl]-1,4-dihydro-5H-1-benzazepin-5-one | C23H18N4O | 366.424 | 4 / 1 | 3.9 | Yes |
| 519716 | 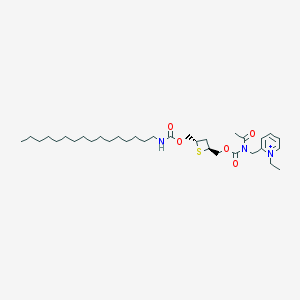 CHEMBL12183 CHEMBL12183 | C33H56N3O5S+ | 606.887 | 6 / 1 | 9.2 | No |
| 4343 | 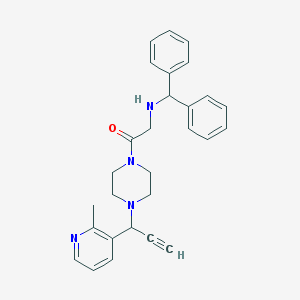 CHEMBL101957 CHEMBL101957 | C28H30N4O | 438.575 | 4 / 1 | 3.5 | Yes |
| 4669 | 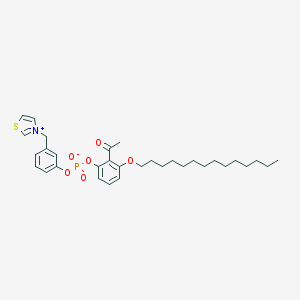 CHEMBL46870 CHEMBL46870 | C32H44NO6PS | 601.739 | 7 / 0 | 9.6 | No |
| 519718 | 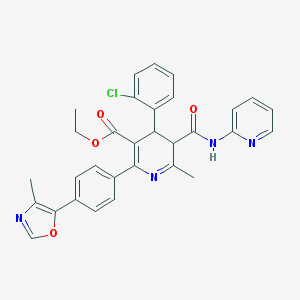 BDBM50004624 BDBM50004624 | C31H27ClN4O4 | 555.031 | 7 / 1 | 5.0 | No |
| 5079 | 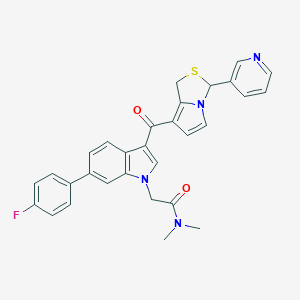 CHEMBL99053 CHEMBL99053 | C30H25FN4O2S | 524.614 | 5 / 0 | 4.4 | No |
| 5416 | 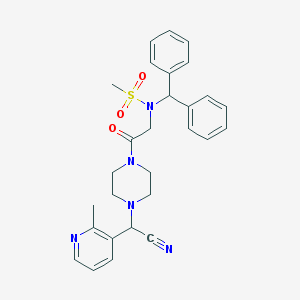 CHEMBL418989 CHEMBL418989 | C28H31N5O3S | 517.648 | 7 / 0 | 2.9 | No |
| 5784 | 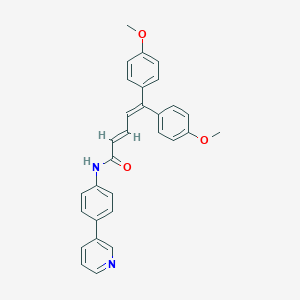 CHEMBL53014 CHEMBL53014 | C30H26N2O3 | 462.549 | 4 / 1 | 6.2 | No |
| 519724 | 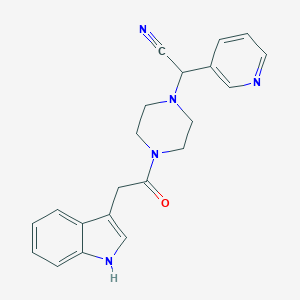 CHEMBL559013 CHEMBL559013 | C21H21N5O | 359.433 | 4 / 1 | 1.7 | Yes |
| 6169 | 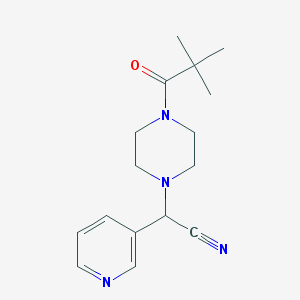 CHEMBL134025 CHEMBL134025 | C16H22N4O | 286.379 | 4 / 0 | 1.3 | Yes |
| 6932 | 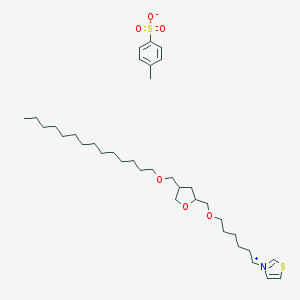 CHEMBL127766 CHEMBL127766 | C36H61NO6S2 | 668.005 | 7 / 0 | N/A | No |
