

You can:
| Name | Glucose-dependent insulinotropic receptor |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Gpr119 |
| Synonym | G protein-coupled receptor 119 G-protein coupled receptor 119 G-protein coupled receptor 2 glucose-dependent insulinotropic receptor GPCR2 [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 468 |
| Amino acid sequence | MESSFSFGVILAVLTILIIAVNALVVVAMLLSIYKNDGVGLCFTLNLAVADTLIGVAISGLVTDQLSSSAQHTQKTLCSLRMAFVTSSAAASVLTVMLIAFDRYLAIKQPLRYFQIMNGLVAGGCIAGLWLISYLIGFLPLGVSIFQQTTYHGPCTFFAVFHPRFVLTLSCAGFFPAVLLFVFFYCDMLKIASVHSQHIRKMEHAGAMVGACRPPRPVNDFKAVRTVSVLIGSFTLSWSPFLITSIVQVACHKCCLYQVLEKYLWLLGVGNSLLNPLIYAYWQREVRQQLCHMALGLLADGSTQPQIETLKGKEERKKVGRKTLYTCDAQTLYTCDAQTLYTCDAQTLYTCDACDTQTLYTCDAQTLYTCDAQTLYTCDAQTLYTCDAQTLYTCDAQTLYTCDTQTLYTCDAQTLYTCDAQTLYTCDAQTLYTCDAQTLYTSSLVTGQTEQTPLKRANMSDPLRTCRG |
| UniProt | Q7TQN8 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL5262 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 441712 | 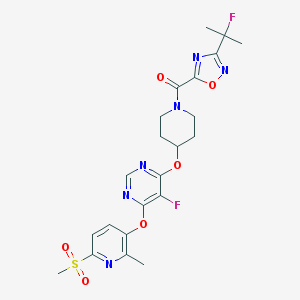 CHEMBL3326687 CHEMBL3326687 | C22H24F2N6O6S | 538.527 | 13 / 0 | 2.4 | No |
| 5711 | 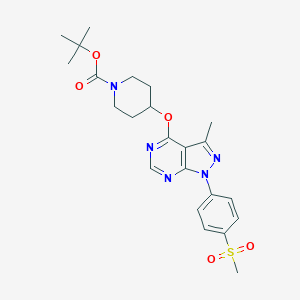 CHEMBL1773288 CHEMBL1773288 | C23H29N5O5S | 487.575 | 8 / 0 | 3.2 | Yes |
| 6164 | 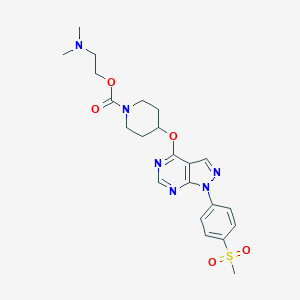 CHEMBL1775177 CHEMBL1775177 | C22H28N6O5S | 488.563 | 9 / 0 | 1.9 | Yes |
| 10216 | 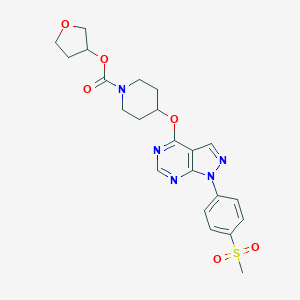 CHEMBL1775175 CHEMBL1775175 | C22H25N5O6S | 487.531 | 9 / 0 | 1.9 | Yes |
| 11415 | 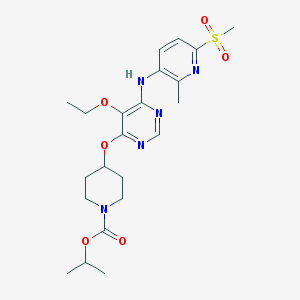 CHEMBL1951033 CHEMBL1951033 | C22H31N5O6S | 493.579 | 10 / 1 | 2.9 | Yes |
