

You can:
| Name | Growth hormone secretagogue receptor type 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GHSR |
| Synonym | ghrelin receptor GH-releasing peptide receptor GHRP GHS-R growth hormone-releasing peptide receptor [ Show all ] |
| Disease | N/A |
| Length | 366 |
| Amino acid sequence | MWNATPSEEPGFNLTLADLDWDASPGNDSLGDELLQLFPAPLLAGVTATCVALFVVGIAGNLLTMLVVSRFRELRTTTNLYLSSMAFSDLLIFLCMPLDLVRLWQYRPWNFGDLLCKLFQFVSESCTYATVLTITALSVERYFAICFPLRAKVVVTKGRVKLVIFVIWAVAFCSAGPIFVLVGVEHENGTDPWDTNECRPTEFAVRSGLLTVMVWVSSIFFFLPVFCLTVLYSLIGRKLWRRRRGDAVVGASLRDQNHKQTVKMLAVVVFAFILCWLPFHVGRYLFSKSFEPGSLEIAQISQYCNLVSFVLFYLSAAINPILYNIMSKKYRVAVFRLLGFEPFSQRKLSTLKDESSRAWTESSINT |
| UniProt | Q92847 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q92847 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q92847. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL4616 |
| IUPHAR | 246 |
| DrugBank | BE0003383 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 249 | 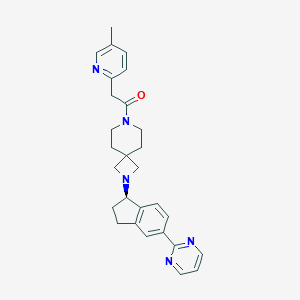 CHEMBL2426667 CHEMBL2426667 | C28H31N5O | 453.59 | 5 / 0 | 2.9 | Yes |
| 547901 | 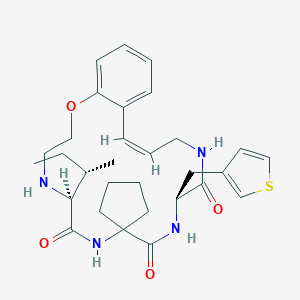 CHEMBL3908156 CHEMBL3908156 | C30H40N4O4S | 552.734 | 6 / 4 | 4.3 | No |
| 740 | 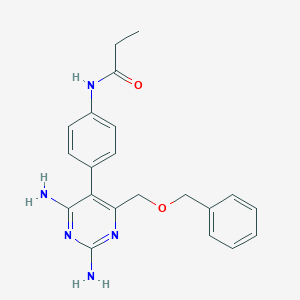 CHEMBL386290 CHEMBL386290 | C21H23N5O2 | 377.448 | 6 / 3 | 2.0 | Yes |
| 547912 | 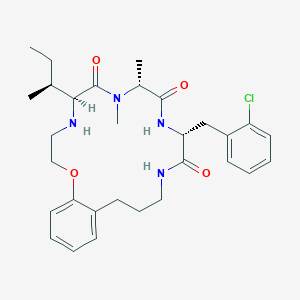 CHEMBL3898794 CHEMBL3898794 | C30H41ClN4O4 | 557.132 | 5 / 3 | 4.8 | No |
| 1305 | 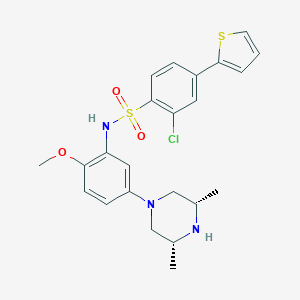 CHEMBL272993 CHEMBL272993 | C23H26ClN3O3S2 | 492.049 | 7 / 2 | 4.7 | Yes |
