

You can:
| Name | Melanocortin receptor 4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | MC4R |
| Synonym | MC4-R MC4 receptor |
| Disease | Obesity; Sexual dysfunction Obesity; Diabetes Obesity Metabolic disorders Sexual dysfunction [ Show all ] |
| Length | 332 |
| Amino acid sequence | MVNSTHRGMHTSLHLWNRSSYRLHSNASESLGKGYSDGGCYEQLFVSPEVFVTLGVISLLENILVIVAIAKNKNLHSPMYFFICSLAVADMLVSVSNGSETIVITLLNSTDTDAQSFTVNIDNVIDSVICSSLLASICSLLSIAVDRYFTIFYALQYHNIMTVKRVGIIISCIWAACTVSGILFIIYSDSSAVIICLITMFFTMLALMASLYVHMFLMARLHIKRIAVLPGTGAIRQGANMKGAITLTILIGVFVVCWAPFFLHLIFYISCPQNPYCVCFMSHFNLYLILIMCNSIIDPLIYALRSQELRKTFKEIICCYPLGGLCDLSSRY |
| UniProt | P32245 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P32245 |
| 3D structure model | This predicted structure model is from GPCR-EXP P32245. |
| BioLiP | N/A |
| Therapeutic Target Database | T72458 |
| ChEMBL | CHEMBL259 |
| IUPHAR | 285 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 206 | 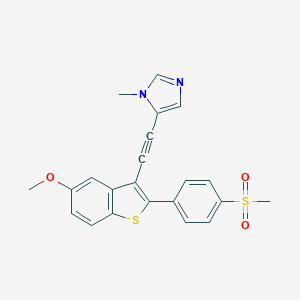 MLS002473569 MLS002473569 | C22H18N2O3S2 | 422.517 | 5 / 0 | 4.0 | Yes |
| 241 | 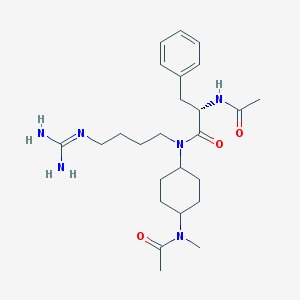 CHEMBL75475 CHEMBL75475 | C25H40N6O3 | 472.634 | 4 / 3 | 1.1 | Yes |
| 447 | 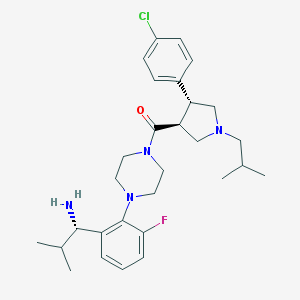 CHEMBL394160 CHEMBL394160 | C29H40ClFN4O | 515.114 | 5 / 1 | 5.1 | No |
| 1346 | 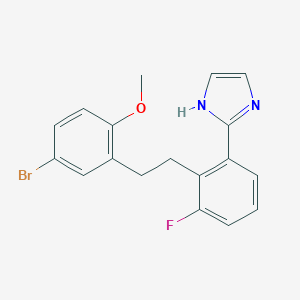 CHEMBL184736 CHEMBL184736 | C18H16BrFN2O | 375.241 | 3 / 1 | 4.7 | Yes |
| 1367 |  CHEMBL596804 CHEMBL596804 | C30H28FN5O3 | 525.584 | 5 / 1 | 4.3 | No |
| 1368 |  CHEMBL598021 CHEMBL598021 | C30H28FN5O3 | 525.584 | 5 / 1 | 4.3 | No |
| 1371 |  CHEMBL596602 CHEMBL596602 | C30H28FN5O3 | 525.584 | 5 / 1 | 4.3 | No |
| 441725 |  CHEMBL2035935 CHEMBL2035935 | C20H23N3O2 | 337.423 | 3 / 3 | 1.1 | Yes |