| 7576 | 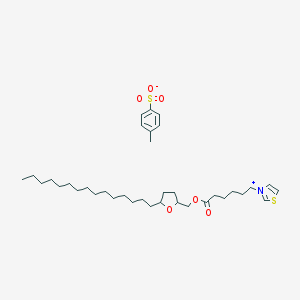 CHEMBL339676 CHEMBL339676 | C36H59NO6S2 | 665.989 | 7 / 0 | N/A | No |
| 7908 | 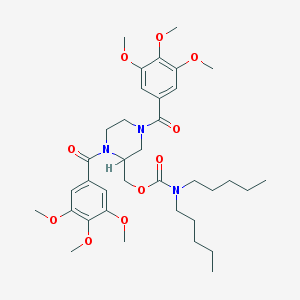 CHEMBL6263 CHEMBL6263 | C36H53N3O10 | 687.831 | 10 / 0 | 5.7 | No |
| 7929 | 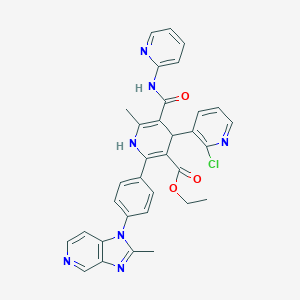 2-[4-(2-Methyl-1H-imidazo[4,5-c]pyridin-1-yl)phenyl]-4-(2-chloro-3-pyridinyl)-5-(2-pyridinylcarbamoyl)-6-methyl-1,4-dihydropyridine-3-carboxylic acid ethyl ester 2-[4-(2-Methyl-1H-imidazo[4,5-c]pyridin-1-yl)phenyl]-4-(2-chloro-3-pyridinyl)-5-(2-pyridinylcarbamoyl)-6-methyl-1,4-dihydropyridine-3-carboxylic acid ethyl ester | C33H28ClN7O3 | 606.083 | 8 / 2 | 4.8 | No |
| 519731 | 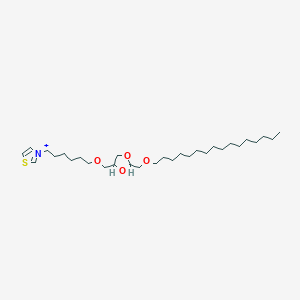 CHEMBL127875 CHEMBL127875 | C30H56NO4S+ | 526.841 | 5 / 0 | 9.6 | No |
| 519732 | 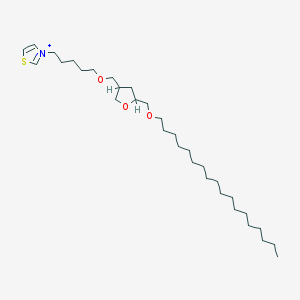 CHEMBL127009 CHEMBL127009 | C32H60NO3S+ | 538.896 | 4 / 0 | 10.9 | No |
| 8370 | 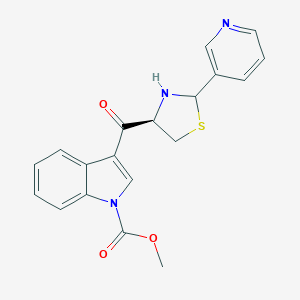 CHEMBL417563 CHEMBL417563 | C19H17N3O3S | 367.423 | 6 / 1 | 2.6 | Yes |
| 8445 | 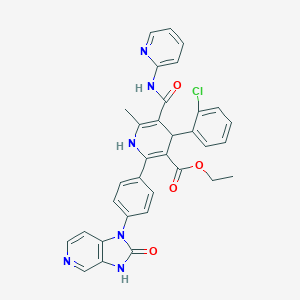 CID 15005360 CID 15005360 | C33H27ClN6O4 | 607.067 | 7 / 3 | 4.4 | No |
| 442060 | 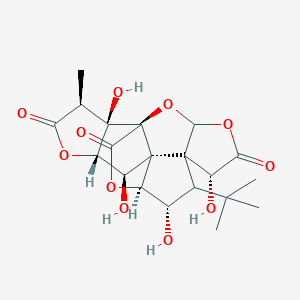 Ginkgolide C Ginkgolide C | C20H24O11 | 440.401 | 11 / 4 | -1.4 | No |
| 9089 | 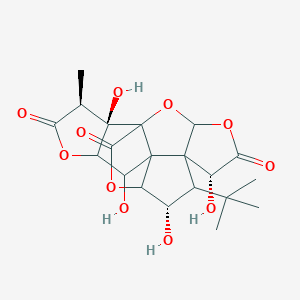 CHEMBL347488 CHEMBL347488 | C20H24O11 | 440.401 | 11 / 4 | -1.4 | No |
