

You can:
| Name | Adenosine receptor A3 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Adora3 |
| Synonym | A3 receptor A3AR Adenosine receptor A3 ARA3 TGPCR1 |
| Disease | N/A for non-human GPCRs |
| Length | 320 |
| Amino acid sequence | MKANNTTTSALWLQITYITMEAAIGLCAVVGNMLVIWVVKLNRTLRTTTFYFIVSLALADIAVGVLVIPLAIAVSLEVQMHFYACLFMSCVLLVFTHASIMSLLAIAVDRYLRVKLTVRYRTVTTQRRIWLFLGLCWLVSFLVGLTPMFGWNRKVTLELSQNSSTLSCHFRSVVGLDYMVFFSFITWILIPLVVMCIIYLDIFYIIRNKLSQNLTGFRETRAFYGREFKTAKSLFLVLFLFALCWLPLSIINFVSYFNVKIPEIAMCLGILLSHANSMMNPIVYACKIKKFKETYFVILRACRLCQTSDSLDSNLEQTTE |
| UniProt | P28647 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3360 |
| IUPHAR | 21 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 653 | 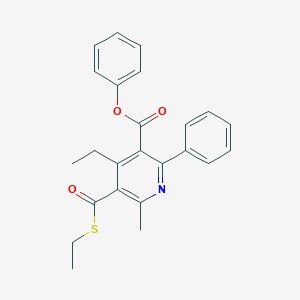 CHEMBL323403 CHEMBL323403 | C24H23NO3S | 405.512 | 5 / 0 | 5.9 | No |
| 521464 | 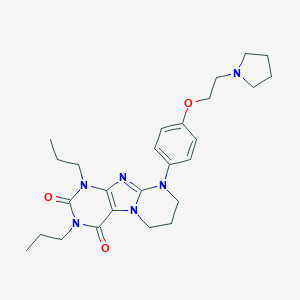 CHEMBL3827623 CHEMBL3827623 | C26H36N6O3 | 480.613 | 6 / 0 | 3.9 | Yes |
| 1205 | 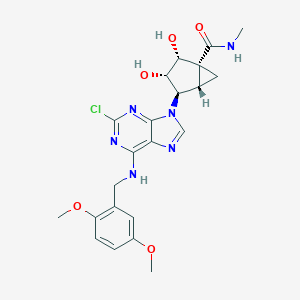 CHEMBL178846 CHEMBL178846 | C22H25ClN6O5 | 488.929 | 9 / 4 | 1.3 | Yes |
| 3857 | 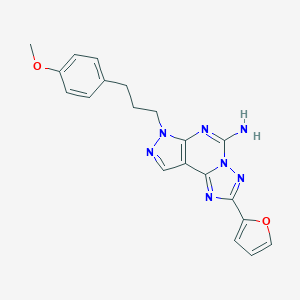 316173-57-6 316173-57-6 | C20H19N7O2 | 389.419 | 7 / 1 | 2.7 | Yes |
| 4603 | 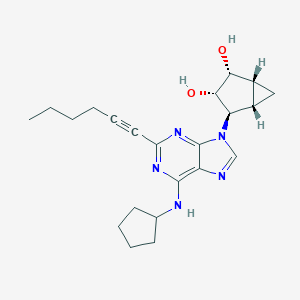 CHEMBL3125715 CHEMBL3125715 | C22H29N5O2 | 395.507 | 6 / 3 | 3.1 | Yes |
| 4752 | 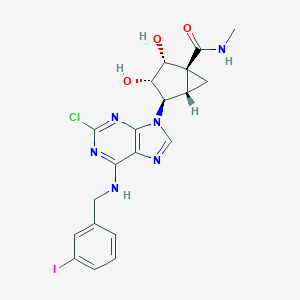 CHEMBL27376 CHEMBL27376 | C20H20ClIN6O3 | 554.773 | 7 / 4 | 2.0 | No |
| 6721 | 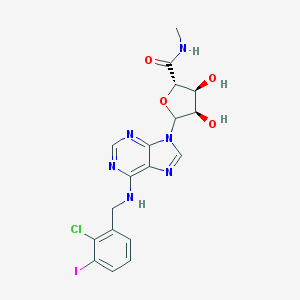 CHEMBL608327 CHEMBL608327 | C18H18ClIN6O4 | 544.734 | 8 / 4 | 1.5 | No |
