

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MGNHTWEGCHVDSRVDHLFPPSLYIFVIGVGLPTNCLALWAAYRQVQQRNELGVYLMNLSIADLLYICTLPLWVDYFLHHDNWIHGPGSCKLFGFIFYTNIYISIAFLCCISVDRYLAVAHPLRFARLRRVKTAVAVSSVVWATELGANSAPLFHDELFRDRYNHTFCFEKFPMEGWVAWMNLYRVFVGFLFPWALMLLSYRGILRAVRGSVSTERQEKAKIKRLALSLIAIVLVCFAPYHVLLLSRSAIYLGRPWDCGFEERVFSAYHSSLAFTSLNCVADPILYCLVNEGARSDVAKALHNLLRFLASDKPQEMANASLTLETPLTSKRNSTAKAMTGSWAATPPSQGDQVQLKMLPPAQ | |
| CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHCSSCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99999999998741688879999999999999989998898663267886989999999999999999879999999579998888567999999999999999999999999723787512000136675011001489999999999899998701457999557608798768999999999999999999999999999999980765556421332899999999999998568999999999998478898169999999999999999999989899998048989999999999873366777777777888762278877788877777788889998788889855699998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MGNHTWEGCHVDSRVDHLFPPSLYIFVIGVGLPTNCLALWAAYRQVQQRNELGVYLMNLSIADLLYICTLPLWVDYFLHHDNWIHGPGSCKLFGFIFYTNIYISIAFLCCISVDRYLAVAHPLRFARLRRVKTAVAVSSVVWATELGANSAPLFHDELFRDRYNHTFCFEKFPMEGWVAWMNLYRVFVGFLFPWALMLLSYRGILRAVRGSVSTERQEKAKIKRLALSLIAIVLVCFAPYHVLLLSRSAIYLGRPWDCGFEERVFSAYHSSLAFTSLNCVADPILYCLVNEGARSDVAKALHNLLRFLASDKPQEMANASLTLETPLTSKRNSTAKAMTGSWAATPPSQGDQVQLKMLPPAQ | |
| 76142364042544031100020012002302310110000001223433101000000000000000001010000024330200300120012211200100000000002100000010020242233210000000011000000011000120345742211010313471020001121113023312200120000001102324546443110000000000000000132101000010013032344041340020011001000130001102010000450142014002400432445544534444444534432445444444545444434543451336514278 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHCSSCCCCCSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MGNHTWEGCHVDSRVDHLFPPSLYIFVIGVGLPTNCLALWAAYRQVQQRNELGVYLMNLSIADLLYICTLPLWVDYFLHHDNWIHGPGSCKLFGFIFYTNIYISIAFLCCISVDRYLAVAHPLRFARLRRVKTAVAVSSVVWATELGANSAPLFHDELFRDRYNHTFCFEKFPMEGWVAWMNLYRVFVGFLFPWALMLLSYRGILRAVRGSVSTERQEKAKIKRLALSLIAIVLVCFAPYHVLLLSRSAIYLGRPWDCGFEERVFSAYHSSLAFTSLNCVADPILYCLVNEGARSDVAKALHNLLRFLASDKPQEMANASLTLETPLTSKRNSTAKAMTGSWAATPPSQGDQVQLKMLPPAQ | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.24 | 0.22 | 0.80 | 3.30 | Download | -------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG-LHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK-KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------ | |||||||||||||||||||
