

Go to page (83 pages in total): [First page] << < 45 46 47 48 49 50 51 52 53 54 55 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4901 |
PF11309 (DUF3112) |
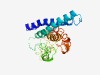 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF3112) (Length=217) PVGGSRPLFWNTMFLLYGVVVLVIVMTIIASAVPYVYLLSEHAYMGYKKTVEASAILIMLYTLTAVSLLGLAYFFKPTRK DENLYTYQPWWIESFSPFYFVKPGAKEEAEQSFMKRGHNHRHAIRVIAATHHHHNMVEGLSRERGDLKHNWSLVIITLTT IFIFIGQLGRTVAVFQGRYQIDSGPVCKPVAMYICWGLFEVIINLLYIIGRVDLRFY |
| 4902 |
PF11299 (DUF3100) |
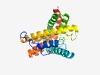 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF3100) (Length=234) VIAEAIGVIKFKFGPGTIVLLPMLYALIIGIFLSPKFLKVVNEKDMNDAGTLITVSLMLLMARYGTLIGPTLPTIIKSSP ALILQEFGNVGTVFLGVPIAVLLGLKRESIGAAHSIAREPNIALISDIYGLDGEEGKGVMGIYICGTVFGTVFFGLLASF CAAYLPFHPYSLAMASGVGSASMMTASVGSLSAMYPQMADTLAAFGAASNMLSGLDGLYMSLWIALPLTEWLYR |
| 4903 |
PF11243 (DUF3045) |
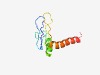 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF3045) (Length=88) MFVVHTIYENEGNTTRDYGHVNQFFRCNPEFRAQKDERIFKKCVEQGFIYIKHWMQGNKVRTTYHRSLTELNDELIYNRA VNQTLKDE |
| 4904 |
PF11190 (DUF2976) |
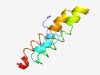 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2976) (Length=87) LPTMEPPSSGGGGGLFGQIKGYSQDGVVLGGLIISAIAFIVVANAAISTFSEIQDGKKTWTQFGAIVVVGVILLVAVIWL LTKSASI |
| 4905 |
PF11166 (DUF2951) |
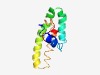 Estimated TM-score=0.50; easy target. |
>Protein of unknown function (DUF2951) (Length=98) MGWFKKHEHEWRIRRLEENDKTMFSTLNEIKLGQKTQEQVNIKLDKTLDAIQKEREIDEKNKKENDKNIRDMKMWVLGLV GTIFGSLIIALLRMLMGI |
| 4906 |
PF11162 (DUF2946) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF2946) (Length=111) LGLLAILMATLAPTVSHTLAAARADDSTLGEGCSMPSMQHQEQDSKSHPDSTMSDGQACGYCGLLAHLPVIPGVQATFAV TVRAIQHPVATRFESVRLVEPLTPAQPRAPP |
| 4907 |
PF11123 (DNA_Packaging_2) |
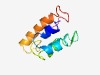 Estimated TM-score=0.50; hard target. |
>DNA packaging protein (Length=82) DKSLITFLEMLDTAMAQRMLADLSDHERRSPQLYNAINKLLDRHKFQIGKLQPDVHILGGLAGALEEYKEKVGDNGLTDD DI |
| 4908 |
PF11093 (Mitochondr_Som1) |
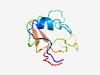 Estimated TM-score=0.50; very hard target. |
>Mitochondrial export protein Som1 (Length=89) MTPPCKVFPVSELELHVQANVKGKKRKLPRGADVDLTKCALKEMTQFRCYVDDSKSRDSPVRCWPIHRLFRQCRDKKGTF TIETTAWEG |
| 4909 |
PF11092 (Alveol-reg_P311) |
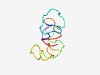 Estimated TM-score=0.50; hard target. |
>Neuronal protein 3.1 (p311) (Length=67) VYYPELSVWVSQEPFPNKEMEGRLPKGRLPVPKEVNRKKNDETEAASLTPLGSNELHSPRISYLHSF |
| 4910 |
PF11058 (Ral) |
 Estimated TM-score=0.50; hard target. |
>Antirestriction protein Ral (Length=66) MTTTIDKNQWCGQFKRCNGCKLQSECMVKPEEMFPVMEDGKYVDKWAIRTTEMIARELGKQNNKAA |
| 4911 |
PF11045 (YbjM) |