| 7419 | 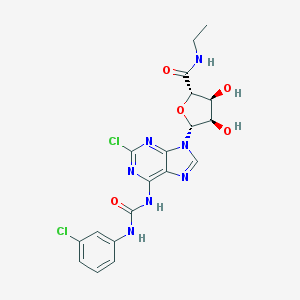 CHEMBL380914 CHEMBL380914 | C19H19Cl2N7O5 | 496.305 | 8 / 5 | 1.3 | Yes |
| 8515 | 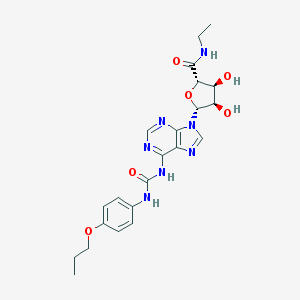 CHEMBL206807 CHEMBL206807 | C22H27N7O6 | 485.501 | 9 / 5 | 0.6 | Yes |
| 8518 | 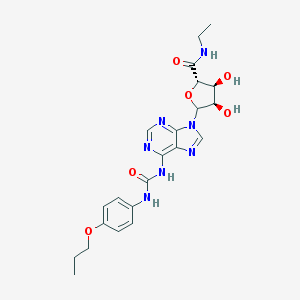 CHEMBL610394 CHEMBL610394 | C22H27N7O6 | 485.501 | 9 / 5 | 0.6 | Yes |
| 10225 | 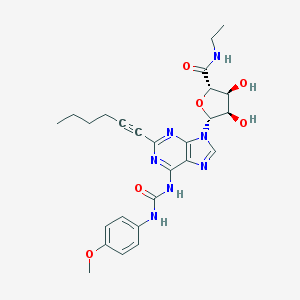 CHEMBL413046 CHEMBL413046 | C26H31N7O6 | 537.577 | 9 / 5 | 2.0 | No |
| 11438 | 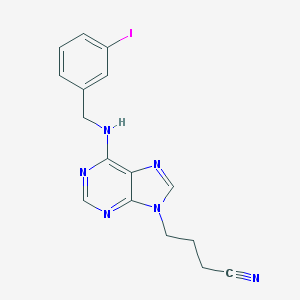 CHEMBL2113704 CHEMBL2113704 | C16H15IN6 | 418.242 | 5 / 1 | 2.7 | Yes |
| 11517 | 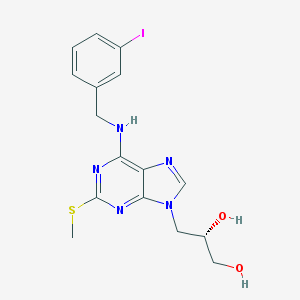 CHEMBL2113685 CHEMBL2113685 | C16H18IN5O2S | 471.317 | 7 / 3 | 2.0 | Yes |
| 14168 | 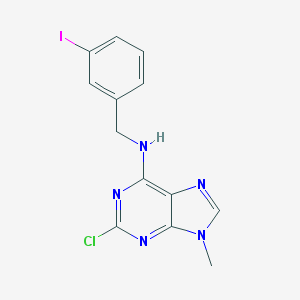 CHEMBL2113702 CHEMBL2113702 | C13H11ClIN5 | 399.62 | 4 / 1 | 3.5 | Yes |
| 16714 | 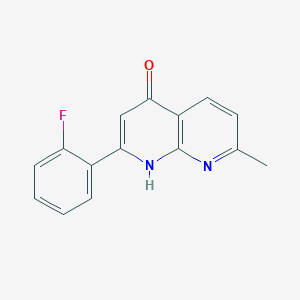 CHEMBL95384 CHEMBL95384 | C15H11FN2O | 254.264 | 4 / 1 | 3.0 | Yes |
| 17483 | 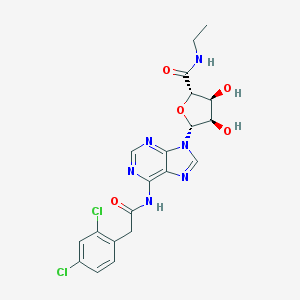 CHEMBL383548 CHEMBL383548 | C20H20Cl2N6O5 | 495.317 | 8 / 4 | 1.2 | Yes |
| 20224 | 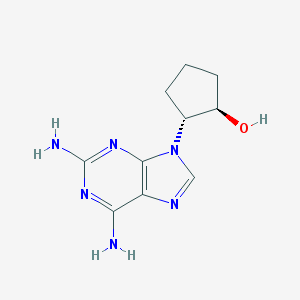 CHEMBL2112597 CHEMBL2112597 | C10H14N6O | 234.263 | 6 / 3 | -0.1 | Yes |
| 522081 | 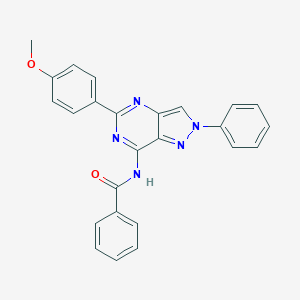 CHEMBL3751909 CHEMBL3751909 | C25H19N5O2 | 421.46 | 5 / 1 | 4.4 | Yes |
| 22453 | 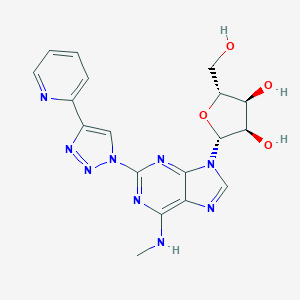 CHEMBL218326 CHEMBL218326 | C18H19N9O4 | 425.409 | 11 / 4 | -0.9 | No |
| 23107 | 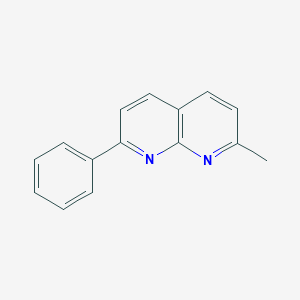 2-methyl-7-phenyl-1,8-naphthyridine 2-methyl-7-phenyl-1,8-naphthyridine | C15H12N2 | 220.275 | 2 / 0 | 3.5 | Yes |