| 2 | 4xnwA | 0.29 | 0.27 | 0.82 | 4.24 | Download | ---SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLQTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR--------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.24 | 0.22 | 0.80 | 3.60 | Download | -------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG-LHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM-KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.24 | 0.25 | 0.77 | 1.56 | Download | ------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVEDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.24 | 0.27 | 0.79 | 1.18 | Download | -----------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE--DSVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.24 | 0.22 | 0.80 | 3.33 | Download | --------CQKIKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKE-GLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEE-EKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------ | |||||||||||||||||||
| 7 | 5t1a | 0.24 | 0.27 | 0.78 | 1.72 | Download | ------------KQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE--DSVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.23 | 0.22 | 0.80 | 4.40 | Download | -------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE-----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------ | |||||||||||||||||||
| 9 | 4mbsA | 0.24 | 0.22 | 0.80 | 3.46 | Download | -------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK-KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.25 | 0.24 | 0.80 | 4.35 | Download | -GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKY-RQGSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI------TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
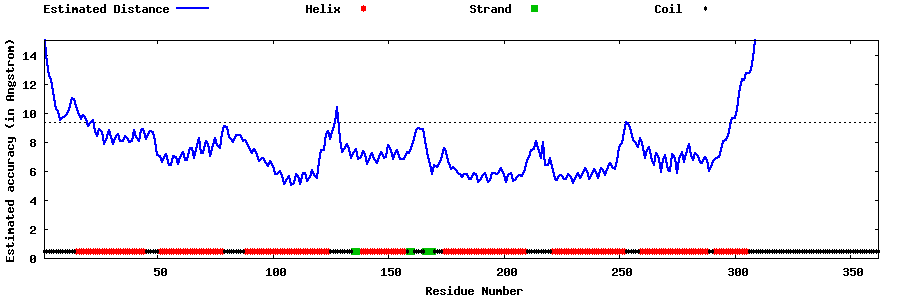
| Generated 3D models | Estimated local accuracy of models | ||
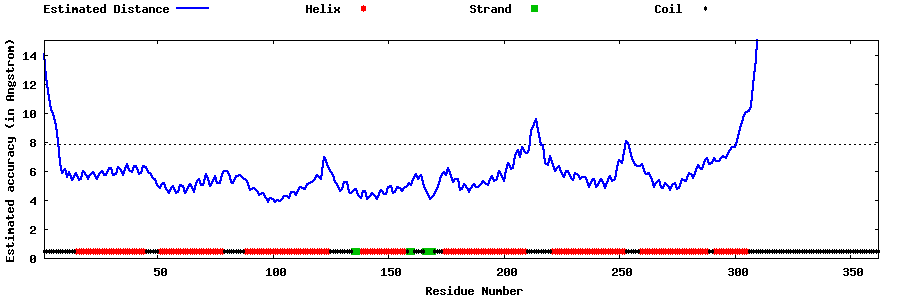 | |||
|
|
|||
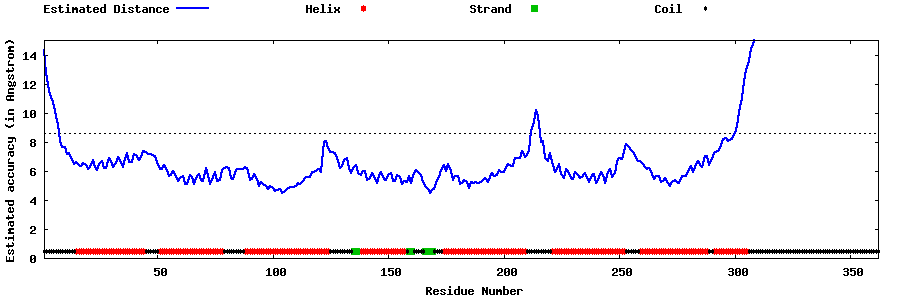 | |||
|
| |||
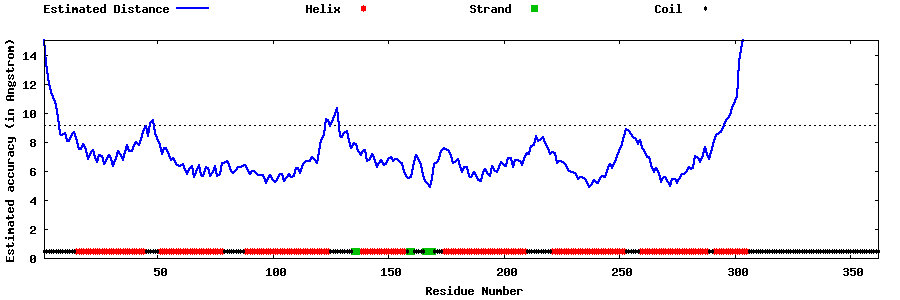 | |||
|
| |||
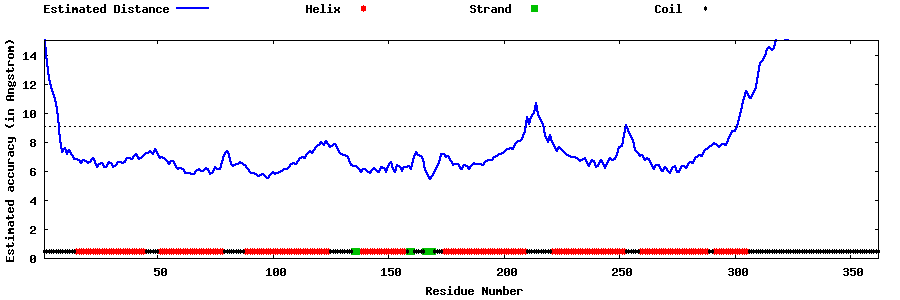 | |||
|
| |||
 | |||
|
| |||