| 442061 | 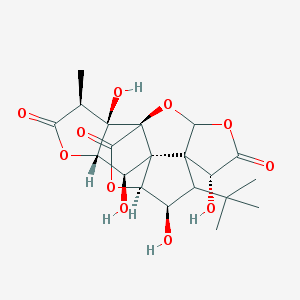 CHEMBL3349895 CHEMBL3349895 | C20H24O11 | 440.401 | 11 / 4 | -1.4 | No |
| 9092 | 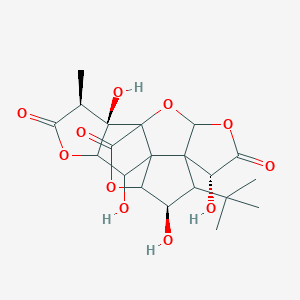 CHEMBL156251 CHEMBL156251 | C20H24O11 | 440.401 | 11 / 4 | -1.4 | No |
| 9373 | 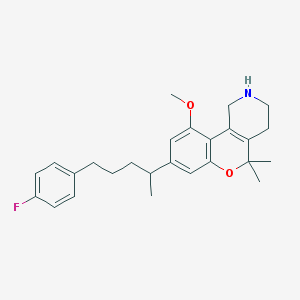 CHEMBL62322 CHEMBL62322 | C26H32FNO2 | 409.545 | 4 / 1 | 5.1 | No |
| 519735 | 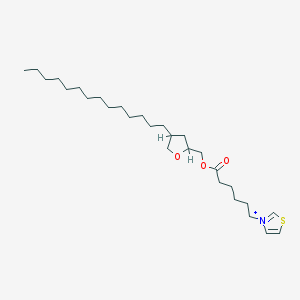 CHEMBL127416 CHEMBL127416 | C28H50NO3S+ | 480.772 | 4 / 0 | 9.6 | No |
| 10074 | 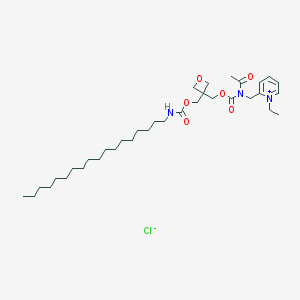 CHEMBL12352 CHEMBL12352 | C35H60ClN3O6 | 654.33 | 7 / 1 | N/A | No |
| 10833 |  CHEMBL3216875 CHEMBL3216875 | C27H29Cl2N3O2 | 498.448 | 5 / 2 | N/A | N/A |
| 11094 |  CHEMBL291177 CHEMBL291177 | C30H42NO5PS | 559.702 | 6 / 0 | 9.9 | No |
| 11371 |  CHEMBL1788193 CHEMBL1788193 | C34H60ClN3O6 | 642.319 | 7 / 1 | N/A | No |
| 11372 |  CHEMBL2093039 CHEMBL2093039 | C34H60ClN3O6 | 642.319 | 7 / 1 | N/A | No |
| 11374 |  CV-6209 CV-6209 | C34H60ClN3O6 | 642.319 | 7 / 1 | N/A | No |
| 12048 |  LEXIPAFANT LEXIPAFANT | C23H30N4O4S | 458.577 | 7 / 0 | 3.5 | Yes |
| 12244 |  CID 15005338 CID 15005338 | C33H29ClN4O3 | 565.07 | 6 / 2 | 6.0 | No |
| 12795 |  CHEMBL174262 CHEMBL174262 | C25H20N4O3 | 424.46 | 5 / 1 | 3.7 | Yes |
| 13750 |  CHEMBL421843 CHEMBL421843 | C32H55IN2O6 | 690.704 | 7 / 0 | N/A | No |