| 24131 | 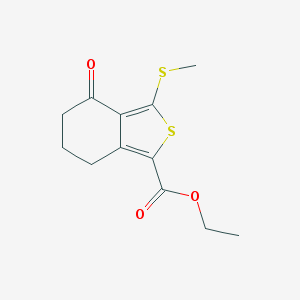 168279-54-7 168279-54-7 | C12H14O3S2 | 270.361 | 5 / 0 | 3.1 | Yes |
| 24604 | 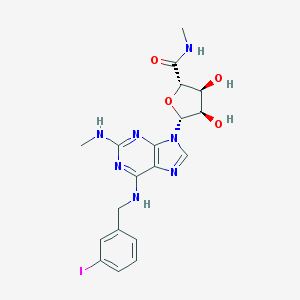 CHEMBL2113562 CHEMBL2113562 | C19H22IN7O4 | 539.334 | 9 / 5 | 1.2 | No |
| 24836 | 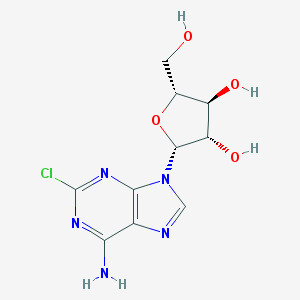 CHEMBL2112076 CHEMBL2112076 | C10H12ClN5O4 | 301.687 | 8 / 4 | -0.1 | Yes |
| 24851 | 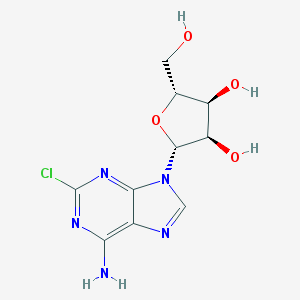 2-Chloroadenosine 2-Chloroadenosine | C10H12ClN5O4 | 301.687 | 8 / 4 | -0.1 | Yes |
| 25398 | 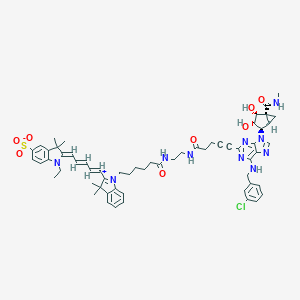 CHEMBL574212 CHEMBL574212 | C60H69ClN10O8S | 1125.78 | 13 / 6 | 6.3 | No |
| 25509 | 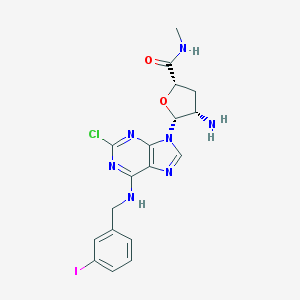 CHEMBL439976 CHEMBL439976 | C18H19ClIN7O2 | 527.751 | 7 / 3 | 2.0 | No |
| 26374 | 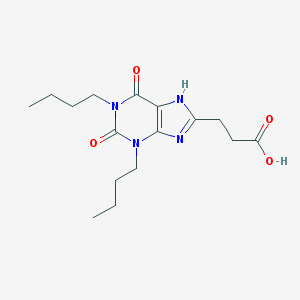 CHEMBL115608 CHEMBL115608 | C16H24N4O4 | 336.392 | 5 / 2 | 2.8 | Yes |
| 26795 | 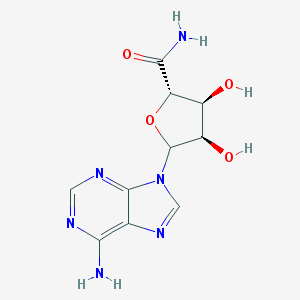 CHEMBL605866 CHEMBL605866 | C10H12N6O4 | 280.244 | 8 / 4 | -1.5 | Yes |
| 27199 | 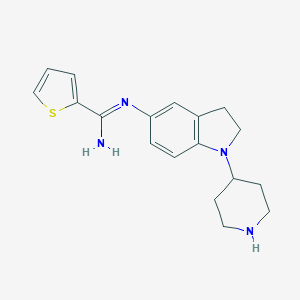 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27778 | 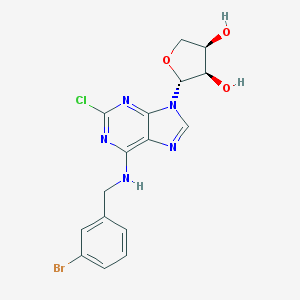 CHEMBL561976 CHEMBL561976 | C16H15BrClN5O3 | 440.682 | 7 / 3 | 2.0 | Yes |
| 28519 | 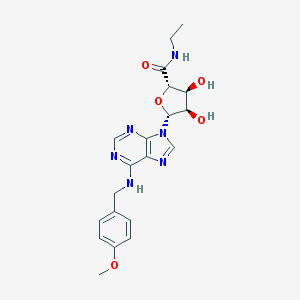 MPC-MECA MPC-MECA | C20H24N6O5 | 428.449 | 9 / 4 | 0.6 | Yes |
| 29532 | 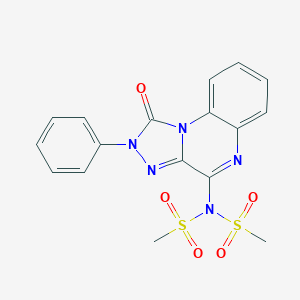 CHEMBL519319 CHEMBL519319 | C17H15N5O5S2 | 433.457 | 7 / 0 | 1.4 | Yes |
| 32046 | 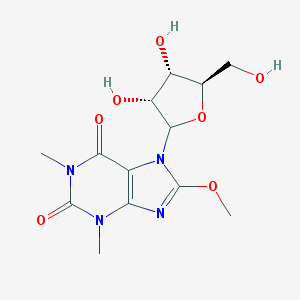 CHEMBL608026 CHEMBL608026 | C13H18N4O7 | 342.308 | 8 / 3 | -1.7 | Yes |
| 33925 | 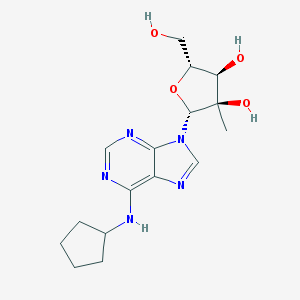 205171-06-8 205171-06-8 | C16H23N5O4 | 349.391 | 8 / 4 | 0.3 | Yes |