| 442204 | 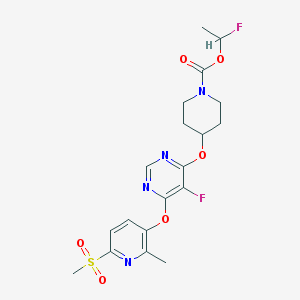 CHEMBL3325849 CHEMBL3325849 | C19H22F2N4O6S | 472.464 | 11 / 0 | 2.7 | No |
| 14295 | 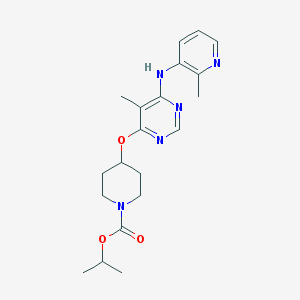 CHEMBL1951015 CHEMBL1951015 | C20H27N5O3 | 385.468 | 7 / 1 | 3.3 | Yes |
| 16830 | 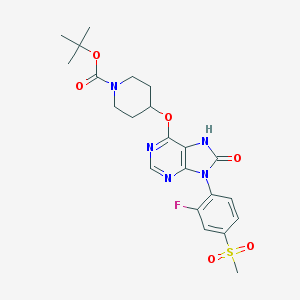 CHEMBL1773292 CHEMBL1773292 | C22H26FN5O6S | 507.537 | 9 / 1 | 2.0 | No |
| 17599 | 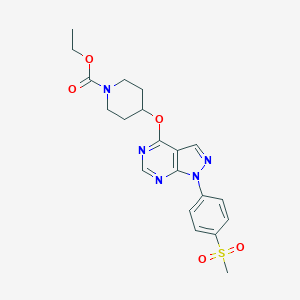 CHEMBL1775170 CHEMBL1775170 | C20H23N5O5S | 445.494 | 8 / 0 | 2.2 | Yes |
| 442681 | 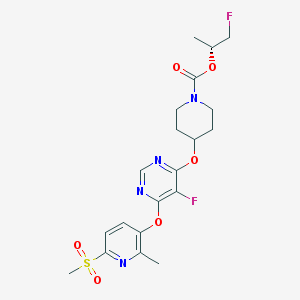 CHEMBL3325850 CHEMBL3325850 | C20H24F2N4O6S | 486.491 | 11 / 0 | 2.6 | No |
| 442682 | 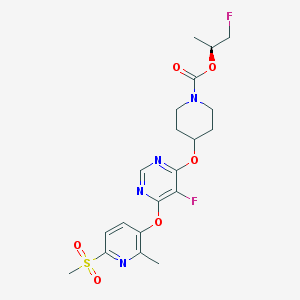 CHEMBL3325851 CHEMBL3325851 | C20H24F2N4O6S | 486.491 | 11 / 0 | 2.6 | No |
| 442835 | 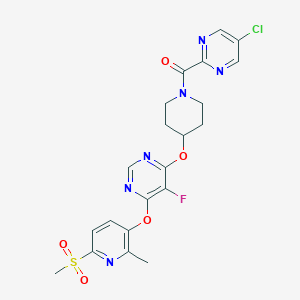 CHEMBL3326689 CHEMBL3326689 | C21H20ClFN6O5S | 522.936 | 11 / 0 | 2.3 | No |
| 32719 | 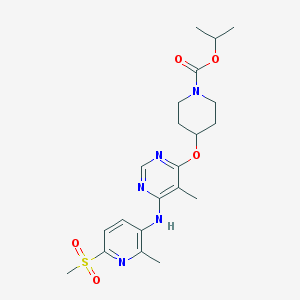 CHEMBL1951022 CHEMBL1951022 | C21H29N5O5S | 463.553 | 9 / 1 | 2.9 | Yes |
| 32816 | 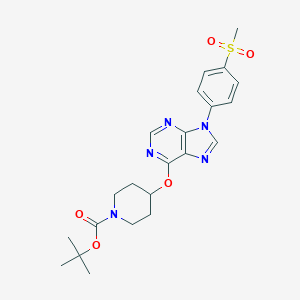 CHEMBL1773290 CHEMBL1773290 | C22H27N5O5S | 473.548 | 8 / 0 | 2.7 | Yes |
| 443058 | 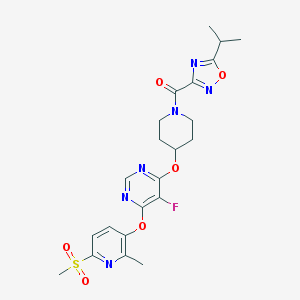 CHEMBL3326685 CHEMBL3326685 | C22H25FN6O6S | 520.536 | 12 / 0 | 2.8 | No |
| 40231 | 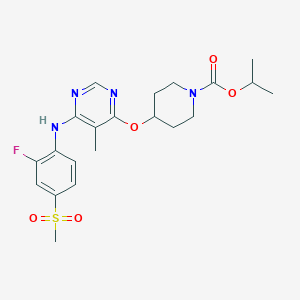 CHEMBL1951012 CHEMBL1951012 | C21H27FN4O5S | 466.528 | 9 / 1 | 3.3 | Yes |
| 443304 | 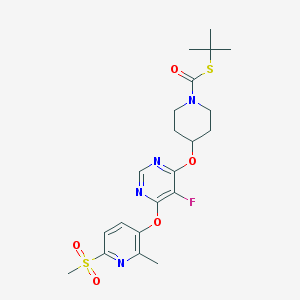 CHEMBL3326673 CHEMBL3326673 | C21H27FN4O5S2 | 498.588 | 10 / 0 | 3.4 | Yes |
