

Go to page (83 pages in total): [First page] << < 46 47 48 49 50 51 52 53 54 55 56 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 5001 |
PF02957 (TT_ORF2) |
 Estimated TM-score=0.50; very hard target. |
>TT viral ORF2 (Length=118) WRPPRRNEFTIQRDWFYSCFHSHSSMCGCADFINHLNHIAAMLGRPEDQNPPPPPGALRPLPALPAAPEAPGDRAPWPMG GGASGEGARGGGGDGAAGDAVGDPADADLVAAIDAAEQ |
| 5002 |
PF02890 (DUF226) |
 Estimated TM-score=0.50; very hard target. |
>Borrelia family of unknown function DUF226 (Length=139) YKLGIDNKRNECRISLRTLFNQMKVEEVRLYPIKEGDKFLGIYYGYRKPIKNIFVKYEINGATKSYGLSKAHYIEFRFKK GSVFCYFKGLFRLLKKEKENTPYNMACIDMFTKLEKHVYEFYGKKYPEKGIITRWIKKN |
| 5003 |
PF02208 (Sorb) |
 Estimated TM-score=0.50; hard target. |
>Sorbin homologous domain (Length=42) PQHMADTSVDDIGIPLRNTDRSKDWYKTMFKQIHKLNRDEDS |
| 5004 |
PF02200 (STE) |
 Estimated TM-score=0.50; hard target. |
>STE like transcription factor (Length=108) FIKRFHLPNGESISCVLWDNVFFISGTDIVRSLTFRFHAFGRPVINPKKFEEGIFSDLRNLKPGHDARLEDPKSELLDIL YKNNCIRTQKKQKVFYWFSVPHDRLFLD |
| 5005 |
PF01677 (Herpes_UL7) |
 Estimated TM-score=0.50; very hard target. |
>Herpesvirus UL7 like (Length=211) MVLEVNRCENLCVACNSPVLVRDGALCVSQTLDYVKQKLVSDTFMGFAMACLLDCEDLVDSINLAPHVFAQRVFVFRPPN SYLLEMCVLSSMLENCETYTRRFVESLVSRARFIYSKSPCIDACFLLHSIEMMASTIIDYFKLDPEGAQPRYPGLMMYKL HGAIEDGSFESKGLLRPIYHESFKLCSDPDASFDEEEETGEAGVTFNVFYC |
| 5006 |
PF01562 (Pep_M12B_propep) |
 Estimated TM-score=0.50; very hard target. |
>Reprolysin family propeptide (Length=155) EIVTPARLNEVGEQLPTGVHFKRRKRSTDPTTANISHHWTSPHAYYQISAFGQDYYLNLTLESGFIAPVYTVTILGASSE GHNSVEGEEEEDTEYQHCFYKGHVNAGQEHTAVISLCSGLLGTFRSPEGEFFVEPLHSYNSEHYEEEHIKPHVVY |
| 5007 |
PF01407 (Gemini_AL3) |
 Estimated TM-score=0.50; hard target. |
>Geminivirus AL3 protein (Length=120) DSRTGENITAHQAENSVFIWEVPNPLYFKIMRVEDPAYTRTRIYHIQIRFNHNLRKALDLHKAFLNFQVWTTSIQASGTT YLNRFRLLVLLYLHRLGVIGINNVIRAVQFATNKSYVNTV |
| 5008 |
PF01104 (Bunya_NS-S) |
 Estimated TM-score=0.50; hard target. |
>Bunyavirus non-structural protein NS-s (Length=91) MYHNGMQLHLTRRSGMWHLLVSMGNNSISVLLESSSSTRRRPRWSYIRRHNQVSILLLVGSNSQWLITIFPNMSQILCQT MPSHSTGCQDT |
| 5009 |
PF01021 (TYA) |
 Estimated TM-score=0.50; very hard target. |
>TYA transposon protein (Length=98) ACASVTSKEVHTNQDPLDVSASKIQEYDKASTKANSQQTTTPASSAVPENPHHASPQPASVPPPQNGPYPQQCMMTQNQA NPSGWSFYGHPSMIPYTP |
| 5010 |
PF01010 (Proton_antipo_C) |
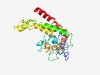 Estimated TM-score=0.50; easy target. |
>NADH-dehyrogenase subunit F, TMs, (complex I) C-terminus (Length=244) RIYLLTFEGHLNVHFQNYSGKKNTPFYSISLWGKVGPKTINQKFRLLTLLTMNNNESASFFSKKTYQIDENIRKMTRPFI TITHFCNKNTYAYPYESDNTMLFPLLVLVLFTLFVGSIGIPFNQKGMDLDILSKWLNPSINLLHQNSNDSMDWYEFVKNA IFSVSIAYFGIFIASFLYKPVYSSLKNLDLINSFVKTGPKRILWDKIINVIYDWSYNRGYIDTFYATFLTGGIRGLAELI HFFD |
| 5011 |
PF00260 (Protamine_P1) |
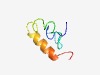 Estimated TM-score=0.50; hard target. |
>Protamine P1 (Length=49) ARYRCCRSQSRSRCYRQRQTSRRRRRRSCQTQRRAMRCCRRRNRLRRRK |
| 5012 |
PF18551 (TackOD1) |
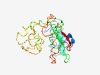 Estimated TM-score=0.49; hard target. |
>Thaumarchaeal output domain 1 (Length=178) FILSYLFTRPKLRLLPQLNHQVTQAFTYPVLDLFGQPQKEDTWEYLQDLVTSGFLKQGAAHSEIQACPSCHAGLLNFTSN CPNCHSVDIQEQQFIHCFTCGHIGPISEFMLREQLRCSRCHTKLRHIGIDYDKPLEDKLCLNCKHSFFEYEVNVTCLVCK KQSMPQDLLTRKLYEYNL |
| 5013 |
PF18550 (NitrOD2) |
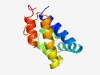 Estimated TM-score=0.49; easy target. |