| 37149 | 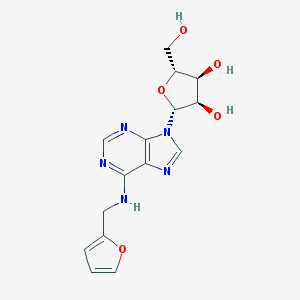 Kinetin riboside Kinetin riboside | C15H17N5O5 | 347.331 | 9 / 4 | 0.2 | Yes |
| 39607 | 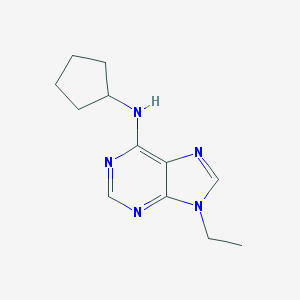 CHEMBL301574 CHEMBL301574 | C12H17N5 | 231.303 | 4 / 1 | 1.7 | Yes |
| 40531 | 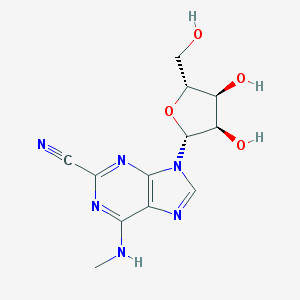 CHEMBL2181972 CHEMBL2181972 | C12H14N6O4 | 306.282 | 9 / 4 | -0.3 | Yes |
| 42380 | 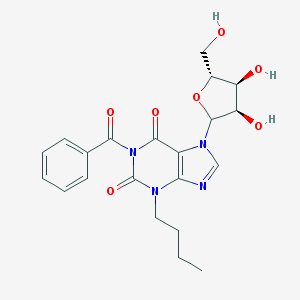 CHEMBL610700 CHEMBL610700 | C21H24N4O7 | 444.444 | 8 / 3 | 0.6 | Yes |
| 553453 | 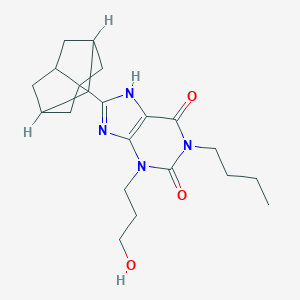 PSB 36 PSB 36 | C21H30N4O3 | 386.496 | 4 / 2 | 4.0 | Yes |
| 43247 | 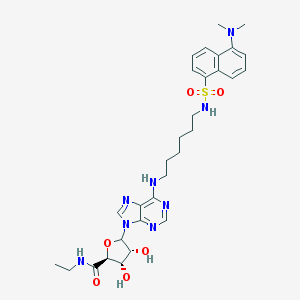 CHEMBL612203 CHEMBL612203 | C30H40N8O6S | 640.76 | 12 / 5 | 2.6 | No |
| 43650 | 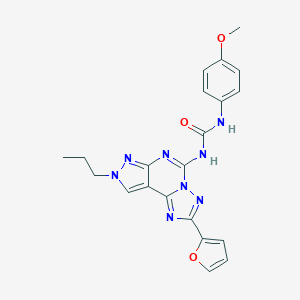 MRE 3008-F20 MRE 3008-F20 | C21H20N8O3 | 432.444 | 7 / 2 | 2.5 | Yes |
| 44276 | 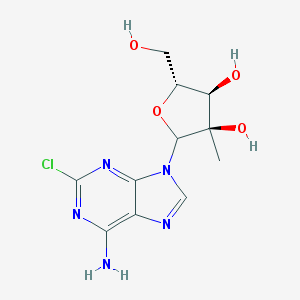 CHEMBL606495 CHEMBL606495 | C11H14ClN5O4 | 315.714 | 8 / 4 | 0.0 | Yes |
| 44498 | 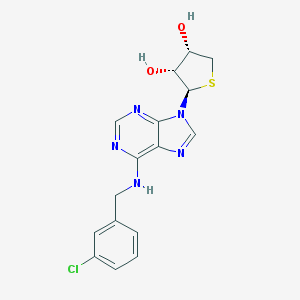 CHEMBL494086 CHEMBL494086 | C16H16ClN5O2S | 377.847 | 7 / 3 | 1.7 | Yes |
| 45035 | 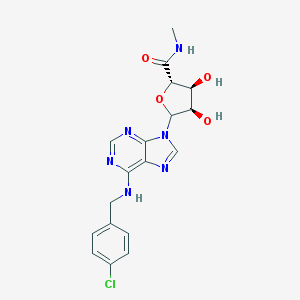 CHEMBL609923 CHEMBL609923 | C18H19ClN6O4 | 418.838 | 8 / 4 | 0.9 | Yes |
| 45651 | 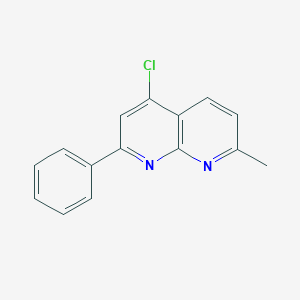 CHEMBL97778 CHEMBL97778 | C15H11ClN2 | 254.717 | 2 / 0 | 4.1 | Yes |
| 46699 | 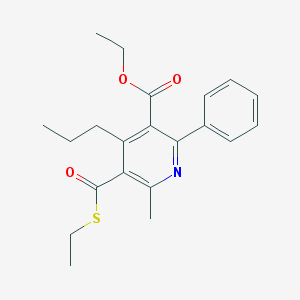 CHEMBL111423 CHEMBL111423 | C21H25NO3S | 371.495 | 5 / 0 | 5.2 | No |
| 48164 | 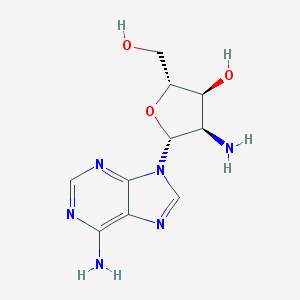 2'-Amino-2'-deoxyadenosine 2'-Amino-2'-deoxyadenosine | C10H14N6O3 | 266.261 | 8 / 4 | -1.3 | Yes |