| 14009 |  CHEMBL68407 CHEMBL68407 | C25H30INO5 | 551.421 | 6 / 0 | N/A | No |
| 519749 |  BDBM50004615 BDBM50004615 | C33H33ClN4O4 | 585.101 | 7 / 0 | 5.8 | No |
| 14653 |  CHEMBL125576 CHEMBL125576 | C33H56ClN3O6 | 626.276 | 8 / 1 | N/A | No |
| 14865 |  CHEMBL318908 CHEMBL318908 | C27H27N5O2 | 453.546 | 6 / 0 | 4.0 | Yes |
| 519754 |  CHEMBL340480 CHEMBL340480 | C31H55N2O4+ | 519.791 | 4 / 1 | 8.7 | No |
| 15058 |  CHEMBL47192 CHEMBL47192 | C28H38NO5PS | 531.648 | 6 / 0 | 8.8 | No |
| 15251 |  CHEMBL421954 CHEMBL421954 | C27H36O5 | 440.58 | 5 / 0 | 6.1 | No |
| 15778 |  CHEMBL118182 CHEMBL118182 | C21H16N6O | 368.4 | 5 / 1 | 2.2 | Yes |
| 16526 |  CHEMBL64209 CHEMBL64209 | C20H21N3O3S2 | 415.526 | 6 / 1 | 2.8 | Yes |
| 519765 |  CHEMBL349327 CHEMBL349327 | C33H51N2O4+ | 539.781 | 4 / 0 | 9.2 | No |
| 17822 |  CHEMBL300742 CHEMBL300742 | C27H26F2N2O | 432.515 | 4 / 1 | 6.4 | No |
| 18141 |  CHEMBL103571 CHEMBL103571 | C28H29N5O2 | 467.573 | 5 / 0 | 3.1 | Yes |
| 519775 |  CHEMBL554536 CHEMBL554536 | C17H14ClN3OS | 343.829 | 4 / 1 | 3.1 | Yes |
| 519783 |  BDBM50004634 BDBM50004634 | C35H29N7O3 | 595.663 | 8 / 1 | 4.1 | No |
| 20161 |  CHEMBL443811 CHEMBL443811 | C27H44N4O | 440.676 | 4 / 0 | 7.3 | No |
| 519787 |  CHEMBL338883 CHEMBL338883 | C28H50NO4S+ | 496.771 | 5 / 0 | 9.4 | No |
