

You can:
| Name | G-protein coupled receptor 4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | GPR4 |
| Synonym | G-protein coupled receptor 19 GPR19 GPR4 |
| Disease | N/A |
| Length | 362 |
| Amino acid sequence | MGNHTWEGCHVDSRVDHLFPPSLYIFVIGVGLPTNCLALWAAYRQVQQRNELGVYLMNLSIADLLYICTLPLWVDYFLHHDNWIHGPGSCKLFGFIFYTNIYISIAFLCCISVDRYLAVAHPLRFARLRRVKTAVAVSSVVWATELGANSAPLFHDELFRDRYNHTFCFEKFPMEGWVAWMNLYRVFVGFLFPWALMLLSYRGILRAVRGSVSTERQEKAKIKRLALSLIAIVLVCFAPYHVLLLSRSAIYLGRPWDCGFEERVFSAYHSSLAFTSLNCVADPILYCLVNEGARSDVAKALHNLLRFLASDKPQEMANASLTLETPLTSKRNSTAKAMTGSWAATPPSQGDQVQLKMLPPAQ |
| UniProt | P46093 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P46093 |
| 3D structure model | This predicted structure model is from GPCR-EXP P46093. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL3638324 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 1605 | 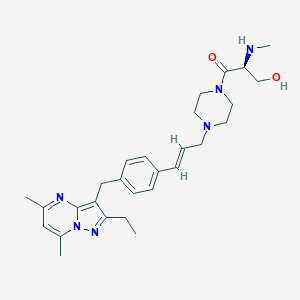 CHEMBL3675725 CHEMBL3675725 | C28H38N6O2 | 490.652 | 6 / 2 | 2.6 | Yes |
| 521493 | 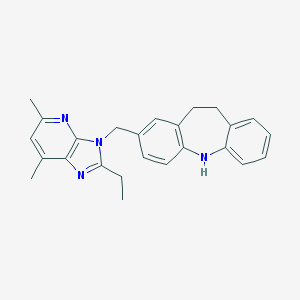 CHEMBL3810032 CHEMBL3810032 | C25H26N4 | 382.511 | 3 / 1 | 5.7 | No |
| 521626 | 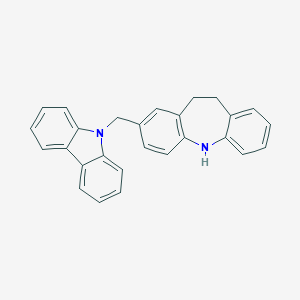 CHEMBL3810295 CHEMBL3810295 | C27H22N2 | 374.487 | 1 / 1 | 6.8 | No |