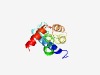 Estimated TM-score=0.50; hard target. |
>Putative inner membrane protein of Enterobacteriaceae (Length=115) NKGWVGATCCFLLFTVVFLSQKIEVSDVAVNDGLRGSPGMLLFLLPGMVACFLSARGRLLYPLFGALAAMPVCLLVLHLW NTPMRSFWQELAYVMSAVFWCVLGALGVMCLRSLY |
| 4912 |
PF11040 (DGF-1_C) |
 Estimated TM-score=0.50; very hard target. |
>Dispersed gene family protein 1 of Trypanosoma cruzi C-terminus (Length=87) WYAEDHHWQELREPRRGGLEALLRDDEESDEETQKPHDMTSSSYASGTTVASSYQPPAPPLQPMAGDTRSDALSLLDRAS SASCSVN |
| 4913 |
PF11029 (DAZAP2) |
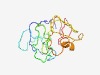 Estimated TM-score=0.50; very hard target. |
>DAZ associated protein 2 (DAZAP2) (Length=135) YSDAPPAYSEIYQPRYMHAPQAAGQLPPMTTAYPATQMYVPMPQSVQVGAMAPNVPMAYYPMGPMYPPGSTVLLDGGFDA GARFGHGTAASIPPPPPGHAPNAAQLAAMQGANVMMTQRKNNFFLGGSGGGYTIW |
| 4914 |
PF10997 (Amj) |
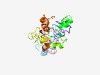 Estimated TM-score=0.50; easy target. |
>Alternate to MurJ (Length=250) AFVTLLTSATVASRPAAVKTGQVAMSITIYQIFFMMTRFANMFYLPILASYVDRASNTGNTDILLLQIRIIIIGSCFGSA IAWLLLPTLVNIFTSGIGALDRHGSMIKVLIKTLKPSSWKNIYKAFAFPSNFGVSLLKLEGVPANFLIFNIFATAIWTVG VLCAMYASAENKDYARTAILLSGLVNALAAIMFSVIVDPKAALITDEVIAGKRPEKHVYIVAVFLMAGNLLGAIISQFFL LPGVKVISWA |
| 4915 |
PF10964 (DUF2766) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2766) (Length=79) MSESLNREQEIESDLVACQLVIKQILDVIEIIAPAEVREKMSHQLKSIDFSKHPAGADPVTKRAIEKAISLIELKFTRH |
| 4916 |
PF10935 (DUF2637) |
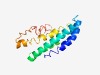 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2637) (Length=154) AALGFVAMAAVMGWGASFVGLHAYGQDQMVGFSYWSAWLIPATFDGAAFGATLITYRASIHGRSAFRGRLLMWIFTAISS WINWIHQMTPEARIVAAGLPVAAVAVFDVVLAELRADYEERHGKRRLRLRPGLLLLRWLVDRGGTSVAFRRQVT |
| 4917 |
PF10907 (DUF2749) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2749) (Length=65) MSRAVIIALVILVAAVSTTATALIVNSRASNPSAPEEQRTAKEKFFGAGKALPPIKDGQEMGPRW |
| 4918 |
PF10856 (DUF2678) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2678) (Length=118) MDDFTTRTYGTSGMDNRPLFGETSARDRIINLVVGGLASLLVLVTIISSFVFPSLPPKPLNIFFAICIMLVCGSVLVLIY WYRQGDLEPKFRNLIYYMLCSIVLLCICANLYFHDVGR |
| 4919 |
PF10830 (DUF2553) |
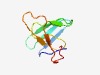 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2553) (Length=73) QKLDVTNDVTGRFQNGRLSLYHDNEMIGQMTSMNEYELKSGYSFENEKFYKTADVVSGDDAKYVDCDYENGWC |
| 4920 |
PF10818 (DUF2547) |
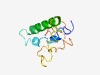 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2547) (Length=96) KTPFWSQLVFGLVAIFALPEIQATLNRENEQNTVINQLVTQYTQVTEDGEQQTLFIAERHLAQMNTTSQAVVFCKILTKS YRFYGDDNHPIRAGPI |
| 4921 |
PF10787 (YfmQ) |
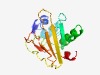 Estimated TM-score=0.50; hard target. |
>Uncharacterised protein from bacillus cereus group (Length=140) MTWAIVMLILMSLVKIVLTCLPTGVIEWLLGKFEVHAKLSDENASLSIDGKRLEGAEKQKVIDQFNEAIFLEKYYIYPGD EERYLHPENGGTPLVIDTKKGKKDVKLFVYRYDDHIDVVKQYKKKVIAYRVLSESLQKES |