>Nitrososphaera output domain 2 (Length=104) MVDAETLAEAILDSLKEIFGPPVFHSLMELIAEDYLGEMDARTAIIERPDLFERAFVGLLGEAGKKILADICEGLCAEFL LDENAADLKTGDLAECMAIIIPKS |
| 5014 |
PF18176 (KptA_kDCL) |
 Estimated TM-score=0.49; hard target. |
>KptA in kinetoplastid DICER domain (Length=152) KYSVRAAWAHKDFFMAQAVLSSHPMMTLKEAIDKFGRGKRLYDFVESLDRWKAELTKGGRSVPVFRPFLLLAVGDVIESR ITGDCTQQAATLLSNKVVSGSAFLPIHEHFLNMHEECRVHVVHSHFVNGVGAVPFEAAPGGESCGVVFLLPV |
| 5015 |
PF18159 (S_4TM) |
 Estimated TM-score=0.49; hard target. |
>SMODS-associating 4TM effector domain (Length=291) NSITQEQLEHKQLERLAAQRQIYSDAKKIQAINMILSVPAVIVWSILVVKFPDLKIWSAFWGITVTFLGFIVFNPFQKEL QEKAAKIQQLFDCDVLQLSWNELNSGKRPEPELICQKSRNYRRNHSDYSKLKDWYPIGVSQLPIHLGRLVCQRTNCWWDA QLRRRYAYYIQLIIIGLSIFVFLIGFINGLTLEKFFLAVFFPLTPVLFFGINQYRDNIEAASRLDRLKDEAESYWRQAIN QKLVPQELTQLSYKLQNSIYDNRRRNPLIFDWIYNRIRDENEEQMNQGAEA |
| 5016 |
PF17589 (DUF5487) |
 Estimated TM-score=0.49; hard target. |
>Family of unknown function (DUF5487) (Length=70) MHRHEFNIGDGYIGIVEWTENYEESTPRFFGTMYVTCEYAPGVVMYAELDEYFADRDDMLKYVEDFIRRE |
| 5017 |
PF17579 (DUF5482) |
 Estimated TM-score=0.49; very hard target. |
>Family of unknown function (DUF5482) (Length=159) MIYNNIDTDLISLGKAKKFTEEEEKWITPIYYEGESLDFTLKNKYVKINKIEENSYGKKFVTIKSKEYSKIIESISEKLG TVSPISSDGSFRAFINNKTKINEKNLDDVSFNACVSLVFPTIYKDTEKKTLQIYIKDLVVVEIIKDELEIELDKLSLAM |
| 5018 |
PF17574 (TA_inhibitor) |
 Estimated TM-score=0.49; hard target. |
>Inhibitor of toxin/antitoxin system (Gp4.5) (Length=89) MSNVAETIRLSDTADQWNRRVHINVRNGKATMVYRWKDSKSSKNHTQRMTLTDEQALRLVNALTKAAVTAIHEAGRVNEA MAILDKIDN |
| 5019 |
PF17566 (DUF5472) |
 Estimated TM-score=0.49; very hard target. |
>Family of unknown function (DUF5472) (Length=73) VMLTCHLNDGDTWLFLWLFTAFVVAVLGLLLLHYRAVHGTEKTKCAKCKSNRNTTVDYVYMSHGDNGDYVYMN |
| 5020 |
PF17565 (DUF5471) |
 Estimated TM-score=0.49; hard target. |
>Family of unknown function (DUF5471) (Length=70) MFKLIKKLGQLLVRMYNVEAKRLNDEARKEATQSRALAIRSNELADSASTKVTEAARVANQAQQLSKFFE |
| 5021 |
PF17525 (DUF5447) |
 Estimated TM-score=0.49; very hard target. |
>Family of unknown function (DUF5447) (Length=91) YLHQPHPEGCDCSVCWSRRETAHPVACQSTPCTECRPVSVLKVDGRWKLTPASICAKHTPSNRPPKYWHVVHDTGKPTPF VPLREPFELVG |
| 5022 |
PF17500 (Colicin_K) |
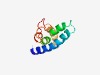 Estimated TM-score=0.49; hard target. |
>Colicin-K (Length=96) MYLKYYLHNLPESLLPWLLYLTLNYNNDISLSFIFFASIHVLLYPYSKLTIFSFIQKTTNIKKEPWYSYNLFYLLYLAMA IPVGLPSFIYYTLKRN |
| 5023 |
PF17474 (U71) |
 Estimated TM-score=0.49; hard target. |
>Tegument protein UL11 homolog (Length=52) MGAKCCKPVSCGMCKKTENTLIDYKGNPILLANEFTVLTDTESEEEGMADLE |
| 5024 |
PF17469 (GP68) |
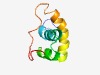 Estimated TM-score=0.49; hard target. |
>Phage protein Gp68 (Length=78) TNWDPNHPSLRSPIAPHETACVLRMHRAGYKGRATMKILKLRGTRLMNQMQRALDAETHAARAGRPIHDALIDPKKVK |
| 5025 |
PF17455 (LtuB) |
 Estimated TM-score=0.49; hard target. |
>Late transcription unit B (Length=79) NYFPASIEGETKKEHKHHYSTASKEKESLRKRAKEFDVLVHSLLDKHVPQNSDQVLIFTYQNGFVETDFHNFGRYSVKL |
| 5026 |
PF17381 (Svs_4_5_6) |
 Estimated TM-score=0.49; very hard target. |
>Seminal vesicle secretory proteins 4/5/6 (Length=101) ARERFSQSAEDPSSSHMGIKIRAGGSGSGSAMEEYSVSENSWSNFKSKHPSSISSESFHEESSSSSEMSSSGGHFGLKMR GSQAGGGMSSFKTRVKSRILK |
| 5027 |
PF17280 (DUF5345) |
 Estimated TM-score=0.49; hard target. |