| 1690 | 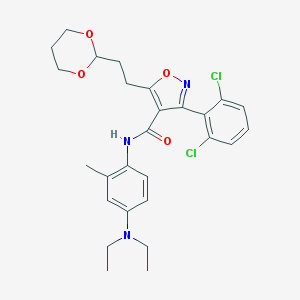 CHEMBL361349 CHEMBL361349 | C27H31Cl2N3O4 | 532.462 | 6 / 1 | 6.0 | No |
| 1711 | 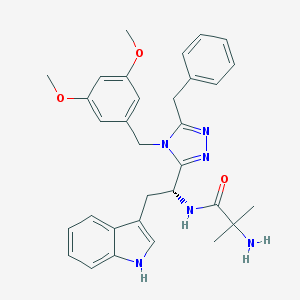 CHEMBL221674 CHEMBL221674 | C32H36N6O3 | 552.679 | 6 / 3 | 3.9 | No |
| 2192 | 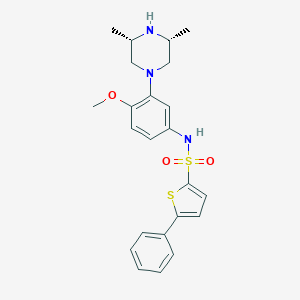 SCHEMBL4801347 SCHEMBL4801347 | C23H27N3O3S2 | 457.607 | 7 / 2 | 4.4 | Yes |
| 535980 |  CHEMBL1923624 CHEMBL1923624 | C29H37FN4O4 | 524.637 | 6 / 3 | 3.3 | No |
| 2378 |  CHEMBL1077716 CHEMBL1077716 | C19H25N3O3S | 375.487 | 4 / 3 | 3.8 | Yes |
| 3008 |  CHEMBL237926 CHEMBL237926 | C27H37N7O4 | 523.638 | 8 / 3 | 2.0 | No |
| 463406 |  CHEMBL3578011 CHEMBL3578011 | C87H143N29O20 | 1915.28 | 25 / 31 | -2.9 | No |
| 547948 | 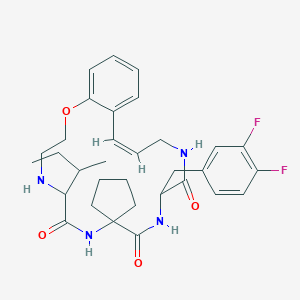 CHEMBL3981960 CHEMBL3981960 | C32H40F2N4O4 | 582.693 | 7 / 4 | 4.8 | No |
| 547949 | 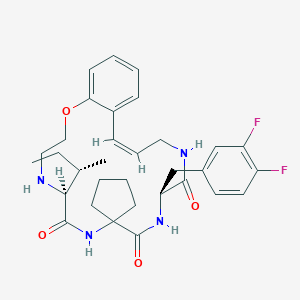 CHEMBL3984464 CHEMBL3984464 | C32H40F2N4O4 | 582.693 | 7 / 4 | 4.8 | No |
| 5231 | 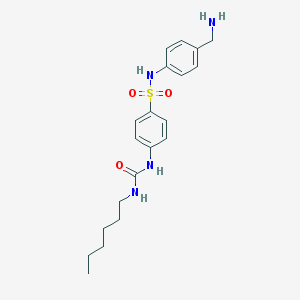 CHEMBL1078533 CHEMBL1078533 | C20H28N4O3S | 404.529 | 5 / 4 | 2.6 | Yes |
| 547958 | 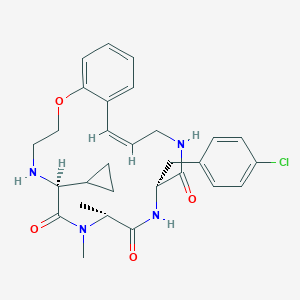 CHEMBL3934367 CHEMBL3934367 | C29H35ClN4O4 | 539.073 | 5 / 3 | 3.8 | No |
| 5417 | 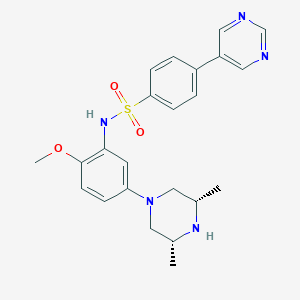 CHEMBL255401 CHEMBL255401 | C23H27N5O3S | 453.561 | 8 / 2 | 2.7 | Yes |
| 5443 | 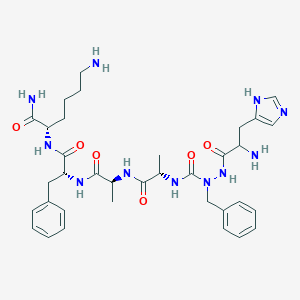 CHEMBL2163437 CHEMBL2163437 | C35H49N11O6 | 719.848 | 9 / 9 | -0.2 | No |