| 4922 |
PF10774 (DUF4226) |
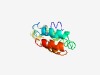 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF4226) (Length=112) QAGSSVAVIQERQALLARQHDAVAEADRELADVLASAHAAMRESVRRLDAIAAEIDRAVPDQDQLAVDTPMGAREFQTFL VAKQREIVAVVAAAHELDRAKSAVLKRLRAQY |
| 4923 |
PF10629 (DUF2475) |
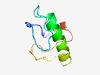 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2475) (Length=67) PPALMPGYRGHVPNVAFSFGDTYGNTTMKYFQDFRNAAMETSHSPYSKGGQFPTLFSRDPSLVLGER |
| 4924 |
PF10616 (DUF2471) |
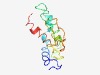 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF2471) (Length=124) PEQPGFNPVHFERAAQRAVVDLQRVVGGPAQRALDLRRRTHPAAVRTMSWQALLNVEELAFSNAGFLNRNDPTVVDAFIR LRDSRLVAADVEEPVDWHRDDDDLPAVYLIVKAMLEAEAEERAE |
| 4925 |
PF10492 (Nrf1_activ_bdg) |
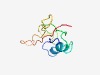 Estimated TM-score=0.50; hard target. |
>Nrf1 activator activation site binding domain (Length=79) SGMITVPVSTQMYQTMVANIQQLPNGDGTVCITPMQMCDLVENGETMETITVGPGMHQMMIQGPPGSEPQVLQVLSLKD |
| 4926 |
PF10333 (Pga1) |
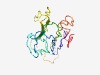 Estimated TM-score=0.50; hard target. |
>GPI-Mannosyltransferase II co-activator (Length=168) LVFWVHWVWANTESFLLHVPRDFPTKQSSSTELIQYPRSISLQDTNINKATFSCGINHSSFVELTHLQQGEAYQIKVCWT ALDPISIDHMDWFVVPHSTLFQGTTSDAARIFVRFDLRADSYPPLEPMTRIPINVSVINTKLAIPVDLYKLIVYIVLVTA GVFLVNRK |
| 4927 |
PF10270 (MMgT) |
 Estimated TM-score=0.50; hard target. |
>Membrane magnesium transporter (Length=102) TFFGLILLAHAGYSAHEHSTLYSNARSSTSPSSALPLDITIETLVAVVLVSVGLVLGAEKLKPISWSEWAGQIEREGGGR NPYRNLEERYGFWDIRAKRKEF |
| 4928 |
PF10117 (McrBC) |
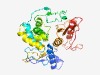 Estimated TM-score=0.50; hard target. |
>McrBC 5-methylcytosine restriction system component (Length=322) LKLDNYVGALETPCGTRIEILPKHFEQGDCIQQSRALLRRMIQKSLDLPTRKVGATALQRFDAPLTEWVINSFLEALDHL IKRGLRFDYQRVEEEQRYLRGQLNTARQMRQPPGRQHQFQMRHDIFQPDRAENRLLKTALDIVCKTAQDPSNWRLSHELR SMLLEIPSSRDTTQDFKQWRRDRLMAHYQPVKPWCELIIQQQTPLAIAGEWHGMSLLFPMEKLFERYVAACLRDSLPPDA TLHTQRSSEYLCTHEGKKVFQLRPDLMITQGEKSWVLDTKWKRLDSELGSKNYGLSQADFYQLFAYGQKYLDGQGDLVLI YP |
| 4929 |
PF09868 (DUF2095) |
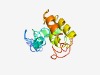 Estimated TM-score=0.50; very hard target. |
>Uncharacterized protein conserved in archaea (DUF2095) (Length=129) EKKKKPIDQLPWQEYDIEEFRQRFPALARELEEDRGIEISGIRLDEYQVLEEEEEEGKIDFSGYNPTIIDFLRRCDTDEQ ALEIINWMEEHGEITPEMAKELRVTLVHKGVRAFGPKKEWGWYERHGKH |
| 4930 |
PF09779 (Ima1_N) |
 Estimated TM-score=0.50; hard target. |
>Ima1 N-terminal domain (Length=126) VNCWFCNQDTVVPYGNRNCWDCPNCEQYNGFQENGDYNKPIPAQYMEHLNHVVSGSPTFCDPTKPQQWVSSQILLCKKCN NHQTMKIKQLASFSPREEGKYDEEIEVYKHHLEQTYKLCRPCQAAV |
| 4931 |
PF09774 (Cid2) |
 Estimated TM-score=0.50; very hard target. |