| 521663 | 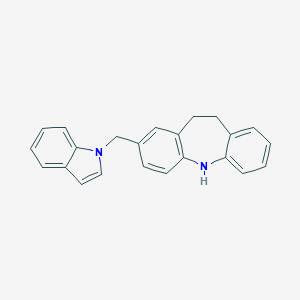 CHEMBL3810132 CHEMBL3810132 | C23H20N2 | 324.427 | 1 / 1 | 5.4 | No |
| 11143 | 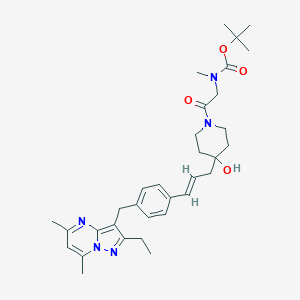 CHEMBL3670936 CHEMBL3670936 | C33H45N5O4 | 575.754 | 6 / 1 | 4.9 | No |
| 11667 | 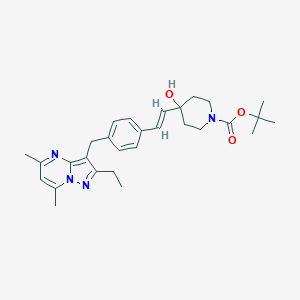 CHEMBL3670930 CHEMBL3670930 | C29H38N4O3 | 490.648 | 5 / 1 | 4.9 | Yes |
| 12167 | 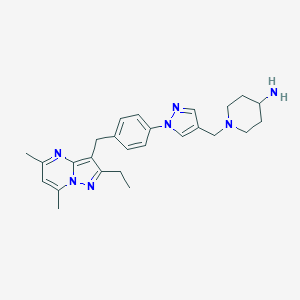 CHEMBL3675739 CHEMBL3675739 | C26H33N7 | 443.599 | 5 / 1 | 3.3 | Yes |
| 13240 | 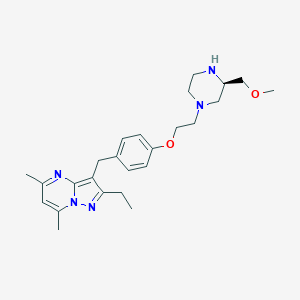 CHEMBL3675736 CHEMBL3675736 | C25H35N5O2 | 437.588 | 6 / 1 | 3.2 | Yes |
| 521900 | 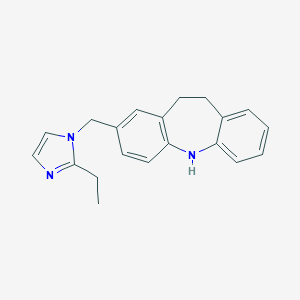 CHEMBL3809759 CHEMBL3809759 | C20H21N3 | 303.409 | 2 / 1 | 4.3 | Yes |
| 16193 | 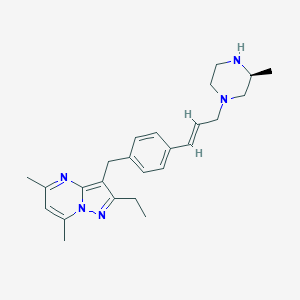 CHEMBL3670938 CHEMBL3670938 | C25H33N5 | 403.574 | 4 / 1 | 4.1 | Yes |
| 521959 | 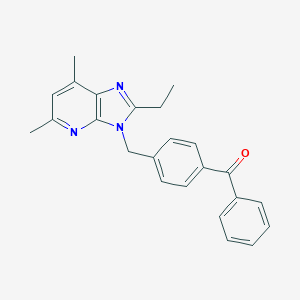 SCHEMBL5426008 SCHEMBL5426008 | C24H23N3O | 369.468 | 3 / 0 | 5.2 | No |
| 522355 | 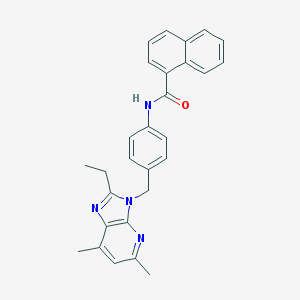 CHEMBL3728144 CHEMBL3728144 | C28H26N4O | 434.543 | 3 / 1 | 6.0 | No |