| 5639 | 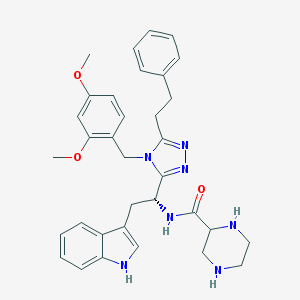 SCHEMBL13018946 SCHEMBL13018946 | C34H39N7O3 | 593.732 | 7 / 4 | 3.5 | No |
| 5913 | 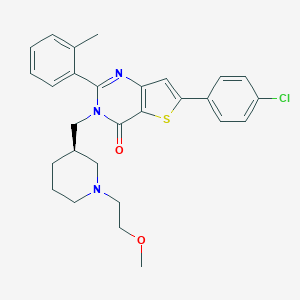 CHEMBL3218648 CHEMBL3218648 | C28H30ClN3O2S | 508.077 | 5 / 0 | 5.8 | No |
| 5934 |  CHEMBL2170701 CHEMBL2170701 | C50H68N10O6 | 905.158 | 8 / 9 | 4.7 | No |
| 5955 |  CHEMBL399652 CHEMBL399652 | C35H35N5O2 | 557.698 | 5 / 2 | 5.9 | No |
| 6652 |  CHEMBL237302 CHEMBL237302 | C19H27N7O2 | 385.472 | 7 / 2 | 0.7 | Yes |
| 6654 |  CHEMBL185876 CHEMBL185876 | C32H41FN4O2 | 532.704 | 4 / 2 | 6.2 | No |
| 6808 | 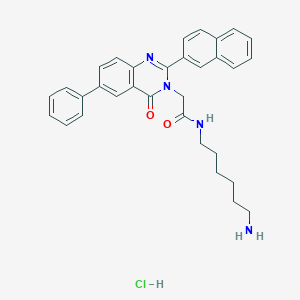 CHEMBL558181 CHEMBL558181 | C32H33ClN4O2 | 541.092 | 4 / 3 | N/A | No |
| 7033 | 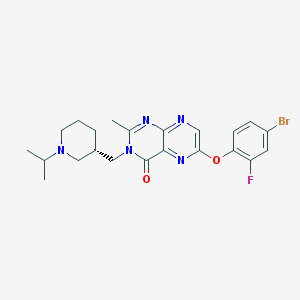 CHEMBL1957010 CHEMBL1957010 | C22H25BrFN5O2 | 490.377 | 7 / 0 | 3.6 | Yes |
| 7040 | 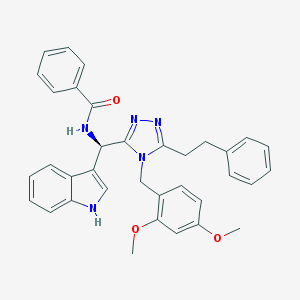 CHEMBL399651 CHEMBL399651 | C35H33N5O3 | 571.681 | 5 / 2 | 5.8 | No |
| 7646 | 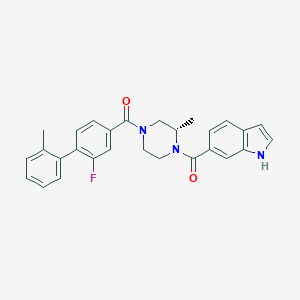 CHEMBL1081612 CHEMBL1081612 | C28H26FN3O2 | 455.533 | 3 / 1 | 5.0 | Yes |
| 7672 |  CHEMBL2163461 CHEMBL2163461 | C39H52N12O6 | 784.923 | 9 / 9 | 0.3 | No |
| 7848 |  CHEMBL490009 CHEMBL490009 | C22H26ClN3O3 | 415.918 | 4 / 2 | 3.3 | Yes |
| 8321 |  CHEMBL2048824 CHEMBL2048824 | C27H27ClN6O | 487.004 | 5 / 0 | 3.7 | Yes |
| 8403 |  CHEMBL202642 CHEMBL202642 | C23H22ClN5OS | 451.973 | 7 / 3 | 4.5 | Yes |
| 8540 |  CHEMBL178610 CHEMBL178610 | C26H29Cl2N3O4 | 518.435 | 6 / 1 | 5.6 | No |
| 8941 |  CHEMBL252255 CHEMBL252255 | C36H33N7O3 | 611.706 | 6 / 3 | 5.9 | No |