| 48679 | 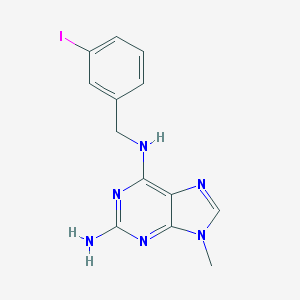 CHEMBL2113700 CHEMBL2113700 | C13H13IN6 | 380.193 | 5 / 2 | 2.2 | Yes |
| 48877 | 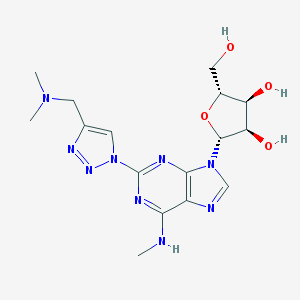 CHEMBL374645 CHEMBL374645 | C16H23N9O4 | 405.419 | 11 / 4 | -0.8 | No |
| 49629 | 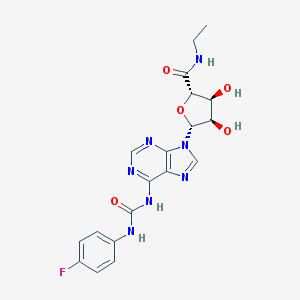 CHEMBL2113635 CHEMBL2113635 | C19H20FN7O5 | 445.411 | 9 / 5 | -0.2 | Yes |
| 52425 | 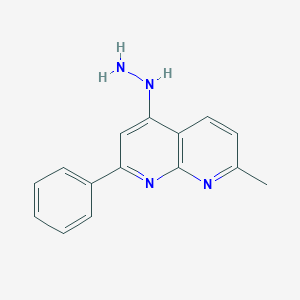 CHEMBL94538 CHEMBL94538 | C15H14N4 | 250.305 | 4 / 2 | 2.8 | Yes |
| 53533 | 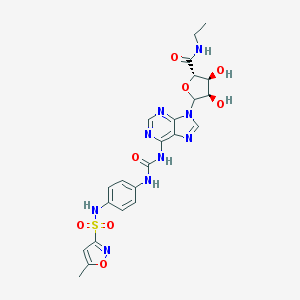 CHEMBL610112 CHEMBL610112 | C23H25N9O8S | 587.568 | 13 / 6 | -0.6 | No |
| 54400 | 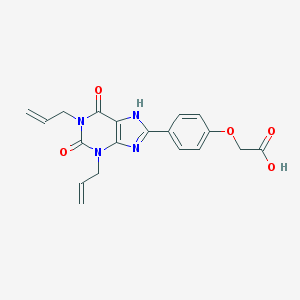 CHEMBL16724 CHEMBL16724 | C19H18N4O5 | 382.376 | 6 / 2 | 3.0 | Yes |
| 54815 | 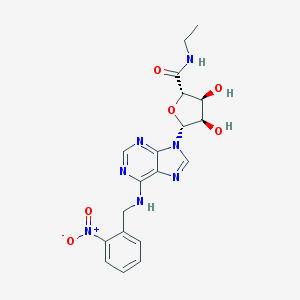 CHEMBL2113570 CHEMBL2113570 | C19H21N7O6 | 443.42 | 10 / 4 | 0.4 | Yes |
| 56160 | 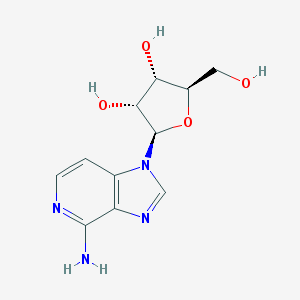 3-DEAZAADENOSINE 3-DEAZAADENOSINE | C11H14N4O4 | 266.257 | 7 / 4 | -1.6 | Yes |
| 56804 | 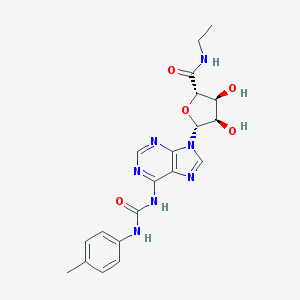 CHEMBL2113626 CHEMBL2113626 | C20H23N7O5 | 441.448 | 8 / 5 | 0.1 | Yes |
| 58231 | 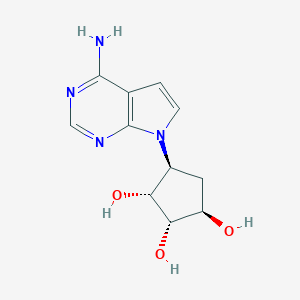 CHEMBL2051757 CHEMBL2051757 | C11H14N4O3 | 250.258 | 6 / 4 | -1.5 | Yes |
| 59359 | 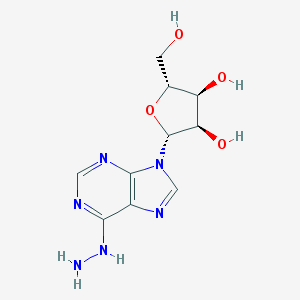 N6-aminoadenosine N6-aminoadenosine | C10H14N6O4 | 282.26 | 9 / 5 | -1.1 | Yes |
| 59762 | 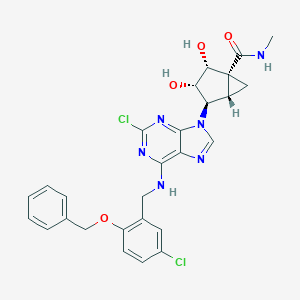 CHEMBL434328 CHEMBL434328 | C27H26Cl2N6O4 | 569.443 | 8 / 4 | 3.5 | No |
| 60972 | 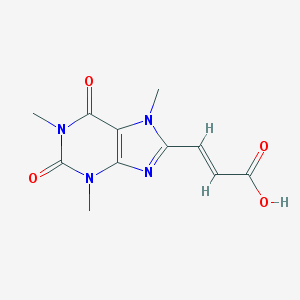 CHEMBL92364 CHEMBL92364 | C11H12N4O4 | 264.241 | 5 / 1 | -0.1 | Yes |