| 53085 | 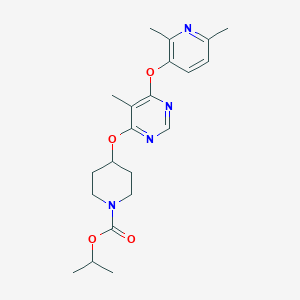 CHEMBL1949674 CHEMBL1949674 | C21H28N4O4 | 400.479 | 7 / 0 | 3.7 | Yes |
| 54122 | 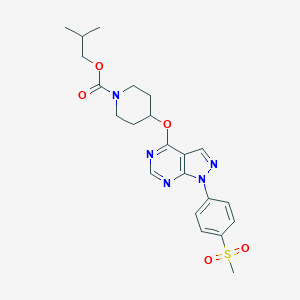 CHEMBL1775173 CHEMBL1775173 | C22H27N5O5S | 473.548 | 8 / 0 | 3.2 | Yes |
| 443890 | 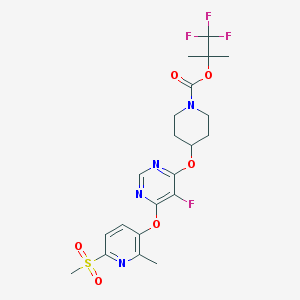 CHEMBL3325846 CHEMBL3325846 | C21H24F4N4O6S | 536.499 | 13 / 0 | 3.6 | No |
| 59099 | 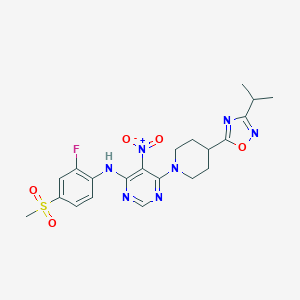 733750-99-7 733750-99-7 | C21H24FN7O5S | 505.525 | 12 / 1 | 3.9 | No |
| 60260 | 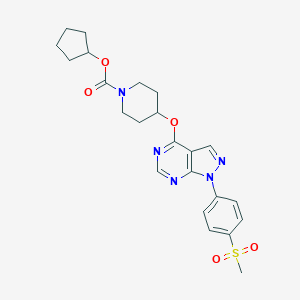 CHEMBL1775174 CHEMBL1775174 | C23H27N5O5S | 485.559 | 8 / 0 | 3.1 | Yes |
| 444084 | 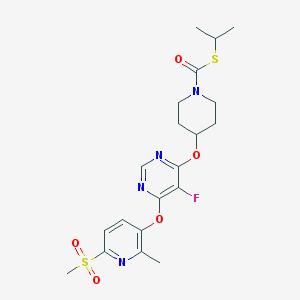 CHEMBL3326672 CHEMBL3326672 | C20H25FN4O5S2 | 484.561 | 10 / 0 | 3.2 | Yes |
| 69336 | 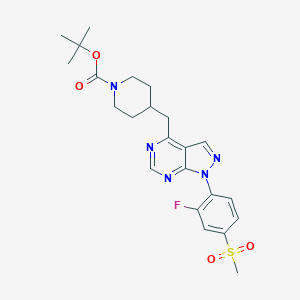 CHEMBL1773287 CHEMBL1773287 | C23H28FN5O4S | 489.566 | 8 / 0 | 3.1 | Yes |
| 76068 | 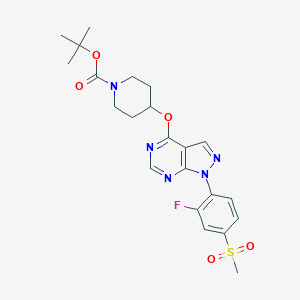 CHEMBL1775178 CHEMBL1775178 | C22H26FN5O5S | 491.538 | 9 / 0 | 2.9 | Yes |
| 76204 | 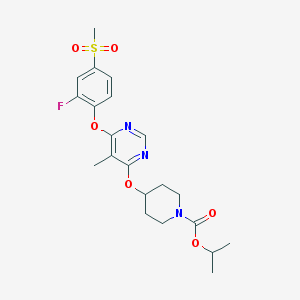 CHEMBL1951011 CHEMBL1951011 | C21H26FN3O6S | 467.512 | 9 / 0 | 3.3 | Yes |
| 90442 | 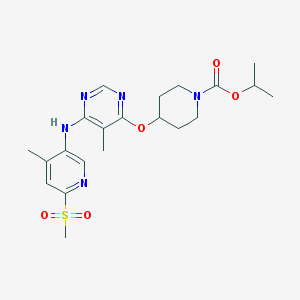 CHEMBL1951025 CHEMBL1951025 | C21H29N5O5S | 463.553 | 9 / 1 | 2.9 | Yes |
| 445414 | 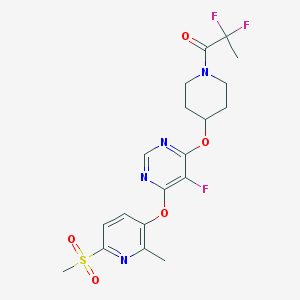 CHEMBL3326680 CHEMBL3326680 | C19H21F3N4O5S | 474.455 | 11 / 0 | 2.5 | No |
| 445816 | 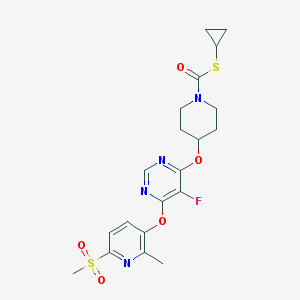 CHEMBL3326674 CHEMBL3326674 | C20H23FN4O5S2 | 482.545 | 10 / 0 | 2.9 | Yes |