>Caffeine-induced death protein 2 (Length=157) CFNERVLRDFLRLSRSAIDDSITQNLNALITPARVGFDPSSTAVRQTGKKTIDPAACQSFKDNVLFPSWQTRSDVLNYCA GVATSPDPEDPDLILRETESAKDRERVVDERLDPYSARFFPKEARTESLAYLIRNERSVEQIIRARTWGLVTERCGD |
| 4932 |
PF09750 (DRY_EERY) |
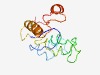 Estimated TM-score=0.50; very hard target. |
>Alternative splicing regulator (Length=127) FIQIHGRPCKIHLDPSVALAADSPSTMMSWQGDRDIMIDRFDVRAHLDFIPEVKHTEVELTEEESQEERFQNYERYRILV QNDFLGIPEDKFFAQLALEEQYGPVTKNEISVEGKKKSQLNAAIGFK |
| 4933 |
PF09593 (Pathogen_betaC1) |
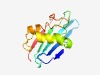 Estimated TM-score=0.50; very hard target. |
>Beta-satellite pathogenicity beta C1 protein (Length=117) TIKYKNQKGMEFIIDVRLKEDNSILVQIQLFSTGSPALATRKFMIPYGHDGIIPPFDFNALEEGIHNMLALMYKESNIGE FKQEDMVECIDMLMMEEAPVIDINVMDAYDVYTHSSV |
| 4934 |
PF09573 (RE_TaqI) |
 Estimated TM-score=0.50; very hard target. |
>TaqI restriction endonuclease (Length=224) LEQFETFLSTVHLATYRTRYLPIKIVEMDLPKDIHAIEKLYEVYWDKKKFLDFDDFYKEYLDSKNSEIDSFRQKITMCSD CFYRGLPARIYRTWASIITQIHAGYVAESVFGEGSVKMSAELDHKGADFQVHYHSRTLNYQVKKKSLGREVRQEKPKAKN QLAGDFVDIKYEVPSSDYFENPKRKDGEYKLPYKRFIENKELKRFPNGFVVFTTYAFERKKLEI |
| 4935 |
PF09354 (HNF_C) |
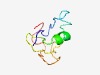 Estimated TM-score=0.50; hard target. |
>HNF3 C-terminal domain (Length=66) NHPFSINNLMSSSEQQQHKLDFKAYEQALQYSSYGAGLPGGLPLGSASMGGRGGIEPSALEPSYYQ |
| 4936 |
PF09350 (DUF1992) |
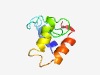 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF1992) (Length=69) LVEDMILQSMKRGDFNNLKGMGKPLKNDESNPYIDVIEYKLNKILINNGFAPPWIMKEMELRTEITSLR |
| 4937 |
PF09316 (Cmyb_C) |
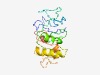 Estimated TM-score=0.50; very hard target. |
>C-myb, C-terminal (Length=160) FSPSQFFNTCSGNEQLNMENPSFTSTPICGQKVLITTPLHKETTPKDQKENVGFRTPTIRRSILGTTPRTPTPFKNALAA QEKKYGPLKIVSQPLAFLEEDIREVLKEETGTDIFLKEEDEPAYKSCKQEHTASVKKVRKSLVLDNWEKEEPDTQLLTED |
| 4938 |
PF08368 (FAST_2) |
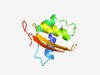 Estimated TM-score=0.50; hard target. |
>FAST kinase-like protein, subdomain 2 (Length=86) RLRLMELNRAVCLECPEFQVPWFHDRFCQQLQRKGNGNASAVQQQIHKMLGEILGGINYAKVSLLTPYFYAVDFECVLDK NKTPLP |
| 4939 |
PF08166 (NUC202) |
 Estimated TM-score=0.50; hard target. |
>NUC202 domain (Length=61) PYSSLRCSVYRVLETWVTTCGISSGVLQGPMHHSDILLANLLSDITPPTDAIKMSTFVQLG |
| 4940 |
PF08074 (CHDCT2) |
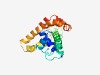 Estimated TM-score=0.50; hard target. |
>CHDCT2 (NUC038) domain (Length=171) YDIWHRRHDYWLLAGIVTHGYARWQDIQNDPRYMILNEPFKSEIHKGNYLEMKNKFLARRFKLLEQALVIEEQLRRAAYL NMTQDPNHPAMALNARLAEVECLAESHQHLSKESLAGNKPANAVLHKVLNQLEELLSDMKADVTRLPSMLSRIPPVAARL QMSERSILSRL |
| 4941 |
PF08058 (NPCC) |
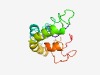 Estimated TM-score=0.50; hard target. |