>Family of unknown function (DUF5345) (Length=78) LSEELDRLDAQYNDITPPSLQELERLMADASVHRRSKERKELLLFWGISLIMISVFLSILGSAPMIYWIIQAIIPIAG |
| 5028 |
PF17095 (CAMSAP_CC1) |
 Estimated TM-score=0.49; hard target. |
>Spectrin-binding region of Ca2+-Calmodulin (Length=58) GTSDPMDNGVMTSQLVNIRLKLEEKRRRIESEKRRMEAQLNRQRQKVGKAAFLQAVTK |
| 5029 |
PF17062 (Osw5) |
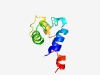 Estimated TM-score=0.49; hard target. |
>Outer spore wall 5 (Length=69) TFYFLTYFIIFMSIGFIASLLILPLLGISFLFATGVVVFGFVSNLTFKMGQYIYFKTDRNLKKILDRMA |
| 5030 |
PF17057 (B3R) |
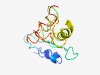 Estimated TM-score=0.49; easy target. |
>Poxviridae B3 protein (Length=123) MKRLETIRHMWSVVYDHFDIVNGKECCYVHTHLSNQNLIPSTVKTNLYMKTMGSCIQMDSMEALEYLSELKESGGWSPRP EMQEFEYPDGVEDTESIERLVEEFFNRSELQAGESVKFGNSIN |
| 5031 |
PF17021 (Mei5_like) |
 Estimated TM-score=0.49; easy target. |
>Putative double-strand recombination repair-like (Length=118) MYFLNVKDNDKHLYNALQVDGITFYSRKNNRSNQKKQRRYSDQFRTNNLNYKDIYRKLKILEKHKDDNKDLDDLTTKWKE CIDKCINILKDVFELPAKEVFKAFHLEKYGFVLEDYGE |
| 5032 |
PF17011 (DUF5093) |
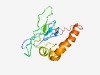 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF5093) (Length=126) INLIFPFDVPGVANKVALSVTTHIKGEIVSFALPFRLLFLTKPPNNSHPYSIKVKKNGDMFVYVLKFESIDLKRFLEKWE LHVLKEVAEEYEEIMKEMDEKMEEYEQREKTGKKRKIVDDDGWVRY |
| 5033 |
PF16660 (PHD20L1_u1) |
 Estimated TM-score=0.49; very hard target. |
>PHD finger protein 20-like protein 1 (Length=97) SQKKNEADISSSANTQKPALLSSTLSSGKARSKKCKHESGDSSGCIKPPKSPLSPELIQVEDLTLVSQLSSSVINKTSPP QPVNPPRPFKHSERRRR |
| 5034 |
PF16643 (cNMPbd_u2) |
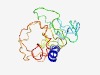 Estimated TM-score=0.49; very hard target. |
>Unstructured region on cNMP-binding protein (Length=162) VRRQVESVAKERLRAAKDDDVRMADAHSIPPNIDDLAIADHIMPRTPRRKKSVPTVTFNDTLDMDNPVTPTRRDEKMMEP YDPDPFLNVDLDNVRSRSRRGSLAPPTPTSIQESPTIPANLDFSKSPGGSPLSPSPSVSISKHASTPPEVPFDFKRPRLV GR |
| 5035 |
PF16597 (Thyroglob_assoc) |
 Estimated TM-score=0.49; very hard target. |
>Thyroglobulin_1 repeat associated disordered domain (Length=61) PGSVNEKLPQREGTGKTDDAAAPVLETQPQGDEEDIASRYPTLWTEQVKSRQNKTNKNSVS |
| 5036 |
PF16327 (CcmF_C) |
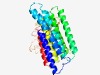 Estimated TM-score=0.49; hard target. |
>Cytochrome c-type biogenesis protein CcmF C-terminal (Length=324) LAFLAIVIGGSLALYAWRAPKVGLGGSFAMLSREGMLLANNVLLVAAMGSVLLGTLYPLFLDALDLGKISVGPPYFDSVF VPLMVPALFLMGIGPLAQWKKANLPDLAKRLKWAAGVSVVSALTLPIVLGNWTPLLSLGLLLAIWIVTTSVVSLRERLTH VTGNSLMERLASVPRSYWGMLVAHCGIAVFIVGVTMVKGFESEKDVRMNVGETASIGDYTFRFDGTQDVMGPNYTAARGA FSISRDGRETTVLYPEKRRYPVQNQIMTEAAIDPGLLRDLYVALGEPLDDGAWSVRLYHKPFIDWIWGGCFIMALGGALA ISDR |
| 5037 |
PF16259 (DUF4913) |
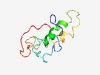 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4913) (Length=99) FGDLMAWVEGYLSPVYRRKVGDAGEAAWCAEWYRHAEAAVRLDAMWRSWEYYRHDRTQGLSLWFLDHADAHMAALLDRAG PFKLCGIRGSHQDAPVPPK |
| 5038 |
PF16228 (DUF4887) |
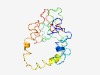 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF4887) (Length=172) IYSFVDSSKKGNERLSDKTTQQEKKKDDKDKKEKKDKKSVEEKKNNTQQTQQVPQTQQTQQTQQTVQTPQRPTTQTPVRQ NPQTGEKRAKEPKTDERKKTRQDSSNSKESPNEKDQSSTQQKPQQNKSNQSSENRQSTQNKSNQTQSTRPQNNGNSGNQN ASSGQNTQKQNN |
| 5039 |
PF16162 (DUF4868) |