| 8942 |  CHEMBL403542 CHEMBL403542 | C36H33N7O3 | 611.706 | 6 / 3 | 5.9 | No |
| 9184 |  CHEMBL270790 CHEMBL270790 | C23H26FN3O3S2 | 475.597 | 8 / 2 | 4.2 | Yes |
| 547985 | 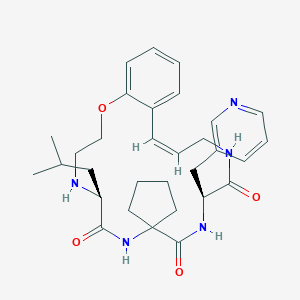 CHEMBL3925748 CHEMBL3925748 | C31H41N5O4 | 547.7 | 6 / 4 | 3.6 | No |
| 547986 | 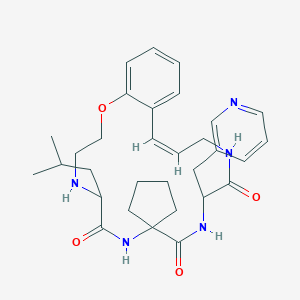 CHEMBL3948571 CHEMBL3948571 | C31H41N5O4 | 547.7 | 6 / 4 | 3.6 | No |
| 9341 | 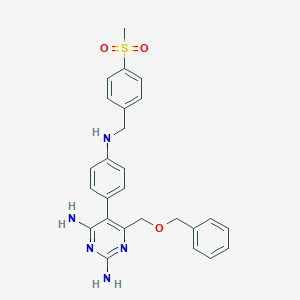 CHEMBL425594 CHEMBL425594 | C26H27N5O3S | 489.594 | 8 / 3 | 3.0 | Yes |
| 10294 | 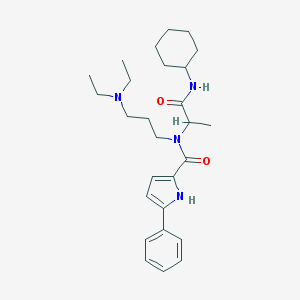 CHEMBL188173 CHEMBL188173 | C27H40N4O2 | 452.643 | 3 / 2 | 4.9 | Yes |
| 442115 |  CHEMBL3394202 CHEMBL3394202 | C30H34N6O3 | 526.641 | 7 / 1 | 2.1 | No |
| 10861 |  CHEMBL1079186 CHEMBL1079186 | C25H32N4O2S | 452.617 | 5 / 2 | 4.7 | Yes |
| 10989 |  CHEMBL1076586 CHEMBL1076586 | C26H23N3O2 | 409.489 | 2 / 1 | 4.1 | Yes |
| 11282 |  CHEMBL1078960 CHEMBL1078960 | C24H28N4O3S | 452.573 | 5 / 4 | 3.3 | Yes |
| 11392 |  BDBM50138207 BDBM50138207 | C32H35N7O2S | 581.739 | 7 / 2 | 4.5 | No |
| 11568 |  CHEMBL2048818 CHEMBL2048818 | C26H34ClN3O4 | 488.025 | 6 / 0 | 4.5 | Yes |
| 12210 |  CHEMBL204474 CHEMBL204474 | C26H23N5O2 | 437.503 | 7 / 2 | 3.5 | Yes |
| 12487 |  CHEMBL222789 CHEMBL222789 | C33H34BrN7O | 624.587 | 4 / 4 | 5.0 | No |
| 12572 | 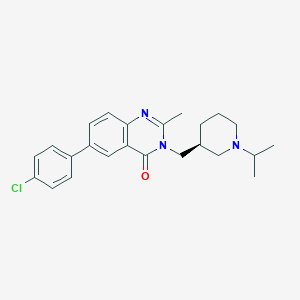 CHEMBL244936 CHEMBL244936 | C24H28ClN3O | 409.958 | 3 / 0 | 4.7 | Yes |
| 12948 | 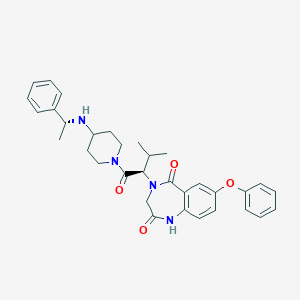 CHEMBL2011832 CHEMBL2011832 | C33H38N4O4 | 554.691 | 5 / 2 | 4.8 | No |