| 1632 |  CHEMBL185094 CHEMBL185094 | C39H43ClFN5O2 | 668.254 | 6 / 3 | 6.3 | No |
| 441737 |  CHEMBL2035943 CHEMBL2035943 | C28H35ClN4O2 | 495.064 | 4 / 2 | 3.2 | Yes |
| 1725 |  CHEMBL1801095 CHEMBL1801095 | C35H46ClF2N3O2 | 614.219 | 5 / 0 | 6.4 | No |
| 1726 |  CHEMBL1801094 CHEMBL1801094 | C35H46ClF2N3O2 | 614.219 | 5 / 0 | 6.4 | No |
| 1838 |  CHEMBL262319 CHEMBL262319 | C33H37Cl2F3N4O | 633.581 | 7 / 1 | 6.7 | No |
| 1898 |  CHEMBL181161 CHEMBL181161 | C37H45ClN10O5 | 745.282 | 6 / 9 | 2.0 | No |
| 2155 |  CHEMBL403967 CHEMBL403967 | C33H47ClFN5O2 | 600.22 | 6 / 1 | 4.8 | No |
| 2352 |  CHEMBL454916 CHEMBL454916 | C28H38Cl2N4O3 | 549.537 | 5 / 2 | 4.8 | No |
| 2390 | 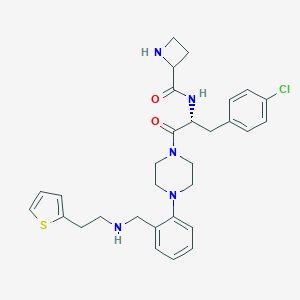 CHEMBL441675 CHEMBL441675 | C30H36ClN5O2S | 566.161 | 6 / 3 | 4.1 | No |
| 2995 | 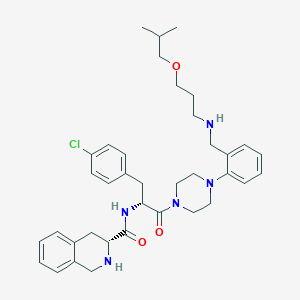 CHEMBL366708 CHEMBL366708 | C37H48ClN5O3 | 646.273 | 6 / 3 | 5.4 | No |
| 3000 | 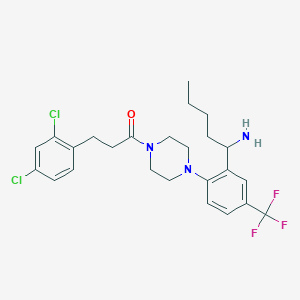 CHEMBL215974 CHEMBL215974 | C25H30Cl2F3N3O | 516.43 | 6 / 1 | 6.0 | No |
| 3269 | 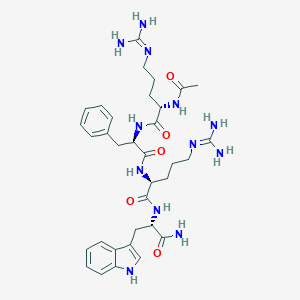 CHEMBL3287059 CHEMBL3287059 | C34H48N12O5 | 704.837 | 7 / 10 | -1.3 | No |
| 3270 |  CHEMBL3287061 CHEMBL3287061 | C34H48N12O5 | 704.837 | 7 / 10 | -1.3 | No |
| 3306 |  CHEMBL212976 CHEMBL212976 | C37H43N5O3 | 605.783 | 4 / 3 | 5.0 | No |
| 3338 |  MLS000544661 MLS000544661 | C18H12Br2N4 | 444.13 | 3 / 0 | 4.6 | Yes |
| 3806 |  CHEMBL235798 CHEMBL235798 | C26H29N5O | 427.552 | 2 / 3 | 3.3 | Yes |
| 3965 |  CHEMBL236014 CHEMBL236014 | C27H36Cl2FN5O2 | 552.516 | 6 / 3 | 3.6 | No |
| 3968 |  CHEMBL236015 CHEMBL236015 | C27H36Cl2FN5O2 | 552.516 | 6 / 3 | 3.6 | No |
| 3992 |  CHEMBL412174 CHEMBL412174 | C52H67BrN12O8 | 1068.09 | 9 / 11 | 3.0 | No |