| 21070 |  CHEMBL48129 CHEMBL48129 | C32H46NO5PS | 587.756 | 6 / 0 | 11.0 | No |
| 21993 |  CHEMBL367418 CHEMBL367418 | C30H30FN5O2 | 511.601 | 5 / 0 | 4.8 | No |
| 22020 |  CHEMBL132028 CHEMBL132028 | C26H34N2O8 | 502.564 | 8 / 0 | 2.4 | No |
| 22224 | 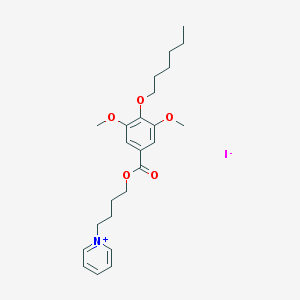 CHEMBL66814 CHEMBL66814 | C24H34INO5 | 543.442 | 6 / 0 | N/A | No |
| 519799 | 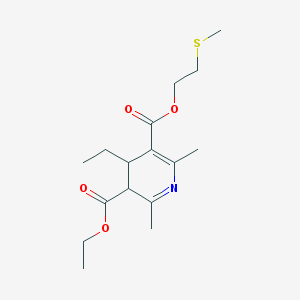 BDBM50014011 BDBM50014011 | C16H25NO4S | 327.439 | 6 / 0 | 2.5 | Yes |
| 22945 | 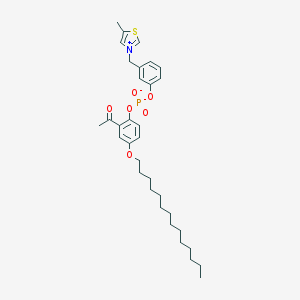 CHEMBL47159 CHEMBL47159 | C33H46NO6PS | 615.766 | 7 / 0 | 10.0 | No |
| 519805 | 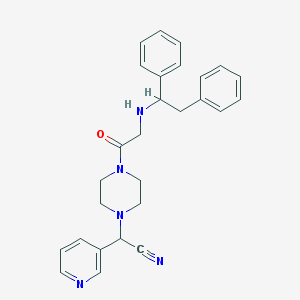 CHEMBL545773 CHEMBL545773 | C27H29N5O | 439.563 | 5 / 1 | 3.0 | Yes |
| 519811 |  CHEMBL124312 CHEMBL124312 | C30H62NO3+ | 484.83 | 3 / 0 | 10.1 | No |
| 519812 |  CHEMBL64493 CHEMBL64493 | C24H21N3O2S | 415.511 | 5 / 1 | 4.0 | Yes |
| 519815 |  CHEMBL154459 CHEMBL154459 | C34H53N2O3+ | 537.809 | 3 / 0 | 10.0 | No |
| 25648 |  CHEMBL118672 CHEMBL118672 | C27H27N7O2 | 481.56 | 6 / 0 | 2.6 | Yes |
| 25840 |  CHEMBL3215984 CHEMBL3215984 | C18H20Cl2N4O | 379.285 | 4 / 2 | N/A | N/A |
| 519816 |  BDBM50006101 BDBM50006101 | C30H54N2O6PS- | 601.804 | 7 / 1 | 8.0 | No |
| 26212 |  CHEMBL1160088 CHEMBL1160088 | C30H55N2O6PS | 602.812 | 7 / 2 | 8.0 | No |
| 26458 |  CHEMBL268597 CHEMBL268597 | C32H42N2O10 | 614.692 | 10 / 0 | 4.2 | No |