| 14050 | 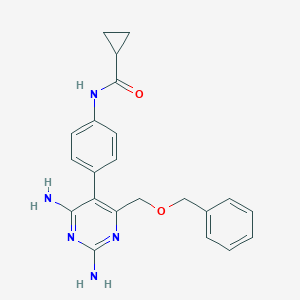 CHEMBL211265 CHEMBL211265 | C22H23N5O2 | 389.459 | 6 / 3 | 1.9 | Yes |
| 14072 | 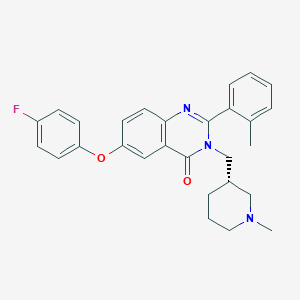 CHEMBL1765654 CHEMBL1765654 | C28H28FN3O2 | 457.549 | 5 / 0 | 5.3 | No |
| 14193 | 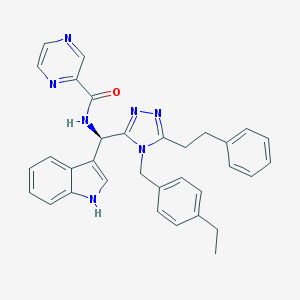 CHEMBL415135 CHEMBL415135 | C33H31N7O | 541.659 | 5 / 2 | 4.9 | No |
| 536357 | 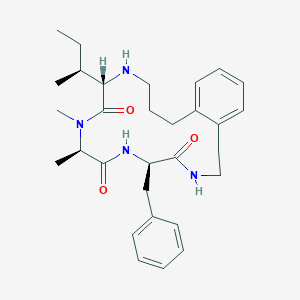 CHEMBL1923633 CHEMBL1923633 | C30H42N4O3 | 506.691 | 4 / 3 | 4.5 | No |
| 14258 | 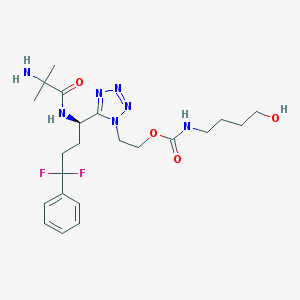 CHEMBL460680 CHEMBL460680 | C22H33F2N7O4 | 497.548 | 10 / 4 | 1.0 | Yes |
| 14908 | 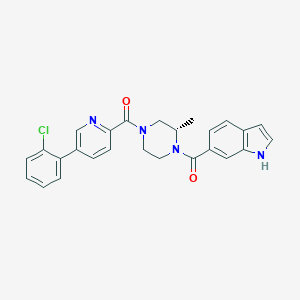 CHEMBL1079622 CHEMBL1079622 | C26H23ClN4O2 | 458.946 | 3 / 1 | 4.4 | Yes |
| 15343 | 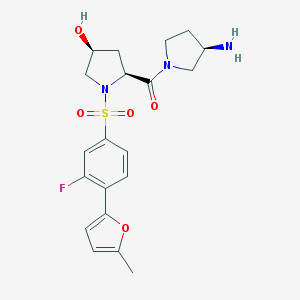 CHEMBL517391 CHEMBL517391 | C20H24FN3O5S | 437.486 | 8 / 2 | 0.6 | Yes |
| 15344 | 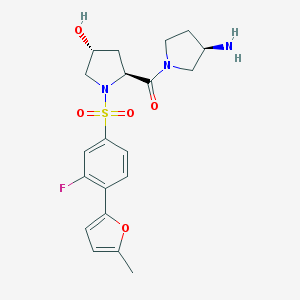 CHEMBL480037 CHEMBL480037 | C20H24FN3O5S | 437.486 | 8 / 2 | 0.6 | Yes |
| 548054 | 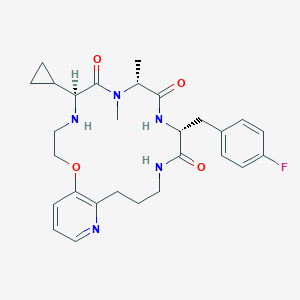 CHEMBL3934402 CHEMBL3934402 | C28H36FN5O4 | 525.625 | 7 / 3 | 2.3 | No |
| 548055 | 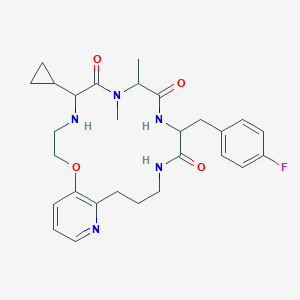 CHEMBL3961456 CHEMBL3961456 | C28H36FN5O4 | 525.625 | 7 / 3 | 2.3 | No |
| 464785 | 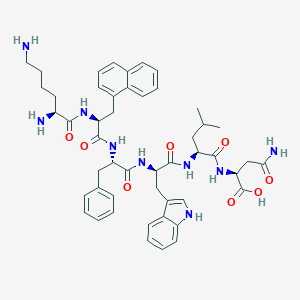 CHEMBL3578006 CHEMBL3578006 | C49H61N9O8 | 904.082 | 10 / 10 | 1.4 | No |
| 15628 | 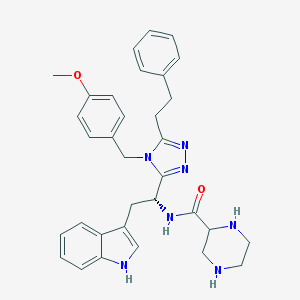 SCHEMBL13018951 SCHEMBL13018951 | C33H37N7O2 | 563.706 | 6 / 4 | 3.5 | No |