| 112854 | 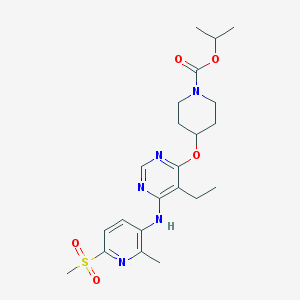 CHEMBL1951028 CHEMBL1951028 | C22H31N5O5S | 477.58 | 9 / 1 | 3.3 | Yes |
| 446310 | 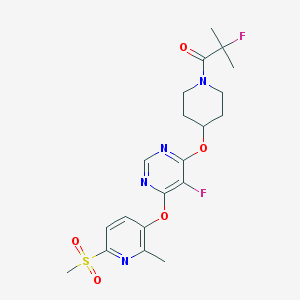 CHEMBL3325461 CHEMBL3325461 | C20H24F2N4O5S | 470.492 | 10 / 0 | 2.4 | Yes |
| 124197 | 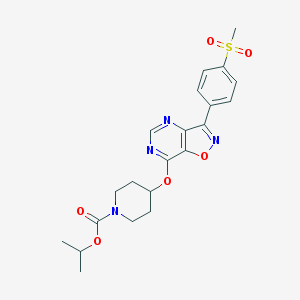 CHEMBL1773294 CHEMBL1773294 | C21H24N4O6S | 460.505 | 9 / 0 | 2.5 | Yes |
| 446686 | 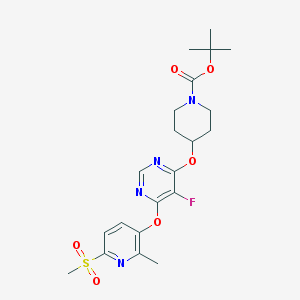 CHEMBL3325844 CHEMBL3325844 | C21H27FN4O6S | 482.527 | 10 / 0 | 2.8 | Yes |
| 131417 | 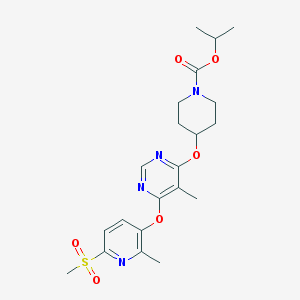 CHEMBL1951021 CHEMBL1951021 | C21H28N4O6S | 464.537 | 9 / 0 | 2.9 | Yes |
| 447188 | 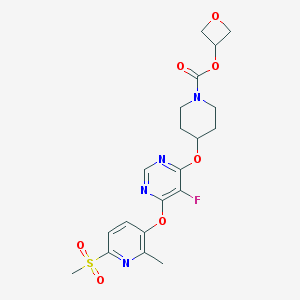 CHEMBL3326669 CHEMBL3326669 | C20H23FN4O7S | 482.483 | 11 / 0 | 1.5 | No |
| 447271 | 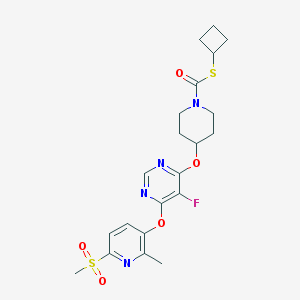 CHEMBL3326676 CHEMBL3326676 | C21H25FN4O5S2 | 496.572 | 10 / 0 | 3.3 | Yes |
| 143825 | 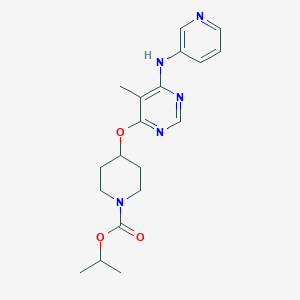 CHEMBL1951016 CHEMBL1951016 | C19H25N5O3 | 371.441 | 7 / 1 | 2.9 | Yes |
| 447414 | 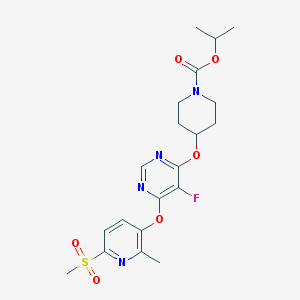 CHEMBL3325843 CHEMBL3325843 | C20H25FN4O6S | 468.5 | 10 / 0 | 2.6 | Yes |
| 148838 | 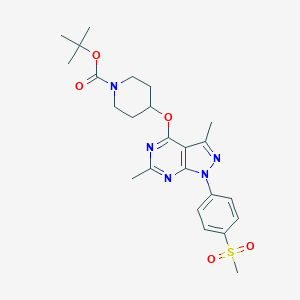 CHEMBL1773289 CHEMBL1773289 | C24H31N5O5S | 501.602 | 8 / 0 | 3.6 | No |
| 448050 | 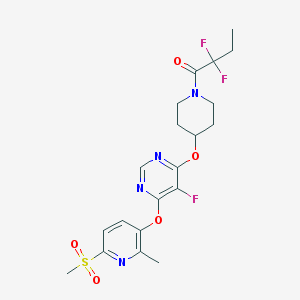 CHEMBL3326681 CHEMBL3326681 | C20H23F3N4O5S | 488.482 | 11 / 0 | 3.0 | No |
| 163313 | 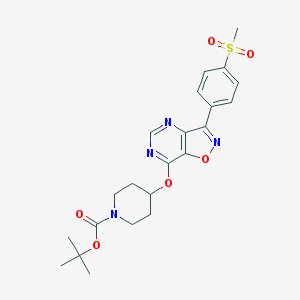 CHEMBL1773293 CHEMBL1773293 | C22H26N4O6S | 474.532 | 9 / 0 | 2.7 | Yes |