| 26732 | 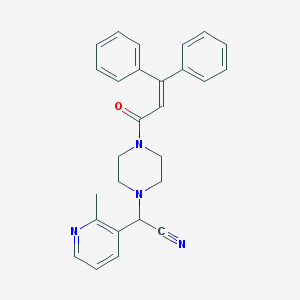 CHEMBL103903 CHEMBL103903 | C27H26N4O | 422.532 | 4 / 0 | 4.3 | Yes |
| 27159 | 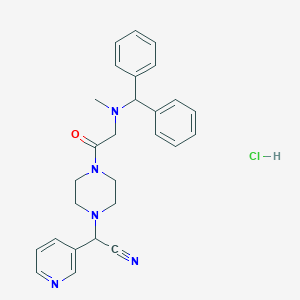 CHEMBL545072 CHEMBL545072 | C27H30ClN5O | 476.021 | 5 / 1 | N/A | N/A |
| 27880 | 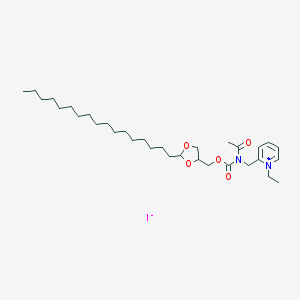 CHEMBL338689 CHEMBL338689 | C32H55IN2O5 | 674.705 | 6 / 0 | N/A | No |
| 27946 | 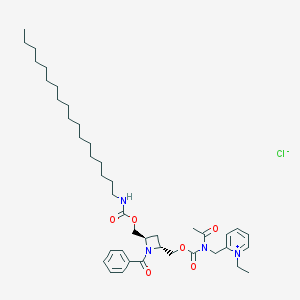 CHEMBL273638 CHEMBL273638 | C42H65ClN4O6 | 757.454 | 7 / 1 | N/A | No |
| 558119 | 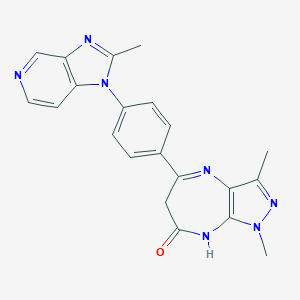 CHEMBL325928 CHEMBL325928 | C21H19N7O | 385.431 | 5 / 1 | 2.0 | Yes |
| 28979 | 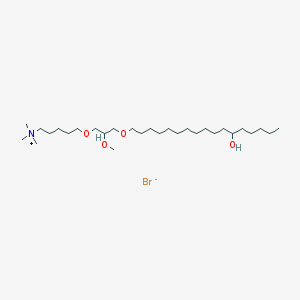 CHEMBL20522 CHEMBL20522 | C29H62BrNO4 | 568.722 | 5 / 1 | N/A | No |
| 519829 | 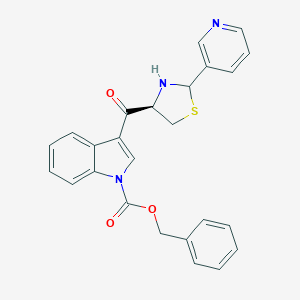 CHEMBL64860 CHEMBL64860 | C25H21N3O3S | 443.521 | 6 / 1 | 4.1 | Yes |
| 519833 | 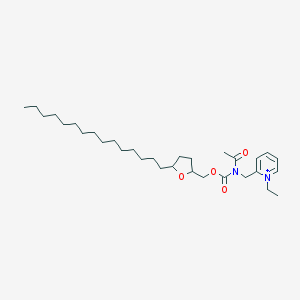 CHEMBL421618 CHEMBL421618 | C31H53N2O4+ | 517.775 | 4 / 0 | 9.0 | No |
| 29714 | 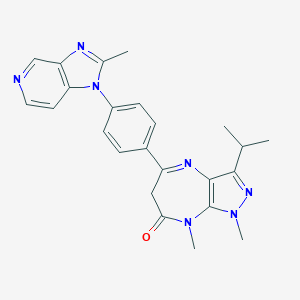 CHEMBL118793 CHEMBL118793 | C24H25N7O | 427.512 | 5 / 0 | 2.9 | Yes |
| 30851 | 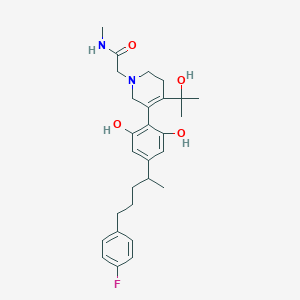 CHEMBL65715 CHEMBL65715 | C28H37FN2O4 | 484.612 | 6 / 4 | 3.8 | Yes |
| 31307 | 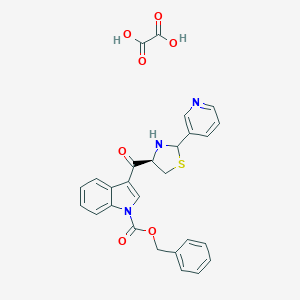 CHEMBL64860 CHEMBL64860 | C27H23N3O7S | 533.555 | 10 / 3 | N/A | No |