| 16016 | 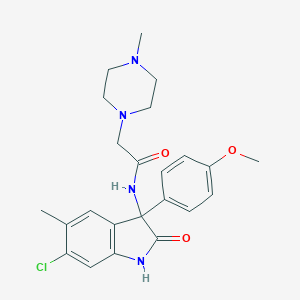 CHEMBL2164835 CHEMBL2164835 | C23H27ClN4O3 | 442.944 | 5 / 2 | 2.5 | Yes |
| 16050 | 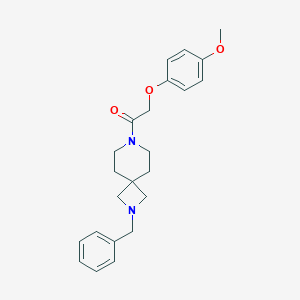 CHEMBL2048808 CHEMBL2048808 | C23H28N2O3 | 380.488 | 4 / 0 | 3.3 | Yes |
| 16156 | 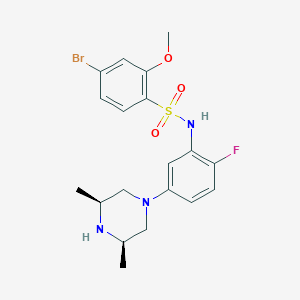 SCHEMBL4805648 SCHEMBL4805648 | C19H23BrFN3O3S | 472.373 | 7 / 2 | 3.5 | Yes |
| 16226 | 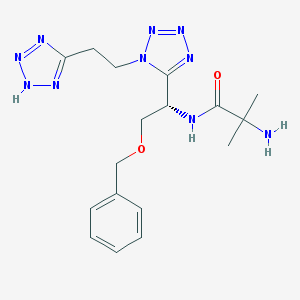 CHEMBL237925 CHEMBL237925 | C17H24N10O2 | 400.447 | 9 / 3 | -0.7 | Yes |
| 536399 | 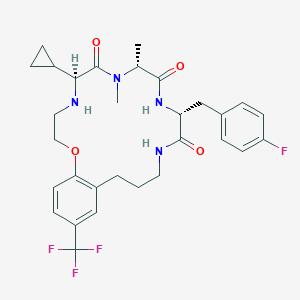 CHEMBL3961160 CHEMBL3961160 | C30H36F4N4O4 | 592.636 | 9 / 3 | 4.2 | No |
| 16452 | 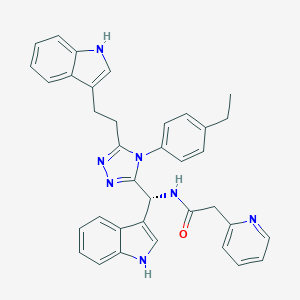 CHEMBL266961 CHEMBL266961 | C36H33N7O | 579.708 | 4 / 3 | 5.8 | No |
| 16485 | 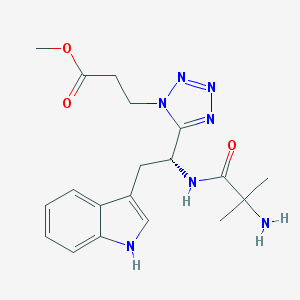 CHEMBL235012 CHEMBL235012 | C19H25N7O3 | 399.455 | 7 / 3 | 0.3 | Yes |
| 16490 | 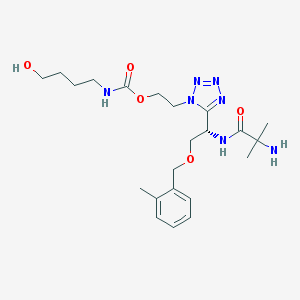 CHEMBL404354 CHEMBL404354 | C22H35N7O5 | 477.566 | 9 / 4 | -0.2 | Yes |
| 16552 | 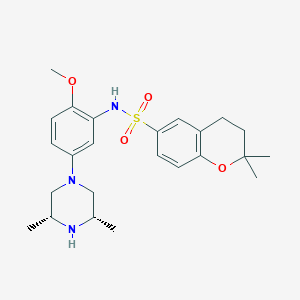 SCHEMBL4802558 SCHEMBL4802558 | C24H33N3O4S | 459.605 | 7 / 2 | 3.7 | Yes |
| 16585 | 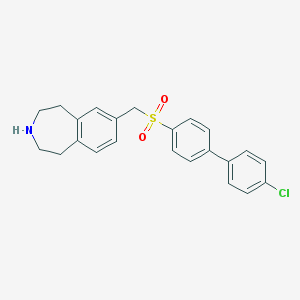 CHEMBL1078344 CHEMBL1078344 | C23H22ClNO2S | 411.944 | 3 / 1 | 4.8 | Yes |
| 16729 | 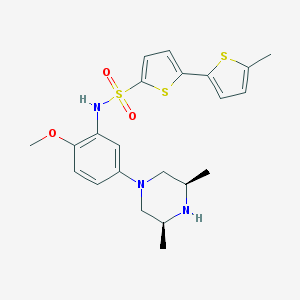 SCHEMBL4802295 SCHEMBL4802295 | C22H27N3O3S3 | 477.656 | 8 / 2 | 4.5 | Yes |
| 17009 | 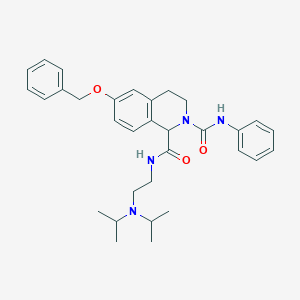 CHEMBL181885 CHEMBL181885 | C32H40N4O3 | 528.697 | 4 / 2 | 5.3 | No |