| 448113 | 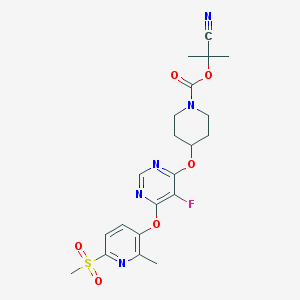 CHEMBL3325847 CHEMBL3325847 | C21H24FN5O6S | 493.51 | 11 / 0 | 2.2 | No |
| 448140 | 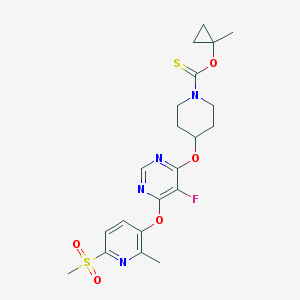 CHEMBL3326678 CHEMBL3326678 | C21H25FN4O5S2 | 496.572 | 10 / 0 | 3.2 | Yes |
| 176125 | 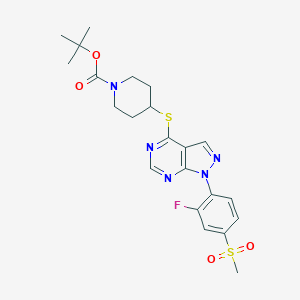 CHEMBL1773283 CHEMBL1773283 | C22H26FN5O4S2 | 507.599 | 9 / 0 | 3.5 | No |
| 179224 | 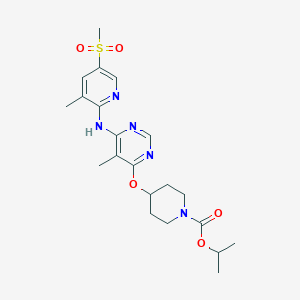 CHEMBL1951013 CHEMBL1951013 | C21H29N5O5S | 463.553 | 9 / 1 | 2.9 | Yes |
| 448946 | 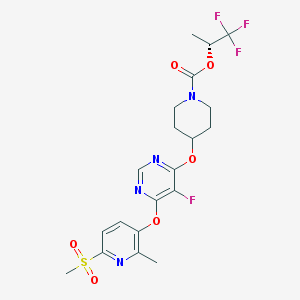 CHEMBL3325852 CHEMBL3325852 | C20H22F4N4O6S | 522.472 | 13 / 0 | 3.4 | No |
| 448949 | 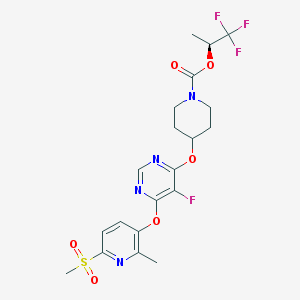 CHEMBL3326666 CHEMBL3326666 | C20H22F4N4O6S | 522.472 | 13 / 0 | 3.4 | No |
| 187695 | 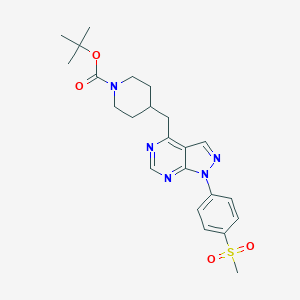 CHEMBL1773286 CHEMBL1773286 | C23H29N5O4S | 471.576 | 7 / 0 | 3.0 | Yes |
| 198441 | 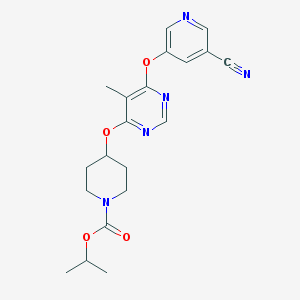 CHEMBL1951019 CHEMBL1951019 | C20H23N5O4 | 397.435 | 8 / 0 | 2.6 | Yes |
| 199906 | 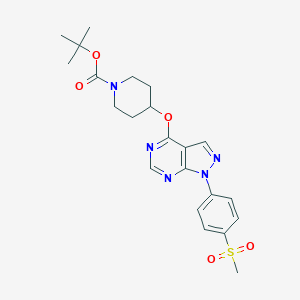 CHEMBL1775169 CHEMBL1775169 | C22H27N5O5S | 473.548 | 8 / 0 | 2.8 | Yes |
| 217225 | 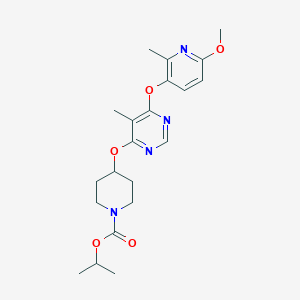 CHEMBL1951020 CHEMBL1951020 | C21H28N4O5 | 416.478 | 8 / 0 | 3.6 | Yes |
| 220130 | 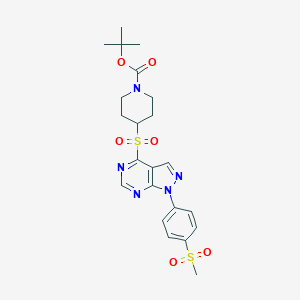 CHEMBL1773285 CHEMBL1773285 | C22H27N5O6S2 | 521.607 | 9 / 0 | 2.1 | No |
| 230198 | 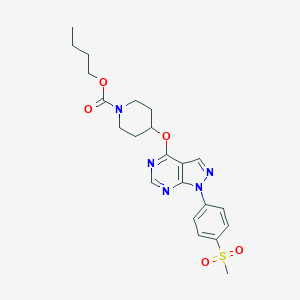 CHEMBL1775172 CHEMBL1775172 | C22H27N5O5S | 473.548 | 8 / 0 | 3.1 | Yes |