| 441866 |  CHEMBL1775057 CHEMBL1775057 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 441868 | 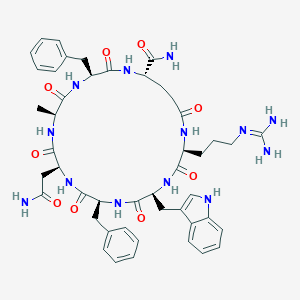 CHEMBL1775063 CHEMBL1775063 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 441873 | 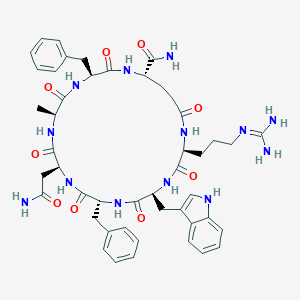 CHEMBL1775064 CHEMBL1775064 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 4249 | 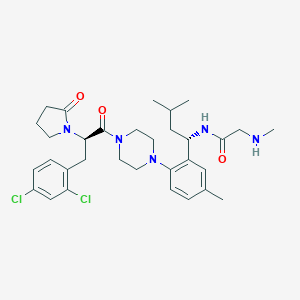 CHEMBL412023 CHEMBL412023 | C32H43Cl2N5O3 | 616.628 | 5 / 2 | 5.0 | No |
| 4622 | 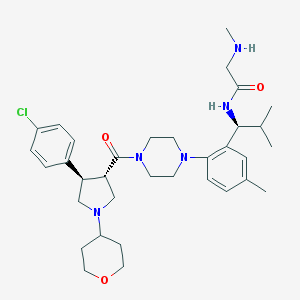 CHEMBL402017 CHEMBL402017 | C34H48ClN5O3 | 610.24 | 6 / 2 | 4.3 | No |
| 4707 | 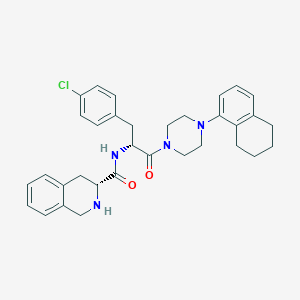 CHEMBL2113144 CHEMBL2113144 | C33H37ClN4O2 | 557.135 | 4 / 2 | 5.8 | No |
| 4806 | 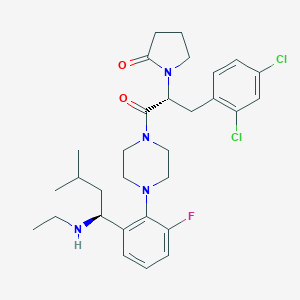 CHEMBL391915 CHEMBL391915 | C30H39Cl2FN4O2 | 577.566 | 5 / 1 | 5.7 | No |
| 4885 | 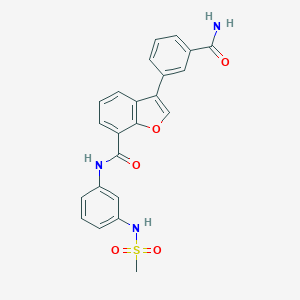 MLS003120640 MLS003120640 | C23H19N3O5S | 449.481 | 6 / 3 | 2.6 | Yes |
| 5004 | 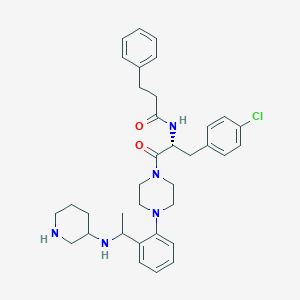 CHEMBL361210 CHEMBL361210 | C35H44ClN5O2 | 602.22 | 5 / 3 | 5.1 | No |
| 5081 |  BDBM50355384 BDBM50355384 | C139H172N48O21 | 2851.22 | 33 / 33 | -2.4 | No |
| 5288 |  MLS000528029 MLS000528029 | C20H20N2O5S | 400.449 | 6 / 2 | 3.7 | Yes |
| 5393 |  MLS003124182 MLS003124182 | C23H22N4O | 370.456 | 3 / 1 | 4.3 | Yes |
| 5474 |  CHEMBL441532 CHEMBL441532 | C29H32Cl2N6O3 | 583.514 | 6 / 3 | 2.9 | No |
| 5836 | 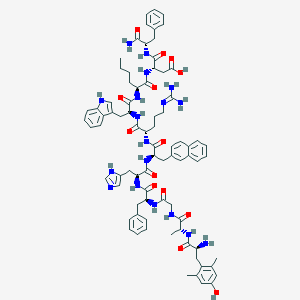 BDBM50321606 BDBM50321606 | C80H98N18O14 | 1535.78 | 17 / 18 | 2.2 | No |
| 5980 | 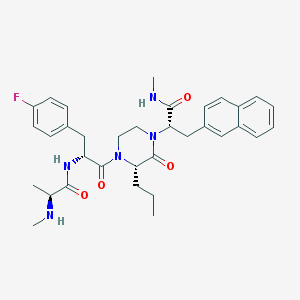 CHEMBL504349 CHEMBL504349 | C34H42FN5O4 | 603.739 | 6 / 3 | 4.3 | No |
| 6070 | 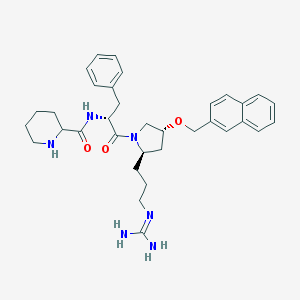 CHEMBL213752 CHEMBL213752 | C34H44N6O3 | 584.765 | 5 / 4 | 3.6 | No |
| 6073 | 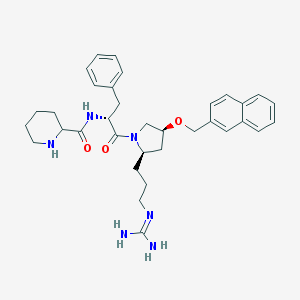 CHEMBL210009 CHEMBL210009 | C34H44N6O3 | 584.765 | 5 / 4 | 3.6 | No |