| 17178 | 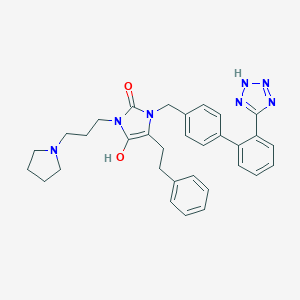 BDBM50138213 BDBM50138213 | C32H35N7O2 | 549.679 | 6 / 2 | 4.8 | No |
| 442360 | 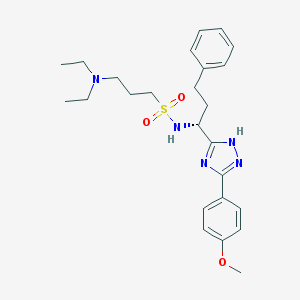 CHEMBL3358450 CHEMBL3358450 | C25H35N5O3S | 485.647 | 7 / 2 | 4.0 | Yes |
| 17734 | 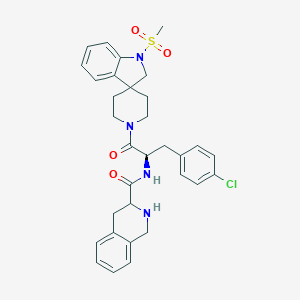 CHEMBL353007 CHEMBL353007 | C32H35ClN4O4S | 607.166 | 6 / 2 | 4.0 | No |
| 17908 | 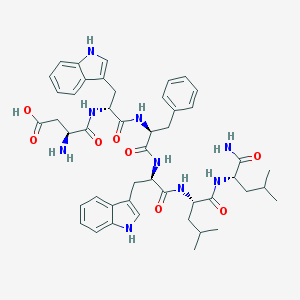 DwFwLL-NH2 DwFwLL-NH2 | C47H59N9O8 | 878.044 | 9 / 10 | 1.9 | No |
| 18017 | 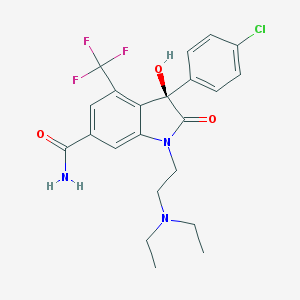 CHEMBL359922 CHEMBL359922 | C22H23ClF3N3O3 | 469.889 | 7 / 2 | 3.0 | Yes |
| 18594 | 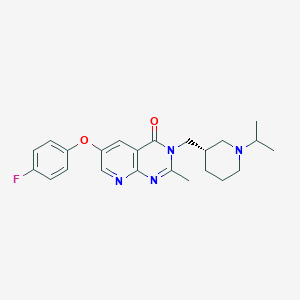 CHEMBL1956989 CHEMBL1956989 | C23H27FN4O2 | 410.493 | 6 / 0 | 3.4 | Yes |
| 18977 | 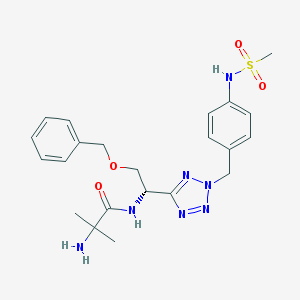 CHEMBL391429 CHEMBL391429 | C22H29N7O4S | 487.579 | 9 / 3 | 0.9 | Yes |
| 522034 | 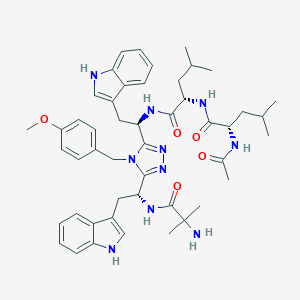 CHEMBL3798559 CHEMBL3798559 | C48H62N10O5 | 859.089 | 8 / 7 | 5.3 | No |
| 536469 | 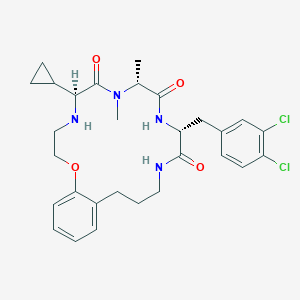 SCHEMBL804623 SCHEMBL804623 | C29H36Cl2N4O4 | 575.531 | 5 / 3 | 4.5 | No |
| 20492 | 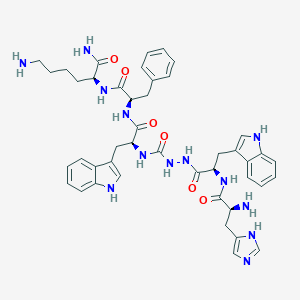 CHEMBL2163476 CHEMBL2163476 | C44H53N13O6 | 859.993 | 9 / 12 | 1.5 | No |
| 21123 |  CHEMBL1161824 CHEMBL1161824 | C27H32N4O4S | 508.637 | 5 / 3 | 2.1 | No |
| 21124 |  BDBM50187327 BDBM50187327 | C27H33N4O4S+ | 509.645 | 4 / 3 | 2.1 | No |
| 21402 |  CHEMBL243888 CHEMBL243888 | C30H32ClN3O2 | 502.055 | 4 / 0 | 5.8 | No |
| 21408 |  SCHEMBL13018947 SCHEMBL13018947 | C36H40N8O3 | 632.769 | 7 / 5 | 3.6 | No |