| 60984 | 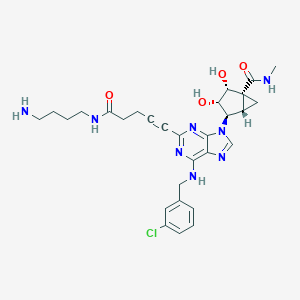 CHEMBL583745 CHEMBL583745 | C29H35ClN8O4 | 595.101 | 9 / 6 | 0.8 | No |
| 523338 | 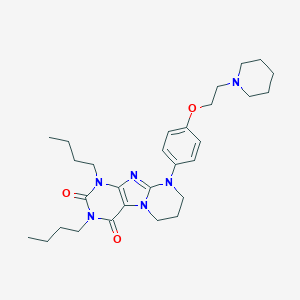 CHEMBL3827201 CHEMBL3827201 | C29H42N6O3 | 522.694 | 6 / 0 | 5.0 | No |
| 61843 | 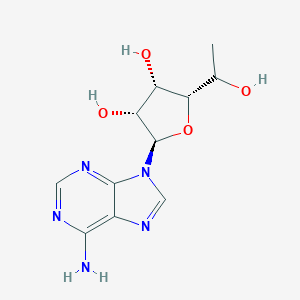 CHEMBL1791403 CHEMBL1791403 | C11H15N5O4 | 281.272 | 8 / 4 | -0.6 | Yes |
| 61898 | 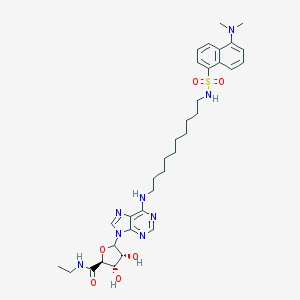 CHEMBL610121 CHEMBL610121 | C34H48N8O6S | 696.868 | 12 / 5 | 4.8 | No |
| 62341 | 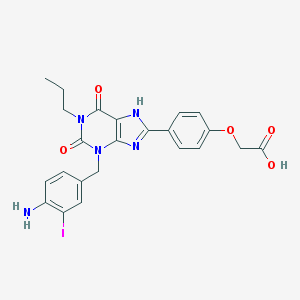 I-Abopx I-Abopx | C23H22IN5O5 | 575.363 | 7 / 3 | 4.4 | No |
| 62973 | 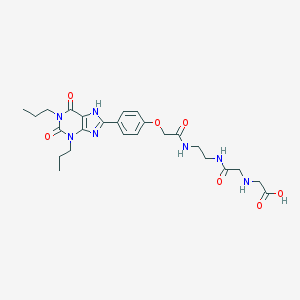 CHEMBL112669 CHEMBL112669 | C25H33N7O7 | 543.581 | 9 / 5 | -0.1 | No |
| 64571 | 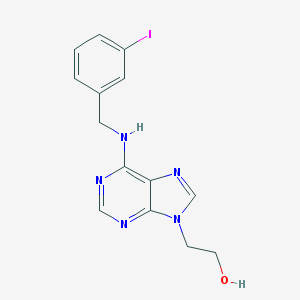 CHEMBL2113689 CHEMBL2113689 | C14H14IN5O | 395.204 | 5 / 2 | 2.2 | Yes |
| 64585 | 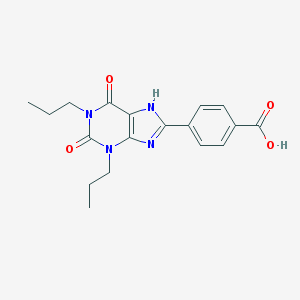 CHEMBL423239 CHEMBL423239 | C18H20N4O4 | 356.382 | 5 / 2 | 3.8 | Yes |
| 67382 | 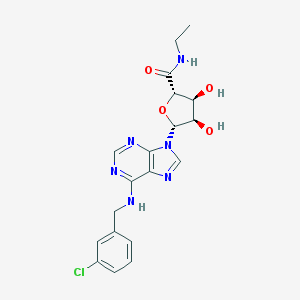 CHEMBL2113565 CHEMBL2113565 | C19H21ClN6O4 | 432.865 | 8 / 4 | 1.2 | Yes |
| 68904 | 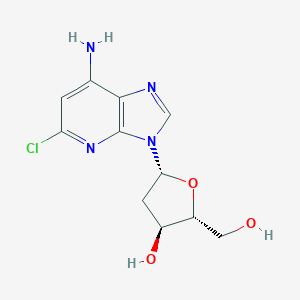 AC1LA9RB AC1LA9RB | C11H13ClN4O3 | 284.7 | 6 / 3 | 1.1 | Yes |
| 69730 | 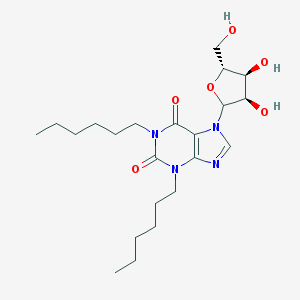 CHEMBL611863 CHEMBL611863 | C22H36N4O6 | 452.552 | 7 / 3 | 2.6 | Yes |
| 69806 | 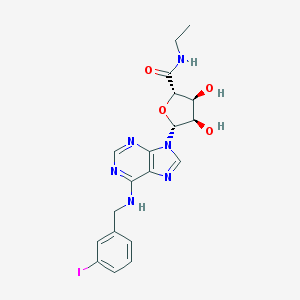 CHEMBL2113613 CHEMBL2113613 | C19H21IN6O4 | 524.319 | 8 / 4 | 1.3 | No |
| 69924 | 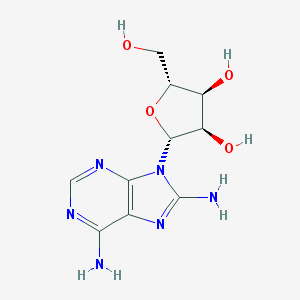 8-Aminoadenosine 8-Aminoadenosine | C10H14N6O4 | 282.26 | 9 / 5 | -1.4 | Yes |