>Nuclear pore complex component (Length=125) GTWRHPRLNEIARRRNATTFSEKNVRQIAYNVIALLGFWSAQLLAKLNIGSQVVPSSFRIYLSWAWFILNLIPFINIGIA CLPLIRPKDDLSDIPLTPAQRKLLGLDPSSAAPTPDAKFSTPPRY |
| 4942 |
PF08004 (DUF1699) |
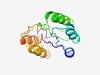 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1699) (Length=130) MKIRVVSSKEEINTLSPNEEIVHLAFRPSNTDIFSLIMKCPQVKALHIPSSYKRTISNSAKMYLKMQGIELLEGDVWGHR KDINEYSEVSQNVYDRIKQYRMDGMQEDDIADKMVRETRLSPDFISFLIK |
| 4943 |
PF07900 (DUF1670) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1670) (Length=208) PKTAEAIIETVKETYELHRFDPDMDGEPGKIKKQVISINAKHCPQLRELPMVTVTLTLDQFMEDQEVRRQFGSRGLRHTR FCRLTDEAIDQGGVLTQEDLADLLEVDVRTIRRDIKHLTQKNIRVQTRGAYHDIGPTLSHKVWIVGLYLQYKTYSEIARK TKHSTTSIKRYIKDFGRVVMCVKKELSVAETAHIAGISERITSEYLDL |
| 4944 |
PF07820 (TraC) |
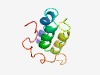 Estimated TM-score=0.50; hard target. |
>TraC-like protein (Length=85) AXDIDAQIEKLRARRRSLIVKSAERFARAATKSGLAEMEITEEELDXXFEEMAARFRXDEKKGVDHATASPRXPAAGATG TAAEV |
| 4945 |
PF07667 (DUF1600) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1600) (Length=113) FQSAWYHVLSPVLFLVFGFCYFYYNPNQQPKVLWKAVLKAMIYPTIYVIYVATIPFVLNANLYNGGATYTVYGDFTNLKD HLGKALPVILGMWLIFFPGSYIAFYYSWIGFNK |
| 4946 |
PF07655 (Secretin_N_2) |
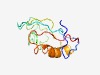 Estimated TM-score=0.50; very hard target. |
>Secretin N-terminal domain (Length=100) TETFPLNYLYMERHGLSLTSVNSGRISDSNNSNNNNNSDNNSSNNSSNNDSSSNNSNETTNGTFIRSKTKTDFWGELKET LVSIVGDTGGGRQVVVTPQA |
| 4947 |
PF07439 (DUF1515) |
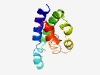 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1515) (Length=113) MIDATVHQQLGTLLAEVKNLREDFRRSEDKSDASRASMHRRMDELVERVGTLEGSTVAMQGDITEMKPITDDVKKWKLMG IGALGVTGIGGAALSVTFADVVKRGLAIFLRGG |
| 4948 |
PF07351 (DUF1480) |
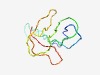 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1480) (Length=79) MNQTKLRISSYEIDDAILSSPSQEDDITISIPCNSDQELCMQLDGWDEHTSIPALLNDKHILLYRKHYDRQKHAWVMRV |
| 4949 |
PF07314 (DUF1461) |
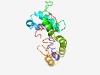 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1461) (Length=187) ILCSFCLIIILLITSVEAVAYWTPGYYEKEYSKYHVTEAVHMEMDDLLTVTYEMMDYLKGQRTDLHITTVVDGQEREFFN EREIAHMEDVRNLFLGGITLRRTCMMAASACLLLLLFLKADIKKVLPRTICLGTGIFFIISAVLGGIISTDFTKYFIIFH KIFFNNDLWILDPSTDLLINIVPEPFF |
| 4950 |
PF07306 (DUF1455) |
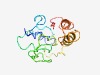 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1455) (Length=130) MSCRAFSLKGLLAFTLLPGSLATAARPSLLRVGGEASGQQTSGGGFHAAGGASATRGRLTSASAVSAPQSFGVLGSTVWE DGFLPSVFSPAQELLSAALQPSPTPSSWVFGRTAISGVRGFLERTVFLVF |
| 4951 |
PF07295 (DUF1451) |
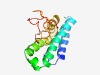 Estimated TM-score=0.50; hard target. |
>Zinc-ribbon containing domain (Length=144) SLTERLKNGERDIDQLVASAEKRLNEAGELTRDEVAQLKQAVRRDLEEFARSYQESREEFSDSVFMRVIKESLWQELADI TDKTQLEWREVFKDVSHHGVYHSGEVVGLGNLVCEKCHHHLAFYTPEVLPRCPKCGHDQFQRRP |