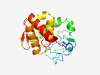 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF4868) (Length=187) RDNALGLLFMFRRGDNKIWGYQHIYTVTIPNKLRKNWLSIQQAETFKEMILPLFPIAKKVNLIIIGQEITTKDISLMQRQ FGFETYIRNSAQTVIGDVSEIGLFENIEKLSSYVERSKLTYAKKMMRIRHSAVLRMSSEQLLNKVQTLPRWSGKFVIENN RIVLKTYNHVENLIDLLDERYTRSDVT |
| 5040 |
PF16125 (DUF4837) |
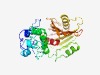 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4837) (Length=293) SSGRAYEILVVVDHALWERPAGRALYDVLDTDVPGLPQSERSFRIMYTSPKDYDSTLKLIRNIIIVDVRDIYTKPAFKYA KDVYAAPQLILTIQAPNEQEFEKFVEENKQQIIDFFTRAEMNRQIAQLQHKHNDYISTKVGSKFDCDIWLPAELASSKEG KDFFWASTNTATGDQNFVIYSYPYTDKNTFTKEYFIHKRDSVMKANIPGAKEGMYMSTDSITVDVRPIDIHGDYTMEARG LWRMKGDFMGGPFVSHTRLDKANQRIITAEIFVYSPDKLKRNLVRMLEASLYT |
| 5041 |
PF16088 (BORCS7) |
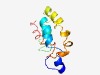 Estimated TM-score=0.49; hard target. |
>BLOC-1-related complex sub-unit 7 (Length=103) MASASSSSAKNLFSDSKRRLAERVAVNVNNASSVARQIVRGSKSNEILMQAAKNFAYHESTVDNSIQNLRKMDIIFQHLN YQYEAIQNAAEKLEYVEEQVGAM |
| 5042 |
PF15994 (DUF4770) |
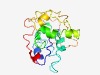 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF4770) (Length=168) LSRLGLIPAVRDKDLKMLMKLSGGNDLAFLWFLMELYYKKPGDEYSSLNEQLILTSIAHLDMITTLRELDNILPPGHQSK RQLEKRRKQKLRDKSWLSPPHKLVKKHSSPYFNKLERPVQYGKPLEVERPDFKVKFFRYEVYKDRNYVPPNEENRWYAKH SFNRAKRI |
| 5043 |
PF15984 (Collagen_mid) |
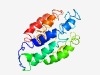 Estimated TM-score=0.49; very hard target. |
>Bacterial collagen, middle region (Length=198) IPGANPATTTALGNVVSNLGNGVTAVGTGVANGLGQLGNSTNPLGTTLGSSGNLVYDAGQAVNSAGQLVTSLGSGPTAPL SALTTPLGSTVSTVGNAVSGLGTQLGSQLTNGPIQQVTQTVSTAITPITSTLAQTTQTVGDTTGLGAPLNGLLSTIGHGL DSAGGKLSGATGNPVGQNLGNVVTKLGDTVTSAGGLLT |
| 5044 |
PF15914 (FAM193_C) |
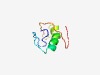 Estimated TM-score=0.49; hard target. |
>FAM193 family C-terminal (Length=57) DDVFLPKDIDLDSTEMDETEREVEYFKRFCLDSARQTRQRLSINWSNFSLKKATFAA |
| 5045 |
PF15852 (DUF4724) |
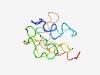 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF4724) (Length=91) MFTRHTLFRQFAVLADTSFNYVKVKPLIVQSKLNLTGVISSSTGEHGSSLDNKYVDIVNDQLSYAPSGREKRKGKEKKNK ENQFSHTSKPE |
| 5046 |
PF15828 (DUF4710) |
 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4710) (Length=78) KRSSRQVHFAFLPDKYEPLVEDDETTGRAKEEKKQQKKQKYKKYRKNVGKALRFGWRCLVIGLQEFAAGYSSPMTVAT |
| 5047 |
PF15822 (MISS) |
 Estimated TM-score=0.49; very hard target. |
>MAPK-interacting and spindle-stabilising protein-like (Length=238) FSLADALPEHSPAKTSAVSNTKPGQPPQGWPGSNPWNNPSAPSSVPSGLPPSATPSTVPFGPAPTGMYPSVPPTGPPPGP PAPFPPSGPSCPPPGGPYPAPTVPGPGPTGPYPTPNMPFPELPRPYGAPTDPAAAGPLGPWGSMSSGPWAPGMGGQYPTP NMPYPSPGPYPAPPPPQAPGAAPPVPWGTVPPGAWGPPAPYPAPTGSYPTPGLYPTPSNPFQVPSGPSGAPPMPGGPH |
| 5048 |
PF15821 (DUF4709) |
 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4709) (Length=110) RLNISDDLKIGFFSTDHATQTDSSEILSVKELSSSTQKLAQMMKSLQVDFGFLKQLLQLKFEDRLKEESLSLFTILHDRI LEIEKHYQQNEDKMRKSFNQQLADAIAVIK |
| 5049 |
PF15808 (BCOR) |
 Estimated TM-score=0.49; very hard target. |
>BCL-6 co-repressor, non-ankyrin-repeat region (Length=218) GLHPKKQRHLQHLRELWEQQVSPPERSPPSKLGRQSRKDLAEAVQPEVTAKVKDCTEEKHPRKRSEVKSNRSWSEESLRT SDNVQGLPVFPVSPHMKSLSSTNANGKRQTQPSCTPASRLAAKQQKIKESRKTDVLCTDEEEDFQPASLLQKYTDCEKPS GKRQCKTKHLTPQERRRRSSLTGDDTTDIENNEDKVMVTRRFRKRPEPASECDSLPSK |
| 5050 |
PF15699 (NPR1_interact) |
 Estimated TM-score=0.49; hard target. |
>NPR1 interacting (Length=118) EEELENKKMEMFFNLIKNYQDAKKRRRRDLTQDSGDTASVPTKRSDYGIVPVFRAEDFSHCMDLNLKPSNSVIPTKNQQE EKQEEEEEEDDDDEGEEDDDEEEEVEKVTIKDNGLDLN |