| 6078 |  CHEMBL213956 CHEMBL213956 | C34H44N6O3 | 584.765 | 5 / 4 | 3.6 | No |
| 6080 |  CHEMBL210011 CHEMBL210011 | C34H44N6O3 | 584.765 | 5 / 4 | 3.6 | No |
| 6154 |  MLS002156773 MLS002156773 | C23H20FN5O2S | 449.504 | 8 / 0 | 3.3 | Yes |
| 6185 |  CHEMBL190551 CHEMBL190551 | C45H67N13O8 | 918.114 | 10 / 10 | -0.2 | No |
| 6466 | 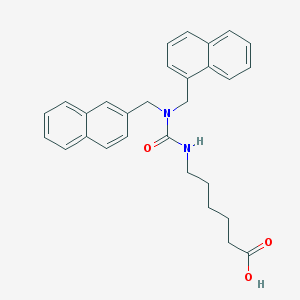 CHEMBL8825 CHEMBL8825 | C29H30N2O3 | 454.57 | 3 / 2 | 5.6 | No |
| 6521 | 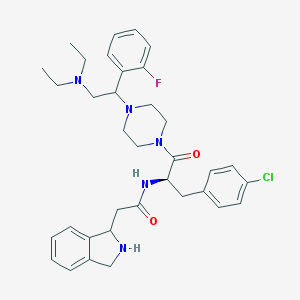 CHEMBL380855 CHEMBL380855 | C35H43ClFN5O2 | 620.21 | 6 / 2 | 4.5 | No |
| 6556 | 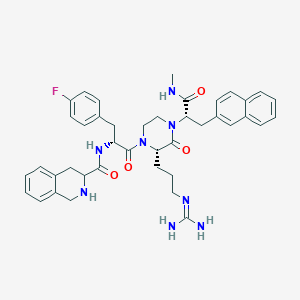 CHEMBL215895 CHEMBL215895 | C41H47FN8O4 | 734.877 | 7 / 5 | 3.4 | No |
| 441955 | 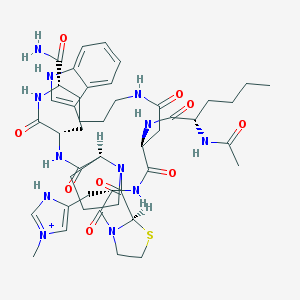 CHEMBL267360 CHEMBL267360 | C45H63N12O9S+ | 948.134 | 10 / 9 | 0.4 | No |
| 6819 |  MLS001004438 MLS001004438 | C18H16Cl2N2O4 | 395.236 | 4 / 1 | 3.5 | Yes |
| 6895 |  CHEMBL539306 CHEMBL539306 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6897 |  CHEMBL553000 CHEMBL553000 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6898 |  CHEMBL542968 CHEMBL542968 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6899 | 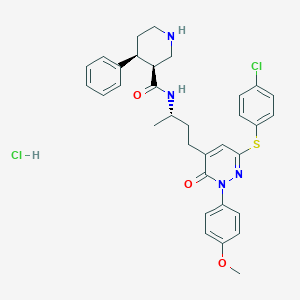 CHEMBL543430 CHEMBL543430 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6900 | 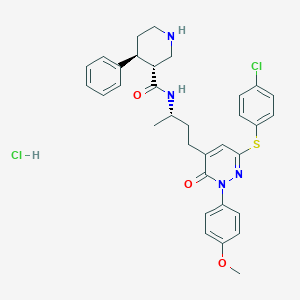 CHEMBL540320 CHEMBL540320 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6901 | 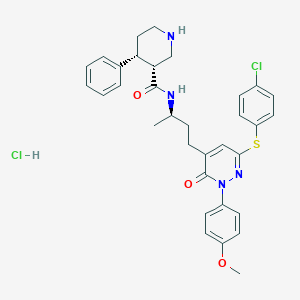 CHEMBL538589 CHEMBL538589 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6902 | 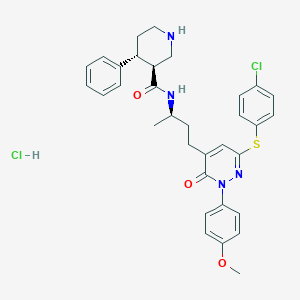 CHEMBL540077 CHEMBL540077 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 6903 | 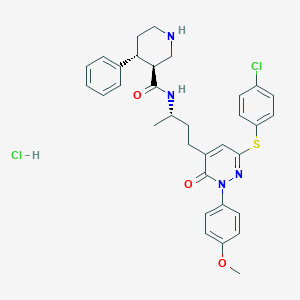 CHEMBL539813 CHEMBL539813 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 7140 | 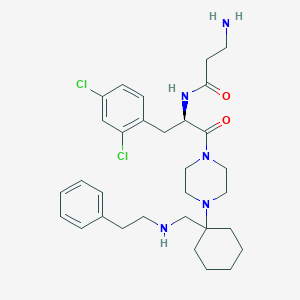 CHEMBL365237 CHEMBL365237 | C31H43Cl2N5O2 | 588.618 | 5 / 3 | 4.3 | No |