| 4952 |
PF07219 (HemY_N) |
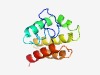 Estimated TM-score=0.50; easy target. |
>HemY protein N-terminus (Length=107) GYVLIQTAGYNIEMSITMLVVFFVILMAVVYLIEWVITRFFRLSSNTYSWFSRRKRLKAQRQTLEGLVKMNEGDYSKAEK LIGKNAKHSDEPVLNLIKAAEAAQNRG |
| 4953 |
PF07185 (DUF1404) |
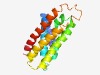 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1404) (Length=168) ILPIVLLAISVNPYTESLEFHYPAIYMISHYLVYFSGIFVGYKYFKGDEISLALGLIPPILWHLPYFFALGAAFTTYRAI LEITLLVGGILVGSSIKYVKFYLKVTLFALWMLGDSALAIIFIVSSPIYSNVAYNFSPYSPSSLPIAGVAMFIAMNVFLG YVIAKYIK |
| 4954 |
PF07125 (DUF1378) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1378) (Length=59) MRFVQLILLYFCTVVCTLYLVSGGYKVIRNYIRKKIDAAAAEKISASQSAGSKPEEPLI |
| 4955 |
PF07037 (DUF1323) |
 Estimated TM-score=0.50; hard target. |
>Putative transcription regulator (DUF1323) (Length=122) MTTEELAECLGVARQTVNRWIREQGWETEKFPGVKGGRARLIHIDARVQEFILNIPAFRNHSAFYQAEEPLAEYSHVVRS HAYRQIIDAVENMSSKEQEKLAQFLSREGIRGFLARLGLDES |
| 4956 |
PF07023 (DUF1315) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1315) (Length=86) QTILASLTPEVVDKFRMAIELGKWPDGRKLTAEQRETCMQAVMVWEHEHLPPAERTGYIHKPVKEDGSIVGAECDVEHEH HYPNMP |
| 4957 |
PF06869 (DUF1258) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1258) (Length=254) EARKLWEYTRNNFSTHTYCNLCGKVLGHINRCNICANAPVATFIRIGGFSQIRELVDSHLNEILQIREELKSGRNVDHVL GSPFFSRGWANESKHHLHLSTLLSIDGVHIPGNNNKLWPVSLLLCDLPIGLMQKSTHVILEGIVECSDTPSTALWNALIP MFMTDIEHHIGRVKNITFSCRITTISADQPAKRAFFGFRHHSAKLSCFFCLAPETYYKHEGPNRKLSRPGHLTIQDSING RNGFTEKKSIIVRH |
| 4958 |
PF06795 (Erythrovirus_X) |
 Estimated TM-score=0.50; hard target. |
>Erythrovirus X protein (Length=81) MDSYLTTPMPYHPVAVMQNLEEKMQYYLVKTYTSLGKLAYNYPVLTMLGLAMSYKLGPRKVLLTVLQGFMTLGIANWLSW E |
| 4959 |
PF06678 (DUF1179) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1179) (Length=106) FPFTTIFKSITLYFGPFLLLIVSIVNCQSKKKFKDDGRVDLVPRNAPVRPDPVKETSAQTCPTAQTPPAKTPMERSGNVA EDTLANVKSLPPEKSDDADKPKKKKK |
| 4960 |
PF06648 (DUF1160) |
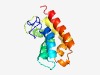 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1160) (Length=121) FVNQVFSNMPVAAKVAMVNLNLKQYLKDLERDETFCSKFTQIIRMFINRDITTHDVCTILDAADGIKLTHGQIDYFCNQV YYNGHILHILQTFIDYQHLTDDEINDLSQFLVKEIDNAIMI |
| 4961 |
PF06531 (DUF1108) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1108) (Length=84) MYYKIGEIKNKIISFNGFEFKVSVMKRHDGISIQIKDMNNVPLKSFHVIDLSELYIATDAMRDVINEWIENNTDEQDRLI NLVM |
| 4962 |
PF06523 (DUF1106) |
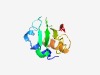 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1106) (Length=91) MASLWKKLFYFPGNRRRYFEQEEHSFSIICGRLRGIVVTFKCSKGIIYLSIKVNPNNSKHILLYNKKEYIFDKLKELFPD EAIEFSIEYEN |
| 4963 |
PF06497 (DUF1098) |
 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF1098) (Length=93) RNRPPARPSAQTQIAAVDMLQTINTAASQTAASLLINDITPNKTESLKILSTQSVGARSLLEPMQANASTIKLNRIETVN VLDFLGNVYDNTI |