| 28369 | 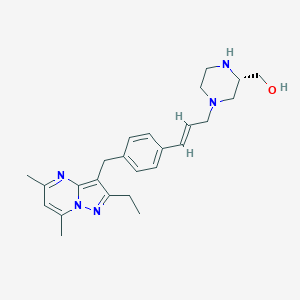 CHEMBL3670937 CHEMBL3670937 | C25H33N5O | 419.573 | 5 / 2 | 3.1 | Yes |
| 37728 | 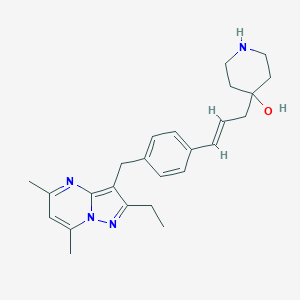 CHEMBL3670932 CHEMBL3670932 | C25H32N4O | 404.558 | 4 / 2 | 4.0 | Yes |
| 42361 | 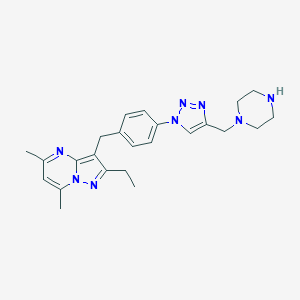 CHEMBL3675741 CHEMBL3675741 | C24H30N8 | 430.56 | 6 / 1 | 2.5 | Yes |
| 51095 | 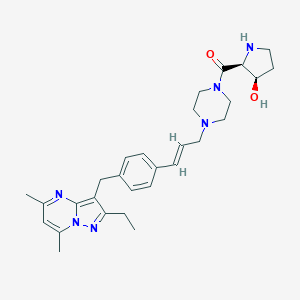 CHEMBL3675726 CHEMBL3675726 | C29H38N6O2 | 502.663 | 6 / 2 | 3.3 | No |
| 58506 | 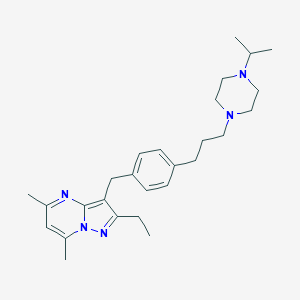 CHEMBL3639746 CHEMBL3639746 | C27H39N5 | 433.644 | 4 / 0 | 5.1 | No |
| 70568 | 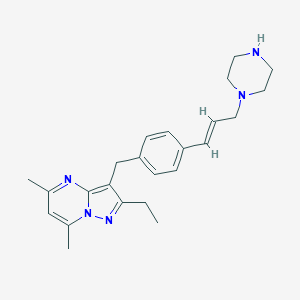 CHEMBL3675724 CHEMBL3675724 | C24H31N5 | 389.547 | 4 / 1 | 3.7 | Yes |
| 524561 | 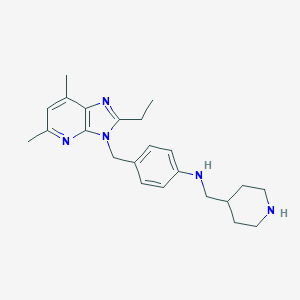 SCHEMBL5417472 SCHEMBL5417472 | C23H31N5 | 377.536 | 4 / 2 | 4.2 | Yes |
| 105355 | 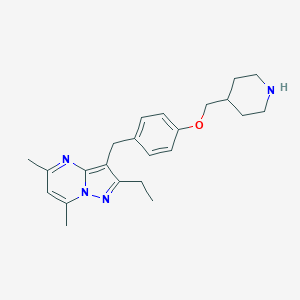 CHEMBL3675730 CHEMBL3675730 | C23H30N4O | 378.52 | 4 / 1 | 4.2 | Yes |
| 126538 | 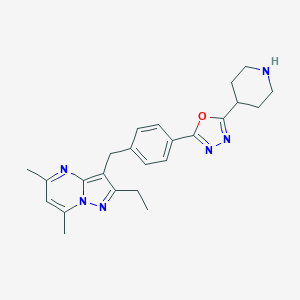 CHEMBL3675743 CHEMBL3675743 | C24H28N6O | 416.529 | 6 / 1 | 3.5 | Yes |
| 131447 | 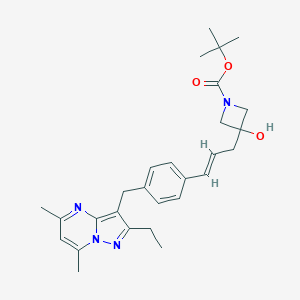 CHEMBL3670931 CHEMBL3670931 | C28H36N4O3 | 476.621 | 5 / 1 | 4.6 | Yes |