| 7550 | 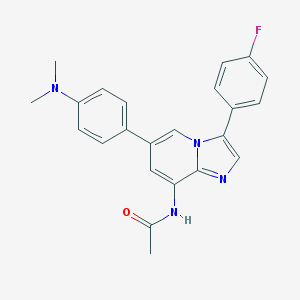 MLS003124195 MLS003124195 | C23H21FN4O | 388.446 | 4 / 1 | 4.5 | Yes |
| 7588 | 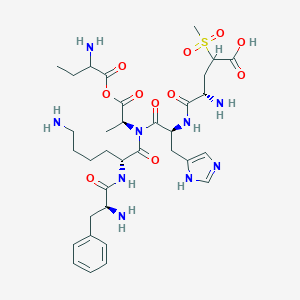 BDBM82410 BDBM82410 | C34H51N9O11S | 793.894 | 16 / 8 | -4.1 | No |
| 7707 |  CHEMBL381739 CHEMBL381739 | C42H57N11O7 | 827.988 | 8 / 9 | 1.2 | No |
| 7801 |  CHEMBL211462 CHEMBL211462 | C25H31F4N3O | 465.537 | 7 / 1 | 4.8 | Yes |
| 7872 |  CHEMBL2113004 CHEMBL2113004 | C34H46ClN5O4S | 656.283 | 7 / 1 | 4.8 | No |
| 463855 |  CHEMBL3644314 CHEMBL3644314 | C47H69N17O9 | 1016.18 | 12 / 15 | -3.1 | No |
| 8093 |  CHEMBL250778 CHEMBL250778 | C26H35ClN4O3S | 519.101 | 6 / 1 | 3.2 | No |
| 8175 |  CHEMBL445669 CHEMBL445669 | C41H51N3O4 | 649.876 | 5 / 3 | 7.7 | No |
| 8289 |  CHEMBL384138 CHEMBL384138 | C22H24Cl2F3N3O | 474.349 | 6 / 1 | 4.6 | Yes |
| 8558 |  MLS000699691 MLS000699691 | C22H21ClN2O5 | 428.869 | 6 / 1 | 4.7 | Yes |
| 8708 |  CHEMBL179837 CHEMBL179837 | C35H37ClN4O4S | 645.215 | 6 / 1 | 6.1 | No |
| 8759 |  CHEMBL2370967 CHEMBL2370967 | C81H105N21O15 | 1612.86 | 19 / 20 | 0.2 | No |
| 8929 |  2,4-Dinitro-benzoic acid 3-(morpholine-4-carbothioyl)-phenyl ester 2,4-Dinitro-benzoic acid 3-(morpholine-4-carbothioyl)-phenyl ester | C18H15N3O7S | 417.392 | 8 / 0 | 2.7 | Yes |
| 8980 |  MLS003124222 MLS003124222 | C25H23N3O5 | 445.475 | 6 / 1 | 3.8 | Yes |
| 8991 |  CHEMBL237070 CHEMBL237070 | C32H42FN3O | 503.706 | 3 / 2 | 6.3 | No |
| 9021 |  CID 44363770 CID 44363770 | C79H109N23O14 | 1604.89 | 18 / 21 | 0.7 | No |
| 9187 |  CHEMBL341055 CHEMBL341055 | C37H44ClN5O5S | 706.299 | 7 / 0 | 4.1 | No |