| 5051 |
PF15664 (TMEM252) |
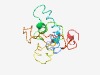 Estimated TM-score=0.49; very hard target. |
>Transmembrane protein 252 family (Length=140) SSGSIFNCRGNLILAYMLLPLGFVILLSGIFWSTYRQASKSKGVFTHVLRRHLSHGALPLATVDRPDFYPPAYEESLDAE KQPCPTEGEVSDIPPPLYTEPGLEFEDAADAHPEAPPSYEESVAGGVPAATPLQDAERQS |
| 5052 |
PF15658 (Latrotoxin_C) |
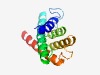 Estimated TM-score=0.49; hard target. |
>Latrotoxin C-terminal domain (Length=137) TFDVHGSLLLANVCIAKKFNCQLYNPGSSESITYDEALVNGINITEAFEVEITRTAKEYGLTGHSLDIDFVEVQNAVIGL IMANKLKEIFELLIIKAEKAYPFNELVPSQIAHKEKFIYRMVELINKFVSKSVEHML |
| 5053 |
PF15581 (Imm58) |
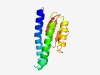 Estimated TM-score=0.49; hard target. |
>Immunity protein 58 (Length=109) MKKLTLFLLLSLSVSIICCAFFAYFWIDRSISLDYLQQSYETERSSVANLQKLIASEWKGLPEGQVQKKLEQVAAKSPER RIVVKKEGSIIWFDQVPFNIEQGRLDSVG |
| 5054 |
PF15562 (Imm17) |
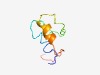 Estimated TM-score=0.49; hard target. |
>Immunity protein 17 (Length=62) LLFTFFLGIYFSAILLNKDWVLFPGDGKYNFDYFIKIFGRTIVRIFIGTLTLIGIGITFSLF |
| 5055 |
PF15503 (PPP1R35_C) |
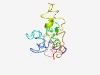 Estimated TM-score=0.49; very hard target. |
>Protein phosphatase 1 regulatory subunit 35 C-terminus (Length=143) PELNTTLAVGVELRGVAEAEFDPKRAVQDQLRKSAHRKNAIEIRATEGVNIPRSQQLYRALVSVSVPEDELISEAVKERL LLAPPPRAHGNKESPTAGPDLLHFYSPSELLRETPFLPGEGVTLPRPRPVPRPSQATFDLYHK |
| 5056 |
PF15500 (Ntox1) |
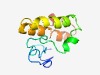 Estimated TM-score=0.49; hard target. |
>Putative RNase-like toxin, toxin_1 (Length=96) IEPNEATARKPADDGHDAVVTPEGVGRCSPAPCPVIQVEYQKEMGDNPDLKQWNDEVQGLRQTDPQRAADEAASLIRACE AARNNAGRAPVEDYVR |
| 5057 |
PF15478 (LKAAEAR) |
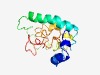 Estimated TM-score=0.49; very hard target. |
>Family of unknown function with LKAAEAR motif (Length=137) PKNWTKKSPQQLKKMGAQQRSRYLAYEEPPKEIQDTVDLTQKRVKDLIQKQKNSTPRMNNEMEIDRQKHSNLIGQLKAAE ARNRIRVMRLQYQAMRAHEVKHLISCQPTSLKALRFEALVPPYTNNRNPGDHLDKLD |
| 5058 |
PF15360 (Apelin) |
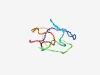 Estimated TM-score=0.49; very hard target. |
>APJ endogenous ligand (Length=55) GPLLQTSDGKEMEEGTIRYLVQPRGARSGPGPWQGGRRKFRRQRPRLSHKGPMPF |
| 5059 |
PF15356 (SPR1) |
 Estimated TM-score=0.49; very hard target. |
>Psoriasis susceptibility locus 2 (Length=114) SEDHSSPPSTEAREEESPPRLPLGPPIPGEPWPGAPPLFEDPPPPGPSHPWGDLPDSGVWPPEAPSTDPPQPPLPDDPWP AGPQPPANPWPPAPEVDHKPHNEPDLDPPREEYR |
| 5060 |
PF15261 (JHY) |
 Estimated TM-score=0.49; very hard target. |
>Jhy protein (Length=126) LGGLGPDYQTAQEKAQKVKRQKEYAQQVREQNKKMCSDPFTSLNKTTLSSENKNSVPRQKALEYAKSIRKPKITPKPRST NQEPSKRNFFEHPQYLEDFDLSQIAMLEMLQRRHEEEKQAVAAFKA |
| 5061 |
PF15219 (TEX12) |
 Estimated TM-score=0.49; hard target. |
>Testis-expressed 12 (Length=96) SQLSSLEKPDFVFSENAVPSFYKAEVLEKVLNDMSKEINLLLTKYAQILSERAAVDASYIEEFDAIFKEASTLENLLKQK RESLRQRFTMIANTLQ |
| 5062 |
PF15206 (FAM209) |
 Estimated TM-score=0.49; very hard target. |
>FAM209 family (Length=149) FMFSSLRDKAKEPPGKVPCGGHFRIRQNLPEHAQGWLGSKWLWLFFVFVLYVILKFRGDSEKNKEQNPPALRGCSLRSPL KKNQNASPSKDYAFNTLTQLEMDLVKFVSKVRNLKVAMATGSNLKLPSSEVPGDPHNNITIYEIWGEED |
| 5063 |
PF15099 (PIRT) |
 Estimated TM-score=0.49; hard target. |
>Phosphoinositide-interacting protein family (Length=127) KSPDESEKSPQSKELLTSNTASTLCISSRSESVWTTAPRSKWDIYHKPVIVVSVGAAVFLFGIVITSLSCIQIKNKNVYK MCGPAFLSLGLMILVCGLVWIPIIRKKQKQRQKSQFLQSLKSFFFNR |