| 237014 | 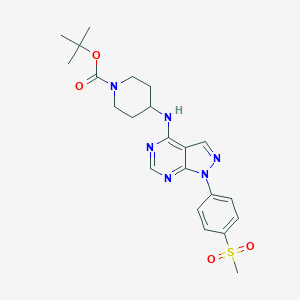 CHEMBL1773277 CHEMBL1773277 | C22H28N6O4S | 472.564 | 8 / 1 | 2.8 | Yes |
| 451326 | 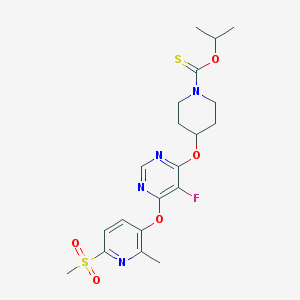 CHEMBL3326677 CHEMBL3326677 | C20H25FN4O5S2 | 484.561 | 10 / 0 | 3.2 | Yes |
| 252753 | 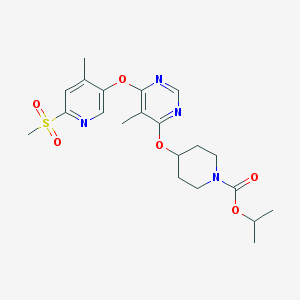 CHEMBL1951024 CHEMBL1951024 | C21H28N4O6S | 464.537 | 9 / 0 | 2.9 | Yes |
| 452135 | 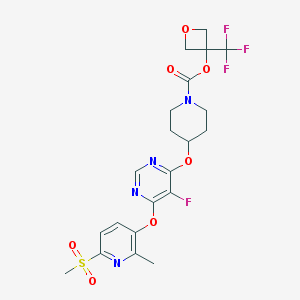 CHEMBL3326671 CHEMBL3326671 | C21H22F4N4O7S | 550.482 | 14 / 0 | 2.5 | No |
| 452139 | 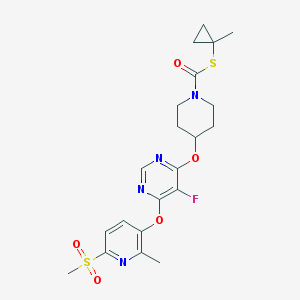 CHEMBL3326675 CHEMBL3326675 | C21H25FN4O5S2 | 496.572 | 10 / 0 | 3.1 | Yes |
| 269670 | 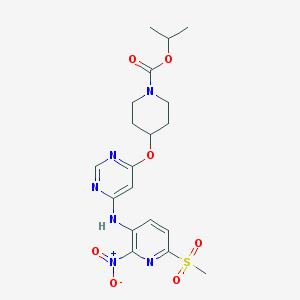 CHEMBL1949688 CHEMBL1949688 | C19H24N6O7S | 480.496 | 11 / 1 | 2.9 | No |
| 452624 | 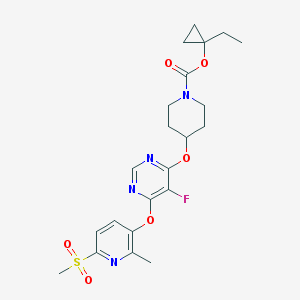 CHEMBL3326668 CHEMBL3326668 | C22H27FN4O6S | 494.538 | 10 / 0 | 3.1 | Yes |
| 282210 | 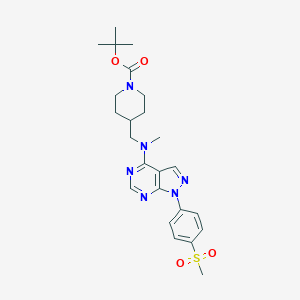 CHEMBL1773279 CHEMBL1773279 | C24H32N6O4S | 500.618 | 8 / 0 | 3.2 | No |
| 290606 | 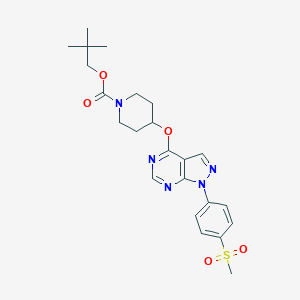 CHEMBL1775176 CHEMBL1775176 | C23H29N5O5S | 487.575 | 8 / 0 | 3.6 | Yes |
| 293256 | 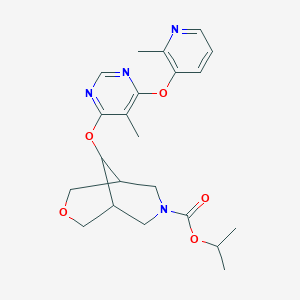 CHEMBL1766082 CHEMBL1766082 | C22H28N4O5 | 428.489 | 8 / 0 | 2.7 | Yes |
| 453436 | 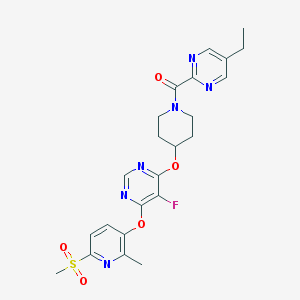 CHEMBL3326688 CHEMBL3326688 | C23H25FN6O5S | 516.548 | 11 / 0 | 2.5 | No |
| 300883 | 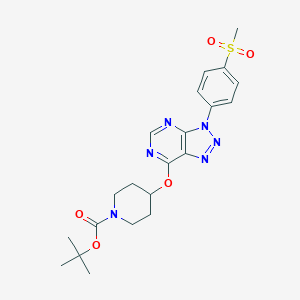 CHEMBL1773291 CHEMBL1773291 | C21H26N6O5S | 474.536 | 9 / 0 | 2.4 | Yes |