| 525457 | 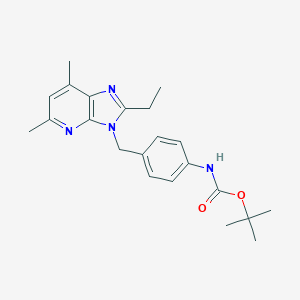 SCHEMBL9129857 SCHEMBL9129857 | C22H28N4O2 | 380.492 | 4 / 1 | 4.5 | Yes |
| 525483 | 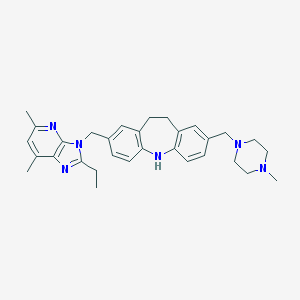 CHEMBL3809413 CHEMBL3809413 | C31H38N6 | 494.687 | 5 / 1 | 5.4 | No |
| 525623 | 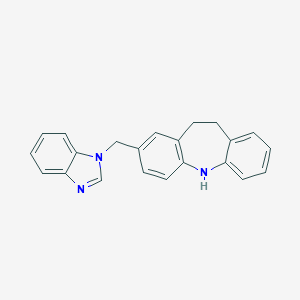 CHEMBL3810150 CHEMBL3810150 | C22H19N3 | 325.415 | 2 / 1 | 4.9 | Yes |
| 525668 | 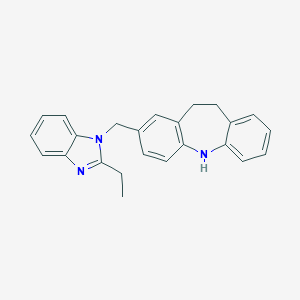 CHEMBL3809249 CHEMBL3809249 | C24H23N3 | 353.469 | 2 / 1 | 5.7 | No |
| 144334 | 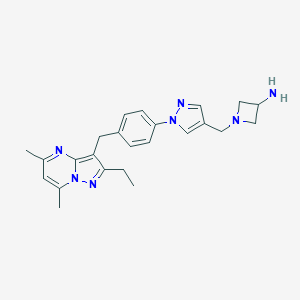 CHEMBL3675738 CHEMBL3675738 | C24H29N7 | 415.545 | 5 / 1 | 2.6 | Yes |
| 155577 | 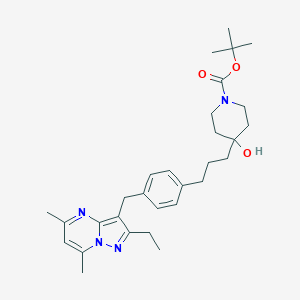 CHEMBL3670933 CHEMBL3670933 | C30H42N4O3 | 506.691 | 5 / 1 | 5.4 | No |
| 168715 | 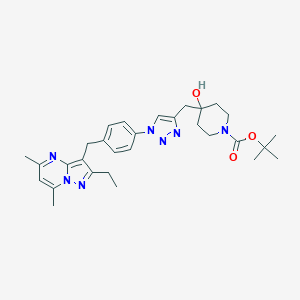 CHEMBL3675742 CHEMBL3675742 | C30H39N7O3 | 545.688 | 7 / 1 | 4.2 | No |
| 169982 | 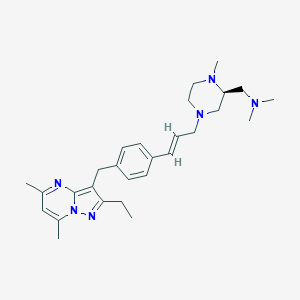 CHEMBL3670942 CHEMBL3670942 | C28H40N6 | 460.67 | 5 / 0 | 4.3 | Yes |
| 185565 | 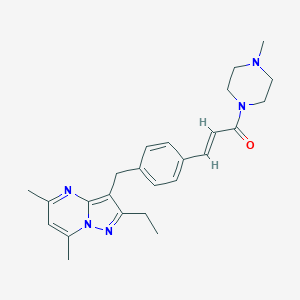 CHEMBL3675728 CHEMBL3675728 | C25H31N5O | 417.557 | 4 / 0 | 3.6 | Yes |