| 5064 |
PF15059 (Speriolin_C) |
 Estimated TM-score=0.49; very hard target. |
>Speriolin C-terminus (Length=148) LVGEIAFQLDRRILSSIFPERVRLYGFTVSNVPEKIIQASLNPVTRKLDEEQCQVLTQRYVSVMNRLHGLGYDSKMHPAL TEQLVNTYGILRERPELTASDSGAYNSMDFLKRVVVETVPPGLLTDALLLLSCLSQLSQDDGKPMFIW |
| 5065 |
PF14984 (CD24) |
 Estimated TM-score=0.49; hard target. |
>CD24 protein (Length=52) SNQTTVVTVSSNSSQSTSTAPNPANATTKAVGGALQSTASLFVVSLSLLHLY |
| 5066 |
PF14983 (DUF4513) |
 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF4513) (Length=132) SGKDTCPILPKLANSCSDESSYKPSNKCNEVHLPRFSLKQGMIPRRYIMPWKENMKFRNMNLKRAEVCGIHAGPLEDSLF LNHSERLCHGEDREVVLKKGPPEIKIADMPLHSPLSRYQSTVISHGFRRRLV |
| 5067 |
PF14959 (GSAP-16) |
 Estimated TM-score=0.49; hard target. |
>gamma-Secretase-activating protein C-term (Length=108) LDLICQILEASWRKHNLHPWVLHFDRRASAAEYAVFHIMTRILEATNSLFLPLPPGFHTLHTILGVRCLPLHNLLHYIDN GVLLLTETAVTRLMKDLDNTEKNEKLKF |
| 5068 |
PF14899 (DUF4492) |
 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4492) (Length=62) RIFRFYVDGFRNMTLGKTLWTIILIKLFIMFFVLKLFFFPNILNANFNTDEERAEHVGMELT |
| 5069 |
PF14886 (FAM183) |
 Estimated TM-score=0.49; very hard target. |
>FAM183A and FAM183B related (Length=103) ILEETIKKEQRHQKLYTTYSVNPFRKMYTLTGKPNSTHDSADGEEDDNFLKIIHRAHQEPVKKYTFPQTEAQEIGWMSKP LIPQDREDRRLHFPRQNSEITKY |
| 5070 |
PF14880 (COX14) |
 Estimated TM-score=0.49; hard target. |
>Cytochrome oxidase c assembly (Length=58) AKYAWYTRITDAAHRLTVLTLLGGTLYMGGGLVYTLYQNGKKYEEEQLLLQEQQQQKQ |
| 5071 |
PF14866 (Toxin_38) |
 Estimated TM-score=0.49; easy target. |
>Potassium channel toxin (Length=55) EILDKVKKVFSKGVADLNNMSELGCPFIEKWCEDHCESKKQVGKCENFDCSCVKL |
| 5072 |
PF14621 (RFX5_DNA_bdg) |
 Estimated TM-score=0.49; very hard target. |
>RFX5 DNA-binding domain (Length=219) PGPGQALPGALTQPQGTENREVGIGGDPGPHDKGVKRTAEVPVSEAIGQDPPAKAAKQDIEDTGSDAKRKRGRPRKKSGG SKERTSPPDKSAPAMDSAQSPRLPRETWASGGESNSAGGSGKPGPVGEAEKGMVLAQGQEDGAVSRGGRGPSSRHAKEAE DKIPLVTSKVSVIKGSRSQKEALHLVKGEVDTAAQGNKDLKGHVLQNSLFHEGKDPKAT |
| 5073 |
PF14576 (SEO_N) |
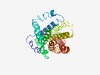 Estimated TM-score=0.49; hard target. |
>Sieve element occlusion N-terminus (Length=270) SDENVMMDQILSTHCPDGREFHVQPLFLVIEDVMHRANAPLPGTIFSATLQGTQAQLDEKAPPNCFSDILKLLGHTIKKT SLEISYKRSEGGNAQATTVSLFDTLSIYPWDVKVVVVLAAFTLIYGEFWLVEELCPTNPLAKAVAHLKQLPEIMGQAESL KPNFESLDNLIKAVLDLTKCIVDFKCLPSQDISPDTPEMVTAPAHIPIAAYWTIRSIVACAFIVINHIGSSHEYFALTTE AWELSSLTHKVNNIHTNLKKQFSLCQQHIA |
| 5074 |
PF14288 (FKS1_dom1) |
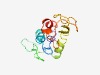 Estimated TM-score=0.49; hard target. |
>1,3-beta-glucan synthase subunit FKS1, domain-1 (Length=123) MTRQLALYLLIWGEANQIRFTAECLCFIYKCALDYLESGSSPSNNSKTNINTYTNSTNELPTLPEGDYLNRVISPLYHFL RDQVYEISDGRYVKREKDHNYVIGYDDVNQLFWYPEGIRKIVL |
| 5075 |
PF14144 (DOG1) |
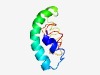 Estimated TM-score=0.49; hard target. |
>Seed dormancy control (Length=75) INDIRSAINSQMGDNELHLLVDGVMVHYDELYKLKSIGAKADVFHILSGLWKTPAERCFMWLGGFRSSELLKIIR |
| 5076 |
PF14071 (YlbD_coat) |
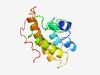 Estimated TM-score=0.49; hard target. |
>Putative coat protein (Length=125) DFKQFVKKHPKLIQNVRKEQRSWQEVYENWVLLGEDDPIWNPFKDEAVQPSSDAETKKEPAEKNDFVSKMVTAVKKMDIN QMNEQINKMSQSISSLQSLLSQFSGSSTKQPRQGSGQHPFSFRKD |
| 5077 |
PF14049 (Dppa2_A) |