| 312873 | 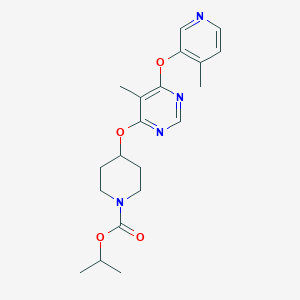 CHEMBL1951017 CHEMBL1951017 | C20H26N4O4 | 386.452 | 7 / 0 | 3.3 | Yes |
| 454723 | 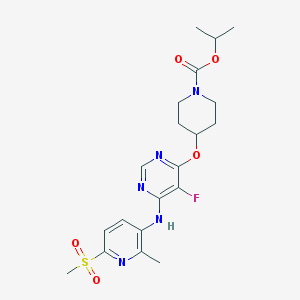 CHEMBL3325842 CHEMBL3325842 | C20H26FN5O5S | 467.516 | 10 / 1 | 2.6 | Yes |
| 454753 | 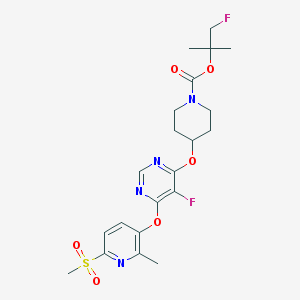 CHEMBL3325848 CHEMBL3325848 | C21H26F2N4O6S | 500.518 | 11 / 0 | 2.8 | No |
| 328719 | 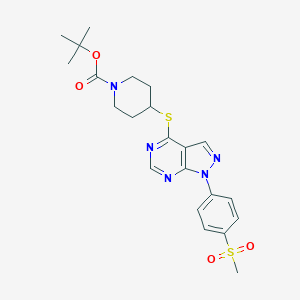 CHEMBL1773282 CHEMBL1773282 | C22H27N5O4S2 | 489.609 | 8 / 0 | 3.4 | Yes |
| 454854 | 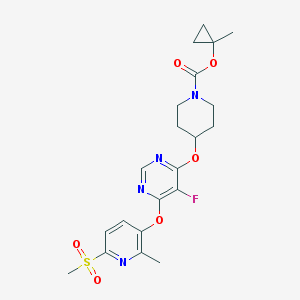 CHEMBL3326667 CHEMBL3326667 | C21H25FN4O6S | 480.511 | 10 / 0 | 2.6 | Yes |
| 455390 | 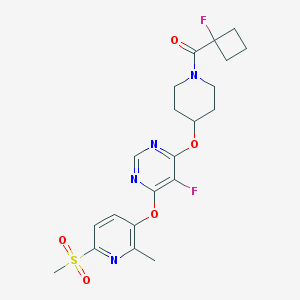 CHEMBL3326682 CHEMBL3326682 | C21H24F2N4O5S | 482.503 | 10 / 0 | 2.5 | Yes |
| 345438 | 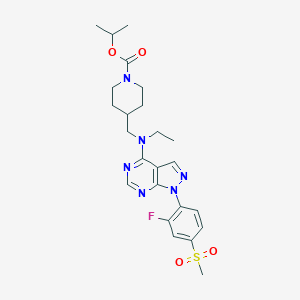 CHEMBL1773281 CHEMBL1773281 | C24H31FN6O4S | 518.608 | 9 / 0 | 3.5 | No |
| 357323 | 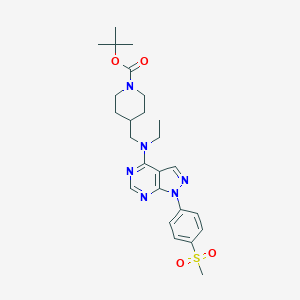 CHEMBL1773280 CHEMBL1773280 | C25H34N6O4S | 514.645 | 8 / 0 | 3.5 | No |
| 362837 | 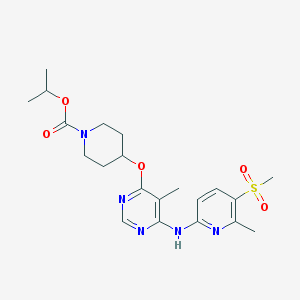 CHEMBL1951014 CHEMBL1951014 | C21H29N5O5S | 463.553 | 9 / 1 | 2.9 | Yes |
| 371007 | 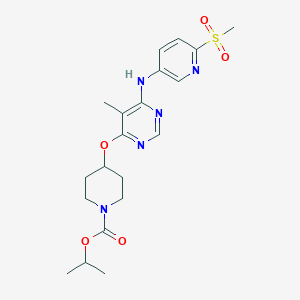 CHEMBL1951023 CHEMBL1951023 | C20H27N5O5S | 449.526 | 9 / 1 | 2.5 | Yes |
| 456470 | 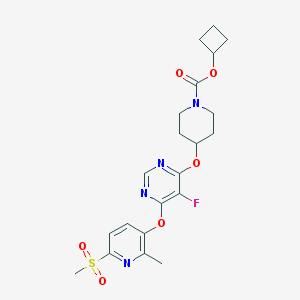 CHEMBL3325845 CHEMBL3325845 | C21H25FN4O6S | 480.511 | 10 / 0 | 2.7 | Yes |
| 376808 | 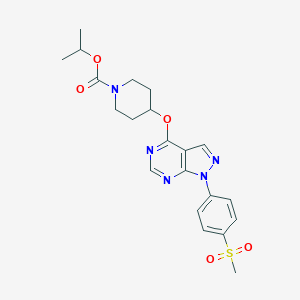 CHEMBL1775171 CHEMBL1775171 | C21H25N5O5S | 459.521 | 8 / 0 | 2.6 | Yes |