| 71367 | 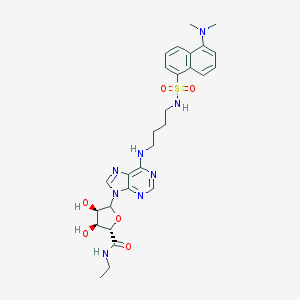 CHEMBL609525 CHEMBL609525 | C28H36N8O6S | 612.706 | 12 / 5 | 1.9 | No |
| 72121 | 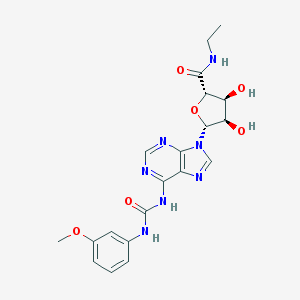 CHEMBL2113633 CHEMBL2113633 | C20H23N7O6 | 457.447 | 9 / 5 | -0.3 | Yes |
| 73230 | 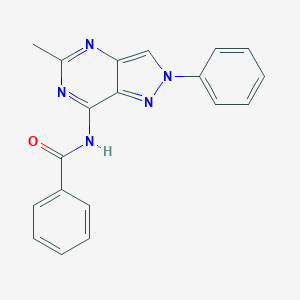 CHEMBL2322923 CHEMBL2322923 | C19H15N5O | 329.363 | 4 / 1 | 3.2 | Yes |
| 73363 | 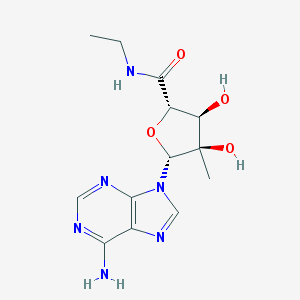 CHEMBL2113418 CHEMBL2113418 | C13H18N6O4 | 322.325 | 8 / 4 | -1.4 | Yes |
| 73384 | 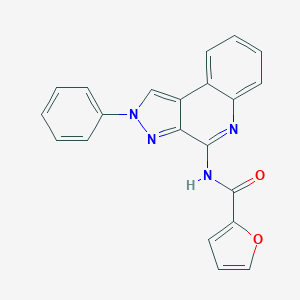 CHEMBL517867 CHEMBL517867 | C21H14N4O2 | 354.369 | 4 / 1 | 4.1 | Yes |
| 74348 | 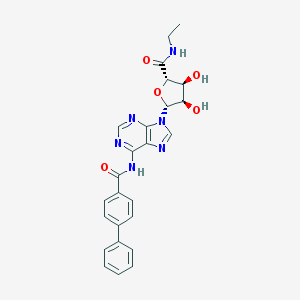 CHEMBL202722 CHEMBL202722 | C25H24N6O5 | 488.504 | 8 / 4 | 1.9 | Yes |
| 75509 | 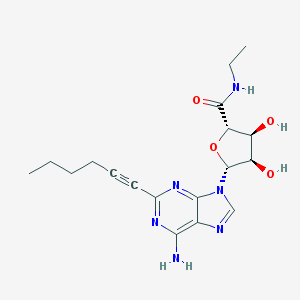 HE-NECA HE-NECA | C18H24N6O4 | 388.428 | 8 / 4 | 0.8 | Yes |
| 76489 | 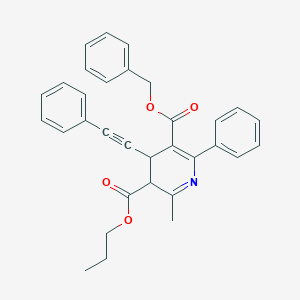 BDBM50059399 BDBM50059399 | C32H29NO4 | 491.587 | 5 / 0 | 6.1 | No |
| 76830 | 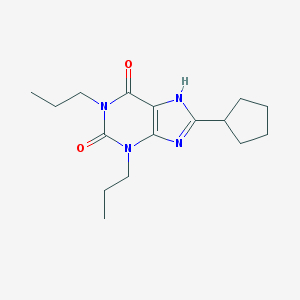 8-Cyclopentyl-1,3-dipropylxanthine 8-Cyclopentyl-1,3-dipropylxanthine | C16H24N4O2 | 304.394 | 3 / 1 | 4.0 | Yes |
| 77493 | 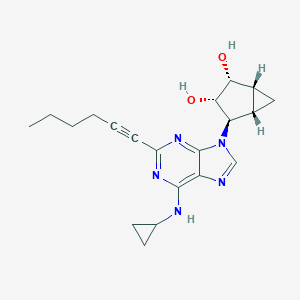 CHEMBL3125714 CHEMBL3125714 | C20H25N5O2 | 367.453 | 6 / 3 | 2.4 | Yes |
| 79062 | 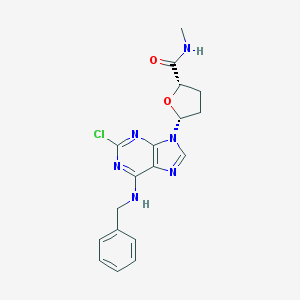 CHEMBL2113696 CHEMBL2113696 | C18H19ClN6O2 | 386.84 | 6 / 2 | 2.6 | Yes |
| 79115 | 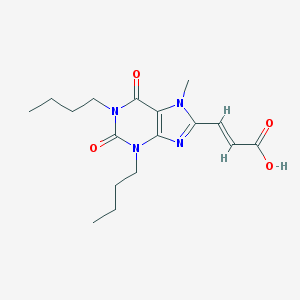 CHEMBL327016 CHEMBL327016 | C17H24N4O4 | 348.403 | 5 / 1 | 2.1 | Yes |
| 79291 | 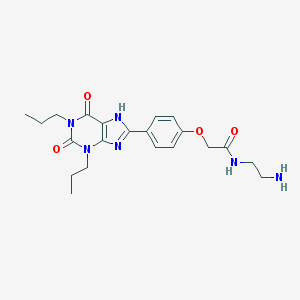 Xanthine amine congener Xanthine amine congener | C21H28N6O4 | 428.493 | 6 / 3 | 2.7 | Yes |