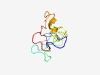 Estimated TM-score=0.49; very hard target. |
>Dppa2/4 conserved region in higher vertebrates (Length=84) KDVPETSQEATLQSSSKKCKMVTKRTRNQKSYKMSEREKGTNTVEVITSAQEAMLAAWARIAARAVQPKTVNSCFIPTSA ENFL |
| 5078 |
PF13916 (Phostensin_N) |
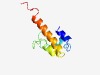 Estimated TM-score=0.49; hard target. |
>PP1-regulatory protein, Phostensin N-terminal (Length=89) AERDRLSQMPAWKRGILERRRAKLGLPPGEGSPVPGNAEAGPPDPDESSVLLEAIGPVHQNRFIQQERQRQQQQQQQRNE VLGDRKAGP |
| 5079 |
PF13834 (DUF4193) |
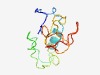 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4193) (Length=99) MATDYDAPRRTDSDDVSEDSLEELKARRNEAQSAVVDVDESDTAESFELPGADLSGEELSVRVVPKQADEFTCSSCFLVH HRSRLASEEGGQMICRDCA |
| 5080 |
PF13663 (DUF4148) |
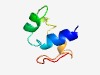 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4148) (Length=51) MKALATAIMVTATLAGMSAAHAEGKTREQVQQELQAAKAAGQVTFGEQDYP |
| 5081 |
PF13494 (DUF4119) |
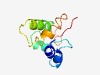 Estimated TM-score=0.49; hard target. |
>Domain of unknown function, B. Theta Gene description (DUF4119) (Length=91) AKTRVKNNAQNRKKSGGRCKKAEIISEEELEKRGGLTGDETALFTVYLQKFDKGDVHMKYSKKLKDLAKHIHEKEHLYVP KQGGYKLMEVS |
| 5082 |
PF13334 (DUF4094) |
 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4094) (Length=93) KVIFMLCTASFLAGSLFTRHTWPYSSINNDFRFTNREQKLSLTTVPDCDRKRKLVEENSVDVMAEVTKTHQAIQSLDKTI STLEMELEAARTS |
| 5083 |
PF13323 (HPIH) |
 Estimated TM-score=0.49; very hard target. |
>N-terminal domain with HPIH motif (Length=147) STPILQFFSSLACSHPIHTVVIVAVLASSSYVGLLQESLFDATTSVRKADWSSLTEGSRRLQVGPETNWKWQNVDQDVVP ASDVEHLALLTLVFPESHSAESANTAPRAHAVPFPQNLSVTPLPQTSNSLTAYSQDSALAFSLPFDQ |
| 5084 |
PF13321 (DUF4084) |
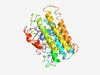 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4084) (Length=304) MINQKKYVHFVTMYIIIFSLWIFLIPKDLNIKEIGILFLFCFATLFSCYCLYKAIKKMKRGDKLFWVLVLCTCLCGLTME ITLFLHSLSIYDQVIFSYKALPFFIVQYILLFSGFAIKFIKHYSIRGLAQFSFDSIFIVIMNIYFTLTFILDISSFRMLT TDTWVLIGYFIAQSLVIYAVISLYRREEYSSSRISLIIGFTIILVYGYIHLFQLNAGIKPSSEVSYLIHTASILLIGLSS ILYILDKPIQHETKTKYYRFDYVRFILPYFSIIITFSFIIFQPWDDKFMLIGLVLSLILLFLRQ |
| 5085 |
PF13319 (DUF4090) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF4090) (Length=84) GADAVDEAIAQGIDFDGTPIPSAKLELYHQVMALEANRQRSGVSNTMRSRIVRIGAKHIPQDELNQKLIDAGFAPLKEKE IAFF |
| 5086 |
PF13310 (Virulence_RhuM) |
 Estimated TM-score=0.49; hard target. |
>Virulence protein RhuM family (Length=250) ATCKNYLQVQNEGAREVKRQSKHYNLEMIIAVGYRVRSHRGTQFRQWATERLNEFMVKGFTMDDERLKEMRNIGADYFDE LLERIRDIRASERRFYQKITDIYATSIDYDPNSTIAREFFATVQNKLHFAIHGHTASELIVERADATKPNMGLTSWKGDK VRKNDVTVAKNYLTQEELSDLNRIVTMYLDYAESQAKKKKPMHMKDWAQKLDAFLEFNEHDILTNAGKISAKLAEQFAKE QYEVFHQQRL |
| 5087 |
PF13293 (DUF4074) |
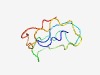 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4074) (Length=62) TGPSMYGLNHLPHHQSASMDYNGATQMPANQHHGPCDPHPTYTDLSAHHPSQGRIQEAPKLT |
| 5088 |
PF13285 (DUF4073) |
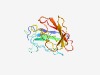 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF4073) (Length=157) LNLPDWAGKKKVKGGDKRGFTVVNTAGIETGWMSAGPDGGEKVTPDGGAFKQGLRLQVYGNDVVIQAYDYQHDKVIKQLS IRYGKIEQLPPDVQADDENNVLVNATEYMEYAVGKQKNWKDYDPEDPPTFPGYQNVYVRHKGEMNLEAGTPIMVKFK |
| 5089 |
PF13226 (DUF4034) |
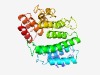 Estimated TM-score=0.49; easy target. |