| 378924 | 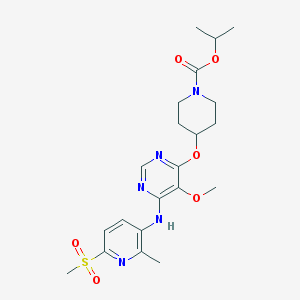 APD597 APD597 | C21H29N5O6S | 479.552 | 10 / 1 | 2.5 | Yes |
| 456971 | 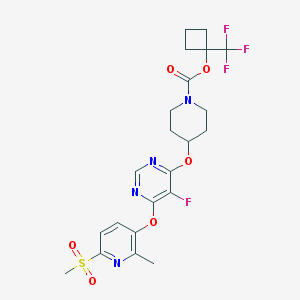 CHEMBL3326670 CHEMBL3326670 | C22H24F4N4O6S | 548.51 | 13 / 0 | 3.7 | No |
| 457066 | 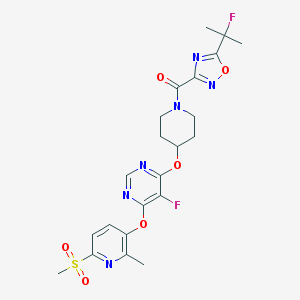 CHEMBL3326686 CHEMBL3326686 | C22H24F2N6O6S | 538.527 | 13 / 0 | 2.4 | No |
| 391101 | 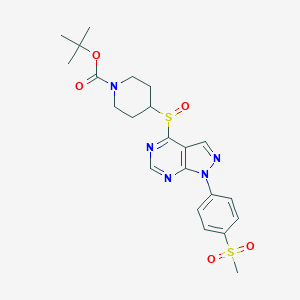 CHEMBL1773284 CHEMBL1773284 | C22H27N5O5S2 | 505.608 | 9 / 0 | 1.9 | No |
| 400657 | 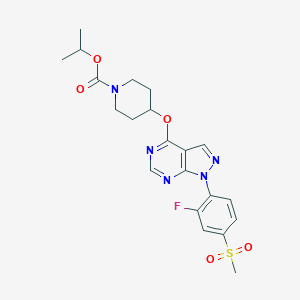 APD668 APD668 | C21H24FN5O5S | 477.511 | 9 / 0 | 2.7 | Yes |
| 457744 | 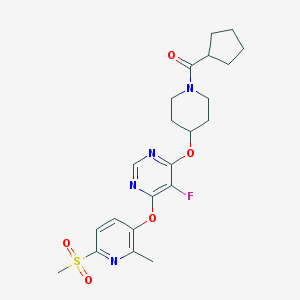 CHEMBL3326683 CHEMBL3326683 | C22H27FN4O5S | 478.539 | 9 / 0 | 3.0 | Yes |
| 410299 | 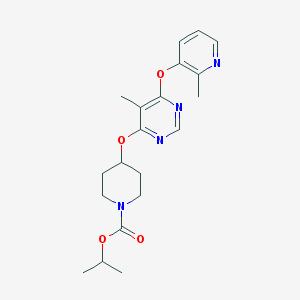 CHEMBL1766081 CHEMBL1766081 | C20H26N4O4 | 386.452 | 7 / 0 | 3.3 | Yes |
| 458582 | 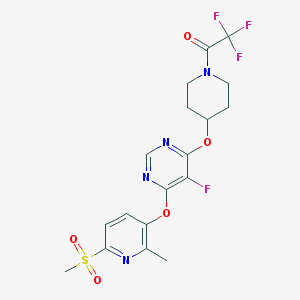 CHEMBL3326679 CHEMBL3326679 | C18H18F4N4O5S | 478.419 | 12 / 0 | 2.5 | No |
| 431265 | 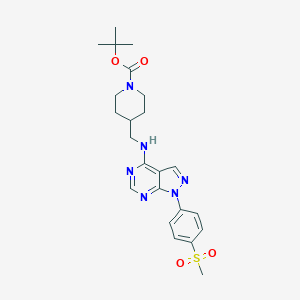 CHEMBL1773278 CHEMBL1773278 | C23H30N6O4S | 486.591 | 8 / 1 | 3.0 | Yes |
| 432538 | 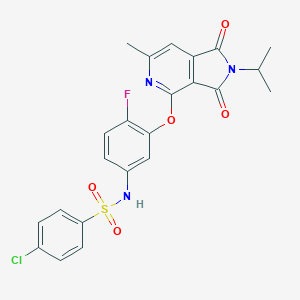 CHEMBL3104879 CHEMBL3104879 | C23H19ClFN3O5S | 503.929 | 8 / 1 | 4.1 | No |
| 438030 | 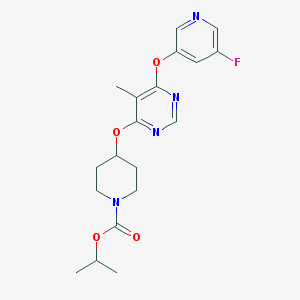 CHEMBL1951018 CHEMBL1951018 | C19H23FN4O4 | 390.415 | 8 / 0 | 3.0 | Yes |
| 438232 | 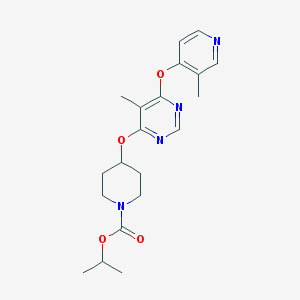 CHEMBL1951026 CHEMBL1951026 | C20H26N4O4 | 386.452 | 7 / 0 | 3.3 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417