| 9335 |  CHEMBL205594 CHEMBL205594 | C25H32BrFN4O2 | 519.459 | 5 / 2 | 5.2 | No |
| 9482 |  CHEMBL232775 CHEMBL232775 | C29H49N5O2 | 499.744 | 5 / 1 | 5.6 | No |
| 9539 |  CHEMBL1923663 CHEMBL1923663 | C44H54N12O6 | 846.994 | 8 / 10 | 1.7 | No |
| 9642 |  CHEMBL248700 CHEMBL248700 | C27H31ClN4O2 | 479.021 | 4 / 2 | 3.7 | Yes |
| 9772 |  CHEMBL237930 CHEMBL237930 | C29H31N3O | 437.587 | 2 / 2 | 4.7 | Yes |
| 9868 |  AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 464076 |  SCHEMBL8369982 SCHEMBL8369982 | C54H73ClN16O10 | 1141.73 | 12 / 14 | -0.8 | No |
| 10113 |  CHEMBL591026 CHEMBL591026 | C32H33N5O4 | 551.647 | 5 / 1 | 4.1 | No |
| 10142 |  CHEMBL508501 CHEMBL508501 | C54H69N15O9 | 1072.24 | 11 / 12 | 0.9 | No |
| 10144 |  CHEMBL509582 CHEMBL509582 | C54H69N15O9 | 1072.24 | 11 / 12 | 0.9 | No |
| 10222 |  MLS000416520 MLS000416520 | C21H19BrN2O6S2 | 539.415 | 8 / 1 | 4.1 | No |
| 10341 |  CHEMBL396429 CHEMBL396429 | C32H42ClF3N4O3 | 623.158 | 8 / 1 | 5.8 | No |
| 10373 |  CID 16151595 CID 16151595 | C77H103N21O15 | 1562.8 | 19 / 22 | -0.3 | No |
| 10800 |  MLS000698493 MLS000698493 | C21H25ClN2O2 | 372.893 | 3 / 2 | 4.9 | Yes |
| 10813 |  CHEMBL191120 CHEMBL191120 | C44H65N13O8 | 904.087 | 10 / 11 | -0.5 | No |
| 11386 |  MLS002166510 MLS002166510 | C24H26O7 | 426.465 | 7 / 0 | 4.9 | Yes |
| 11538 |  CHEMBL383117 CHEMBL383117 | C26H36BrClN4O2 | 551.954 | 4 / 2 | 6.5 | No |
| 536298 |  CHEMBL3895073 CHEMBL3895073 | C50H70N18O9 | 1067.23 | 12 / 15 | -2.6 | No |
| 464291 |  SCHEMBL7863286 SCHEMBL7863286 | C50H70N18O9 | 1067.23 | 12 / 15 | -2.6 | No |
| 11951 |  CID 56672277 CID 56672277 | C163H214N54O27 | 3361.86 | 39 / 42 | -1.9 | No |
| 459326 |  CHEMBL1196971 CHEMBL1196971 | C31H48FN5O3 | 557.755 | 6 / 3 | 3.9 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417