| 31499 | 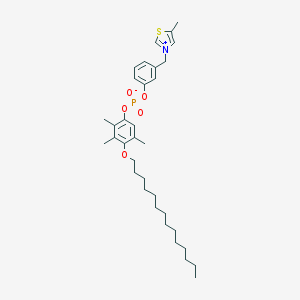 CHEMBL295148 CHEMBL295148 | C34H50NO5PS | 615.81 | 6 / 0 | 11.4 | No |
| 31575 |  CHEMBL24879 CHEMBL24879 | C34H49N3O10 | 659.777 | 10 / 1 | 5.3 | No |
| 519846 |  CHEMBL66185 CHEMBL66185 | C26H46NO5+ | 452.656 | 5 / 0 | 7.0 | No |
| 31795 |  CHEMBL103561 CHEMBL103561 | C28H31N5O | 453.59 | 5 / 1 | 3.3 | Yes |
| 519850 |  CHEMBL12554 CHEMBL12554 | C35H60N3O6+ | 618.88 | 6 / 1 | 9.5 | No |
| 519851 |  CHEMBL12882 CHEMBL12882 | C35H60N3O6+ | 618.88 | 6 / 1 | 9.5 | No |
| 519852 |  CHEMBL340164 CHEMBL340164 | C31H58NO3S+ | 524.869 | 4 / 0 | 10.2 | No |
| 32522 |  CHEMBL64636 CHEMBL64636 | C23H18FN3O2S | 419.474 | 6 / 2 | 4.0 | Yes |
| 33119 |  CHEMBL2114378 CHEMBL2114378 | C22H24O6 | 384.428 | 6 / 0 | 3.2 | Yes |
| 33120 | 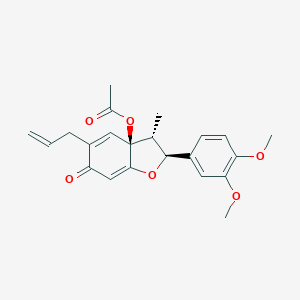 CHEMBL2115333 CHEMBL2115333 | C22H24O6 | 384.428 | 6 / 0 | 3.2 | Yes |
| 33162 | 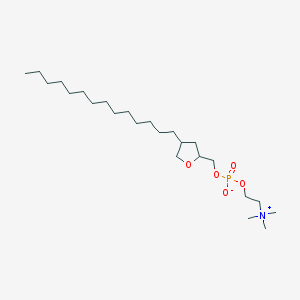 CHEMBL127531 CHEMBL127531 | C24H50NO5P | 463.64 | 5 / 0 | 6.7 | No |
| 33585 | 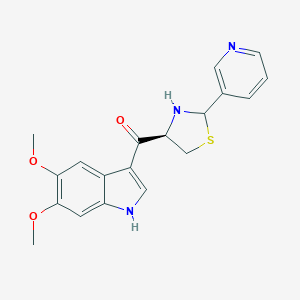 CHEMBL293653 CHEMBL293653 | C19H19N3O3S | 369.439 | 6 / 2 | 2.3 | Yes |
| 33661 | 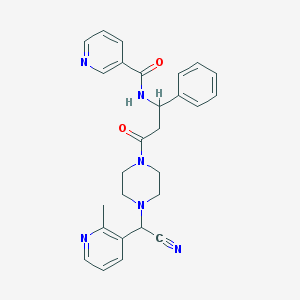 CHEMBL102070 CHEMBL102070 | C27H28N6O2 | 468.561 | 6 / 1 | 1.7 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417