| 4964 |
PF06404 (PSK) |
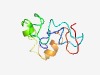 Estimated TM-score=0.50; very hard target. |
>Phytosulfokine precursor protein (PSK) (Length=84) LLLCLLCTRGQAARPTPGSSHQQSQGAGSATSHEKSAAGTGMEMEQVDQEAMRGCEGGEGAEECLMRRTLVAHTDYIYTQ GKHN |
| 4965 |
PF06279 (DUF1033) |
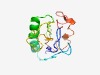 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1033) (Length=118) MYRVIEMYGDFEPWWFLEGWEEDIVASKKFDQYYDALKYYKTCWFRLEQESPLYKSRSDLMTIFWDPEDQRWCDECDDYL QQYHSLALLQDEQVIPDEKLRPGYEKQTGQERHRSCRM |
| 4966 |
PF06231 (DUF1010) |
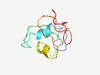 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF1010) (Length=81) MRAATYFSARPCSKGFLAFLASSACQPSATSYHSCSAAPLPWPSGFSWAAHVVKAGRSLLAFGSNSAVKRTGLRPAAYFG R |
| 4967 |
PF06174 (DUF987) |
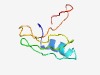 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF987) (Length=65) MKIITRGEAMRIHQQHPTSRLFPFCTGKYRWHGSAEAYTGREVQDIPGVLAVFAERRKDSFGPYV |
| 4968 |
PF06066 (SepZ) |
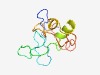 Estimated TM-score=0.50; very hard target. |
>SepZ (Length=100) MDAANLSPSGRVLPLAATINGNVSVDENTGVMKPENGVSRNIRIVAGIALATTALAALGTGIAMACTTNASSQEFLGLGI ATGVLGGVTALGGGLAMKYA |
| 4969 |
PF06053 (DUF929) |
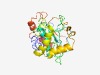 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF929) (Length=240) IIAVIVIVVLVIAFFSIYYISKLSNASTIPAGKFVKISNMDLAPKGEVIVVEQSWYGCPVGAAASWAIYNVLKNYGNITF EFHYSDPDHNPANIPGLIFLNFTPTSIVRFYVAYVYNEYLNASYNGTPIPQNKLVTVGEEILKEEYASMGLNPQVANYII QYETQVPIQQYGKPSAYYVQPPHLNFAILISGPNGTYIITTPIVNPNILSGYSPQYVYSHLDNFQQIIQASQMIQQVILE |
| 4970 |
PF06024 (Orf78) |
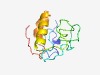 Estimated TM-score=0.50; very hard target. |
>Orf78 (ac78) (Length=112) LDVPYERLGTATKVDYIPLKLALTDLPSENTSDNNDDNQKNNNTQNPKIDINQSNANNYNQHQSVRSKQQFYDILVLGML TVFCILVLLYAIYYFVILRDRQKSNTIRPSYM |
| 4971 |
PF06022 (Cir_Bir_Yir) |
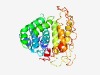 Estimated TM-score=0.50; hard target. |
>Plasmodium variant antigen protein Cir/Yir/Bir (Length=280) CGILISISNSISKKLDKNKNYQIVINESTLNRYCSGNSCDNDLDKINAGCLYLLDSFFKDSSVFRSYAKSNTNIVEYIII WLSYMLNLKEQFGKTNLQYFYEIYIQGGDQYKNSITGVTGYNSYKDLIDNKIDLINMDMSIVPKLYDAFYTLCMMYLEFD EESRNCSKNSGRAKEFVEKYEYLKKNYSITKNSPYYKLLSTLSKDYDNFKKKCSDVSCKDIPPLQSIEKTEINVNLSEHS KLTSVQTSEDASSSSSITNNLFIVLSIFGAIAFFLGISYK |
| 4972 |
PF05754 (DUF834) |
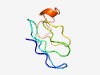 Estimated TM-score=0.50; very hard target. |
>Domain of unknown function (DUF834) (Length=53) SGWRCGNGGLPRVNGDEGLPGAFVGREMEAGLWLAVAVPTVEVAQLGDARGDD |
| 4973 |
PF05564 (Auxin_repressed) |
 Estimated TM-score=0.50; very hard target. |
>Dormancy/auxin associated protein (Length=117) LWDDVVAGPQPDRGLGRLRKITTTPLVVKEGGEAESSSGKFQRSLSMPASPGAPSTPVTPTTPLSARKDNVWRSVFHPGS NLATRGIGAEVFDKPTHPNSPTVYDWLYSGETRSKHH |