| 198887 | 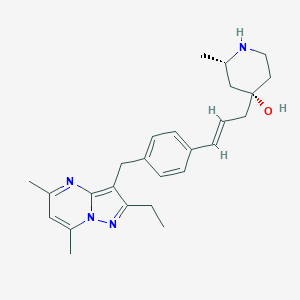 CHEMBL3670934 CHEMBL3670934 | C26H34N4O | 418.585 | 4 / 2 | 4.5 | Yes |
| 527371 | 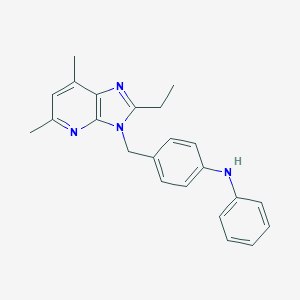 CHEMBL3727521 CHEMBL3727521 | C23H24N4 | 356.473 | 3 / 1 | 5.4 | No |
| 527547 | 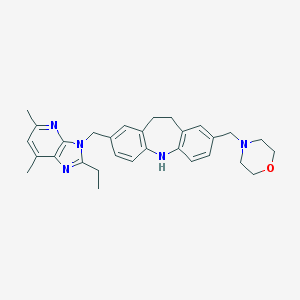 SCHEMBL6046517 SCHEMBL6046517 | C30H35N5O | 481.644 | 5 / 1 | 5.2 | No |
| 221266 | 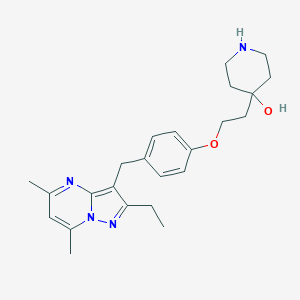 CHEMBL3675734 CHEMBL3675734 | C24H32N4O2 | 408.546 | 5 / 2 | 3.5 | Yes |
| 527981 | 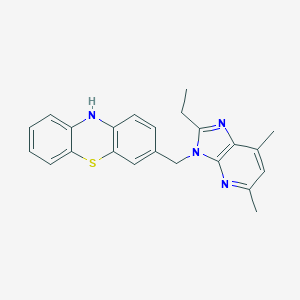 CHEMBL3808576 CHEMBL3808576 | C23H22N4S | 386.517 | 4 / 1 | 5.6 | No |
| 528111 | 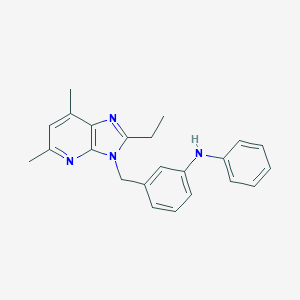 SCHEMBL5780505 SCHEMBL5780505 | C23H24N4 | 356.473 | 3 / 1 | 5.4 | No |
| 528234 | 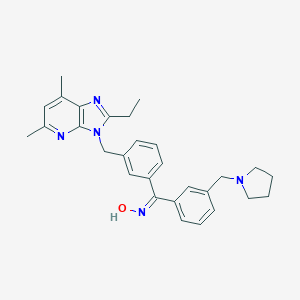 CHEMBL3731852 CHEMBL3731852 | C29H33N5O | 467.617 | 5 / 1 | 5.8 | No |
| 528255 | 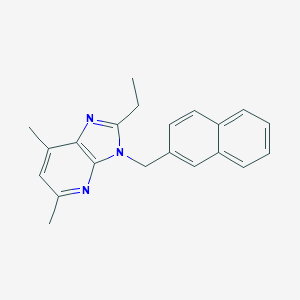 CHEMBL3810343 CHEMBL3810343 | C21H21N3 | 315.42 | 2 / 0 | 5.1 | No |
| 528621 | 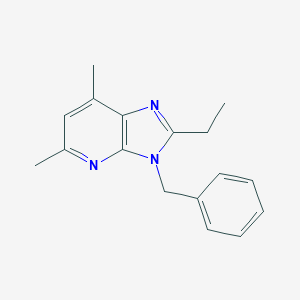 CHEMBL3810339 CHEMBL3810339 | C17H19N3 | 265.36 | 2 / 0 | 3.9 | Yes |
| 252203 | 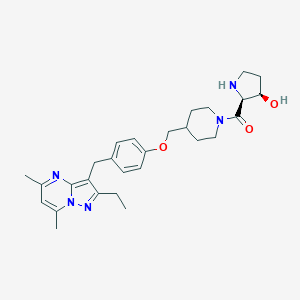 CHEMBL3675731 CHEMBL3675731 | C28H37N5O3 | 491.636 | 6 / 2 | 3.8 | Yes |