| 21881 | 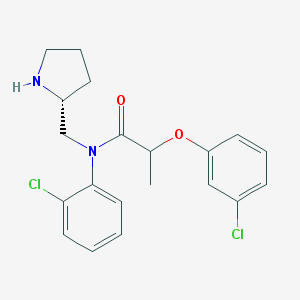 CHEMBL489402 CHEMBL489402 | C20H22Cl2N2O2 | 393.308 | 3 / 1 | 4.8 | Yes |
| 21886 | 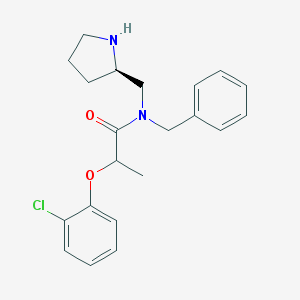 CHEMBL491221 CHEMBL491221 | C21H25ClN2O2 | 372.893 | 3 / 1 | 4.1 | Yes |
| 465644 | 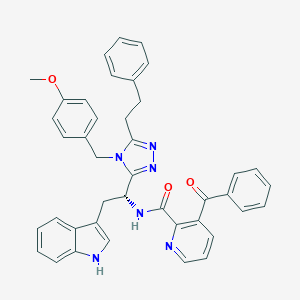 CHEMBL3581047 CHEMBL3581047 | C41H36N6O3 | 660.778 | 6 / 2 | 6.8 | No |
| 548138 | 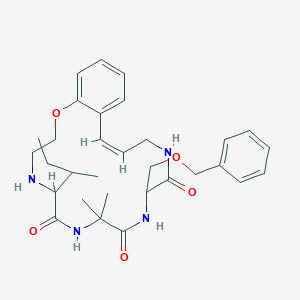 CHEMBL3954124 CHEMBL3954124 | C31H42N4O5 | 550.7 | 6 / 4 | 3.6 | No |
| 548139 | 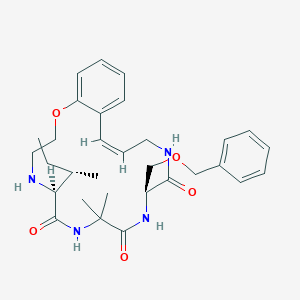 CHEMBL3905495 CHEMBL3905495 | C31H42N4O5 | 550.7 | 6 / 4 | 3.6 | No |
| 548144 | 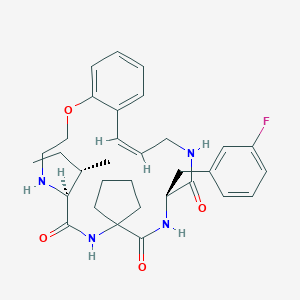 CHEMBL3890889 CHEMBL3890889 | C32H41FN4O4 | 564.702 | 6 / 4 | 4.7 | No |
| 548147 | 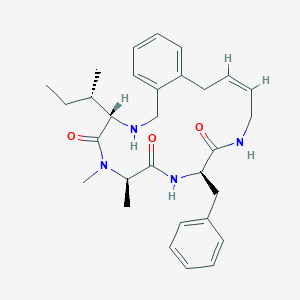 CHEMBL3963078 CHEMBL3963078 | C30H40N4O3 | 504.675 | 4 / 3 | 4.3 | No |
| 22955 | 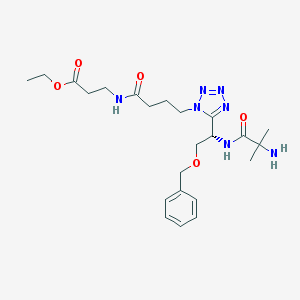 CHEMBL263141 CHEMBL263141 | C23H35N7O5 | 489.577 | 9 / 3 | -0.5 | Yes |
| 23020 | 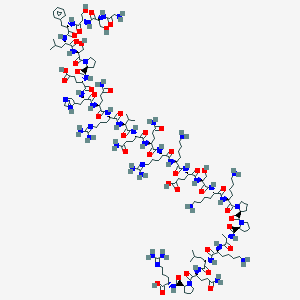 BDBM50163757 BDBM50163757 | C141H235N47O41 | 3244.72 | 50 / 46 | -23.0 | No |
| 23028 | 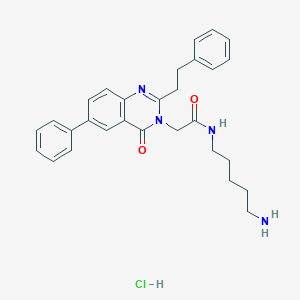 CHEMBL536706 CHEMBL536706 | C29H33ClN4O2 | 505.059 | 4 / 3 | N/A | No |
| 23037 | 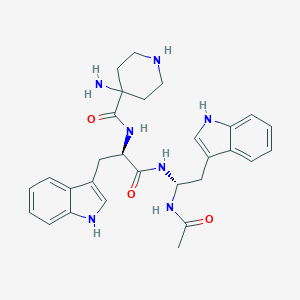 CHEMBL434391 CHEMBL434391 | C29H35N7O3 | 529.645 | 5 / 7 | 1.6 | No |
| 548149 | 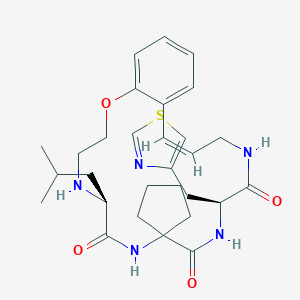 CHEMBL3947631 CHEMBL3947631 | C29H39N5O4S | 553.722 | 7 / 4 | 3.7 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417