| 79582 | 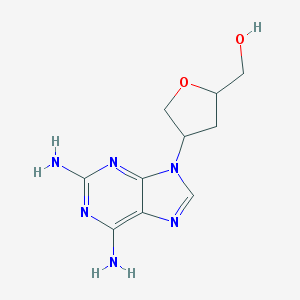 CHEMBL416540 CHEMBL416540 | C10H14N6O2 | 250.262 | 7 / 3 | -0.9 | Yes |
| 81037 | 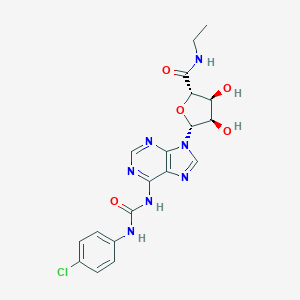 CHEMBL2113631 CHEMBL2113631 | C19H20ClN7O5 | 461.863 | 8 / 5 | 0.3 | Yes |
| 83542 | 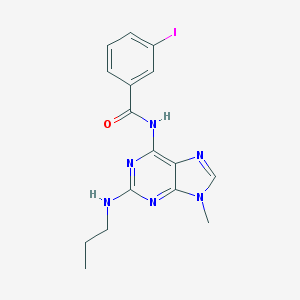 CHEMBL47437 CHEMBL47437 | C16H17IN6O | 436.257 | 5 / 2 | 3.1 | Yes |
| 83651 | 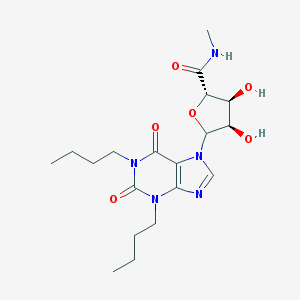 CHEMBL609240 CHEMBL609240 | C19H29N5O6 | 423.47 | 7 / 3 | 0.5 | Yes |
| 83655 | 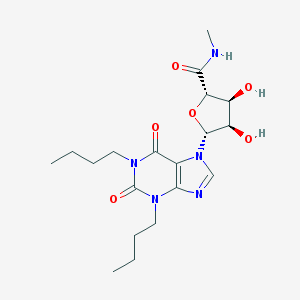 CHEMBL138746 CHEMBL138746 | C19H29N5O6 | 423.47 | 7 / 3 | 0.5 | Yes |
| 84353 | 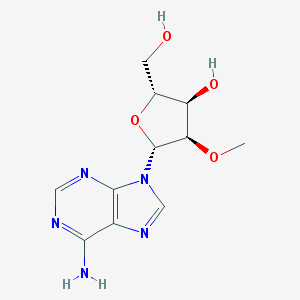 2'-O-Methyladenosine 2'-O-Methyladenosine | C11H15N5O4 | 281.272 | 8 / 3 | -0.5 | Yes |
| 88809 | 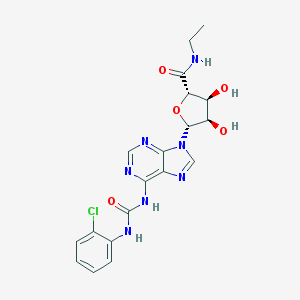 CHEMBL2113637 CHEMBL2113637 | C19H20ClN7O5 | 461.863 | 8 / 5 | 0.3 | Yes |
| 88918 | 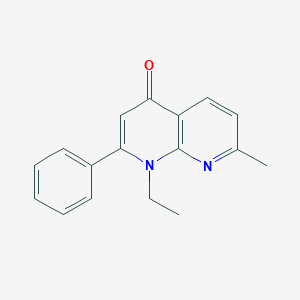 CHEMBL415697 CHEMBL415697 | C17H16N2O | 264.328 | 3 / 0 | 3.4 | Yes |
| 88986 | 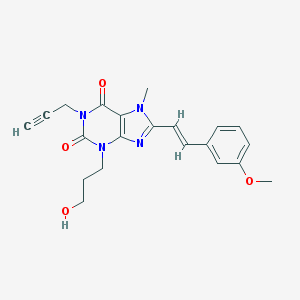 MSX-2 MSX-2 | C21H22N4O4 | 394.431 | 5 / 1 | 1.6 | Yes |
| 89533 | 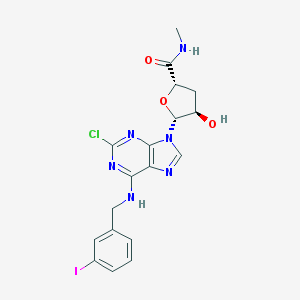 CHEMBL2113703 CHEMBL2113703 | C18H18ClIN6O3 | 528.735 | 7 / 3 | 2.3 | No |
| 91190 |  CHEMBL444550 CHEMBL444550 | C18H20IN5O4 | 497.293 | 8 / 4 | 1.1 | Yes |
| 91660 |  CHEMBL193421 CHEMBL193421 | C19H21N5O4 | 383.408 | 8 / 4 | 1.6 | Yes |
| 92064 |  CHEMBL175543 CHEMBL175543 | C20H20Cl2N6O3 | 463.319 | 7 / 4 | 2.0 | Yes |
| 95073 |  CHEMBL344187 CHEMBL344187 | C19H19IN6O4 | 522.303 | 8 / 4 | 0.3 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417