| 255698 | 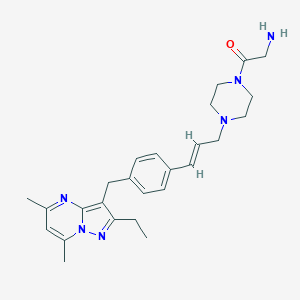 CHEMBL3670940 CHEMBL3670940 | C26H34N6O | 446.599 | 5 / 1 | 2.7 | Yes |
| 528892 | 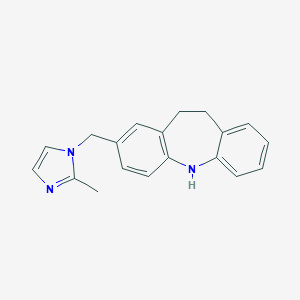 CHEMBL3808954 CHEMBL3808954 | C19H19N3 | 289.382 | 2 / 1 | 3.8 | Yes |
| 529320 | 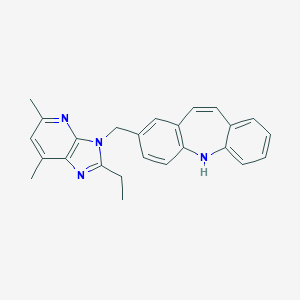 CHEMBL3809977 CHEMBL3809977 | C25H24N4 | 380.495 | 3 / 1 | 5.9 | No |
| 530647 | 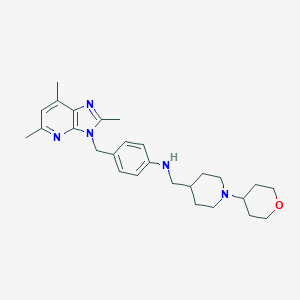 SCHEMBL5428753 SCHEMBL5428753 | C27H37N5O | 447.627 | 5 / 1 | 4.6 | Yes |
| 325980 | 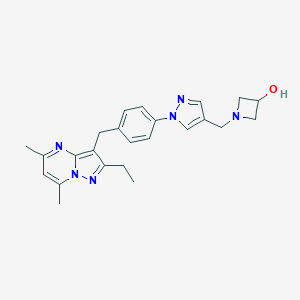 CHEMBL3675737 CHEMBL3675737 | C24H28N6O | 416.529 | 5 / 1 | 2.8 | Yes |
| 332008 | 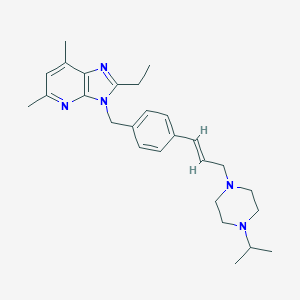 GPR4 antagonist 1 GPR4 antagonist 1 | C27H37N5 | 431.628 | 4 / 0 | 5.0 | Yes |
| 530925 | 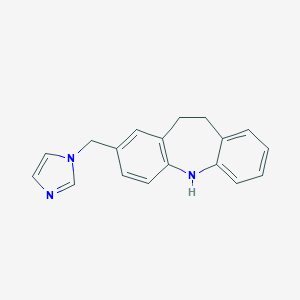 CHEMBL3808552 CHEMBL3808552 | C18H17N3 | 275.355 | 2 / 1 | 3.4 | Yes |
| 340768 | 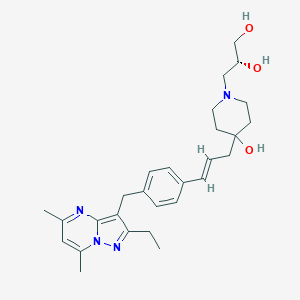 CHEMBL3670935 CHEMBL3670935 | C28H38N4O3 | 478.637 | 6 / 3 | 3.2 | Yes |
| 343424 | 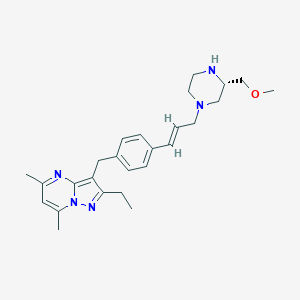 CHEMBL3670939 CHEMBL3670939 | C26H35N5O | 433.6 | 5 / 1 | 3.6 | Yes |