>Domain of unknown function (DUF4034) (Length=279) LCDMRSFLREGRYGELDKQLESALTASFLSRDAEQRYYRGWMDMENTPDLRSVISMGPEGLAQIKAWQHDCPESNHAWLA ETLYWHHCAWRYRSYGWAKKTTAAMWVCAAACNEMMVLAALQALTLEPRQWMAAALIIPAITAFGVPAWLAVIIANEKAD SLPVLGELRDYQQCYPEEMAALMSYSGLNAYAQILAPDALPPGLLRNQTEKALIGPHYWLFASLHIHPTQFYIFTDYIPF QMPRWRGLPQDMLDLIVSPACEHLSLQEKDHLRHLIWWD |
| 5090 |
PF13214 (DUF4022) |
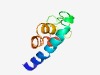 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF4022) (Length=77) MRMDYIMSIITLALLLLAEILVAIILIGVSIEICSYGWKKSNGIKYSCLLLSLLLGTASILGLFAAPAYFFIQLTEK |
| 5091 |
PF13116 (DUF3971) |
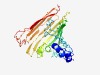 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (Length=280) DDAGEAWRYFPRSLMGASLADYLTHSLIKGKIENATLLFQGNPHDFPFKQNNGQFQVWVPLRHATFQYQPDWPALFDFDI DLNFQNNGLWMQVPKAKLGKVDATQLTAVIPDYGKNKLLIDGKLSGSGKNIHDYFKHSPLAESIGSTLDELQLDGKVDGK LHLDIPLEDGKVNASGEVVLQDTTLFIKPLDSKMEHVSGKFRFDNGKLQSDTLSAQWLGHPLSLRFTTQDLPKHYQVDVD LNAQWAAGKLPEVPAEIRRQLSGTLDWQGKVNIAIPAQNT |
| 5092 |
PF13042 (DUF3902) |
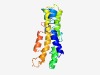 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3902) (Length=161) KSVLKSILISFVFSAVSMCWLLYVLFKGDGDWLLSWIGVLMAYLSLYTLIDLYCKNTYDKKINKWLIKTAVTSFSFAVLG ISFCIIHELLTPWSLSLMVWYWLVMLVLFLTTIISLVSLVFVNRKNHNFTGGYRMLILLNVFLTLGPVLWPLLLSIIGNG M |
| 5093 |
PF13025 (DUF3886) |
 Estimated TM-score=0.49; easy target. |
>Protein of unknown function (DUF3886) (Length=69) ADKPATLKDMLDPQVLAKLKATTDSLKASEAERLEKERQEREAARKAEQKRRDNDFEYLLEHSEMDWKK |
| 5094 |
PF12962 (DUF3851) |
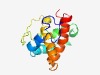 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3851) (Length=125) MNPNILNKNPLMFFDRAVNAQRSQLLTVMADAVSECRTAADQAAELNETGQVGLLRLAEIWSAIRAKEGMGGLVLEGTEA KILSDVVAQFYAYLSGCMFNDPVGMAIYAELHYMMSSLMLGEWFE |
| 5095 |
PF12873 (DUF3825) |
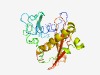 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF3825) (Length=235) LDDLAQEETWEYKHTPTRKPKPILASYFNYTFKRLLEENKVVEQEDKACFNTGLVTDNQEDIFAFFEQNRNNPSQWKFKQ FLKESDYLLRSFNPLPDRASYFDDPTNLIYDIRLGHPRIDYDHIKNRTENRNRFPQKYQSMNDHELQVLLEGAVKLAIKR VMRNYKAAVPQFFWDRNAKVGELQLLLPLCLSSPLKADVALVINRTEYVYSGETILTLDMAYNNARLLAKPDTEW |
| 5096 |
PF12853 (NADH_u_ox_C) |
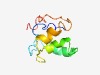 Estimated TM-score=0.49; hard target. |
>C-terminal of NADH-ubiquinone oxidoreductase 21 kDa subunit (Length=89) RYEIKKLLSEGKLPYHDNESKLDDRMKDIANRNSQYSFTTLAILPWFNLAYHPYHGVNIQKYYETRPGEEEWGFTLKPYE EIKAKYAKH |
| 5097 |
PF12606 (RELT) |
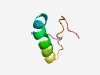 Estimated TM-score=0.49; hard target. |
>Tumour necrosis factor receptor superfamily member 19 (Length=41) YIAYALVPVFFIMGLFGVLICHLLKKKGYRCTTEAEQDIEE |
| 5098 |
PF12542 (CWC25) |
 Estimated TM-score=0.49; hard target. |
>Pre-mRNA splicing factor (Length=98) VDRVDWMYEGPTDSQTGVTEEMEAYLLGKRRIDNLIKGDEHKKLEKGAPVPAGGGAGGFMALQNANSDRDMAAKIREDPL LAIKRQEQAAYEAMMNDP |
| 5099 |
PF12540 (DUF3736) |
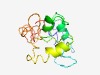 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF3736) (Length=138) RRRLVSKLDLEERRRREAQEKGYYYDLDDSYDESDEEEVRAHLRCVAEQPPLKLDTSSEKLEFLQLFGLTTQQQKEELLT QKRRKRRRMLRERSPSPPTSKRQTPSPRLALTTRYSPDEMNCSPNFEEKRKFLTIFNL |
| 5100 |
PF12523 (DUF3725) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3725) (Length=74) YYSEFDISKGAKILQLVLIGNAEVGRTFLEGNRFVRANIFEIVRKTMVGRLGYDFESELWFCHNCNETSEKYFK |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218