| 531256 | 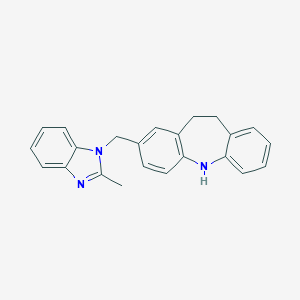 CHEMBL3808563 CHEMBL3808563 | C23H21N3 | 339.442 | 2 / 1 | 5.3 | No |
| 355648 | 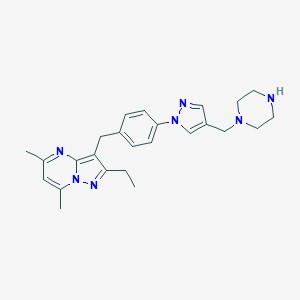 CHEMBL3675740 CHEMBL3675740 | C25H31N7 | 429.572 | 5 / 1 | 3.0 | Yes |
| 363805 | 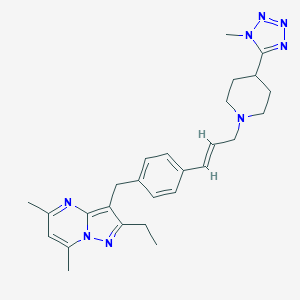 CHEMBL3670941 CHEMBL3670941 | C27H34N8 | 470.625 | 6 / 0 | 4.3 | Yes |
| 531778 | 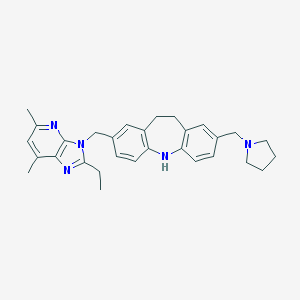 SCHEMBL6046468 SCHEMBL6046468 | C30H35N5 | 465.645 | 4 / 1 | 6.1 | No |
| 369845 | 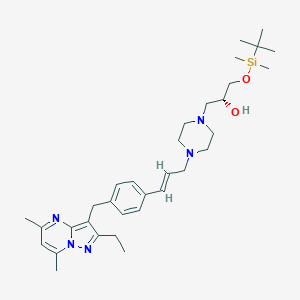 CHEMBL3675727 CHEMBL3675727 | C33H51N5O2Si | 577.889 | 6 / 1 | N/A | No |
| 391336 | 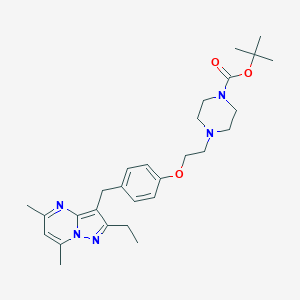 CHEMBL3675735 CHEMBL3675735 | C28H39N5O3 | 493.652 | 6 / 0 | 4.5 | Yes |
| 533083 | 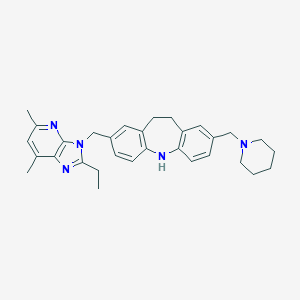 CHEMBL3810385 CHEMBL3810385 | C31H37N5 | 479.672 | 4 / 1 | 6.4 | No |
| 429257 | 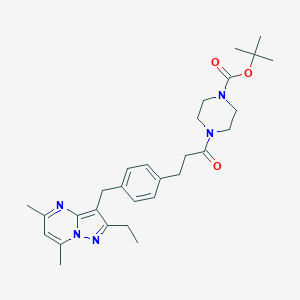 CHEMBL3675729 CHEMBL3675729 | C29H39N5O3 | 505.663 | 5 / 0 | 4.2 | No |
| 438272 | 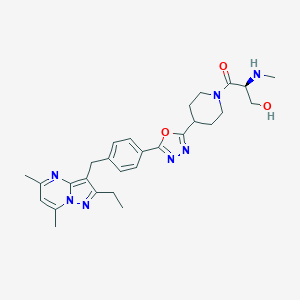 CHEMBL3675745 CHEMBL3675745 | C28H35N7O3 | 517.634 | 8 / 2 | 2.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417