| 4974 |
PF05317 (Thermopsin) |
 Estimated TM-score=0.50; very hard target. |
>Thermopsin (Length=267) YVNPFLYYTSPPAPSGIASFGLYNNSGKVTPYVIETSKVLGYANITSLLAYYKQARKYGVNPYSATLQMNVVLQVNTTQG TFAYWLQDVGSFQTNTNQVTFIDNIWNLTGNPSTLSSSAVSGNGKVASAGSGNTFYYDVGPTFTYSFPFSYVYVVNTSYT SNSVSIWFGYEILQGSQVTSVQYYDKVTISQPGIKSAEITINGNTYTPDGLYYDAELVWGGGGNGAPTQFTHLNSSLGLY YLSSTGVTPIPSLYTFGSNTGESAYNV |
| 4975 |
PF05178 (Kri1) |
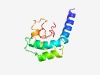 Estimated TM-score=0.50; hard target. |
>KRI1-like family (Length=102) KKEEEKKERETEKNRLRKLKMEELQEKVNHIKEVAGLRASEFTDEEWAKFLDDSWDDQKWEQEMQKRFGEDYYAGEGDGE KKPKKPTWDDDIDIKDIVPDFD |
| 4976 |
PF05081 (DUF682) |
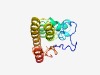 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF682) (Length=153) TTINLFTYKPPPNTVCDDDIDKQNFFIQGVLEALNDNSKSKQACFLELKREQCLLLRKIKHDLLYSCEGNYCQNHVLLDA LHLYKLYKEEFDDTQSAFDIDLMQTCREIVYLTFELFSCAANIVVFVHASANYKDNVLAQLLVELQSKNFITL |
| 4977 |
PF05080 (DUF681) |
 Estimated TM-score=0.50; very hard target. |
>Protein of unknown function (DUF681) (Length=101) MGGACIASRYWLVPARVILRLRLTSFTGHRGVARVDGPMSSLGPNIIKCAVNGGMDMMGKMSSSWTTFMGGYLIARCSAS ATVTHIKCQLRALLWSLRARG |
| 4978 |
PF05022 (SRP40_C) |
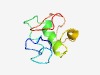 Estimated TM-score=0.50; hard target. |
>SRP40, C-terminal domain (Length=74) PFKRIKDEDVVFLDERLKDNSFMAKGGAIGSYGEKAHRDLIVTRGKGFRNEKNKKKRGSYRGGKIDLESHSIKF |
| 4979 |
PF04867 (DUF643) |
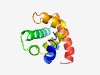 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF643) (Length=114) INEISDFYDCLSPGTKKEISKLYGVKQLTLEQKKDFYRGFVSIQEYKRKTGKSIDEIINGIINPAKNFIKDVLKDKHIIE KYKNFQNMKFDCSYKKGMLEKCLKKMGEKYSDRF |
| 4980 |
PF04866 (Rota_NS6) |
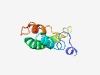 Estimated TM-score=0.50; hard target. |
>Rotavirus non-structural protein 6 (Length=92) MNRLQQRQLFLENLLVGVNSTFHQMQKHSINTCCRSLQRILDHLILLQTIHSPVFRLDRMQLRQMQTLACLWIHQHNHDL QVMSDAIKWISP |
| 4981 |
PF04831 (Popeye) |
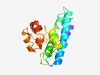 Estimated TM-score=0.50; easy target. |
>Popeye protein conserved region (Length=147) PQHVMYQLANICFIISYLAPGGEKGLLFMHSSLIIGFFIFAMWAWNVVCSPSMFSWNFLFVLMNMGQLLHILYNMRKVKF NPELEDVYRHMFLPLQVSIIATTPSRCIVWQRQSLEYLLIKETHLSTVLALLLSRDITRKLYAMNEK |
| 4982 |
PF04822 (Takusan) |
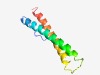 Estimated TM-score=0.50; easy target. |
>Takusan (Length=83) EPSSQSTPFTKKQVKKEIERMTTELQLMTSQRNELRDRLVSISEGTVDNRPYHKPNPLYKKLKLEHKQVMWELRVFENEN TQV |
| 4983 |
PF04761 (Phage_Treg) |
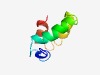 Estimated TM-score=0.50; hard target. |
>Lactococcus bacteriophage putative transcription regulator (Length=57) MEQEKTINHLGQVVYQESVEFYKEKLSVHSKDFLQNSLIPQLYEWSNAYKAAVELTK |
| 4984 |
PF04724 (Glyco_transf_17) |
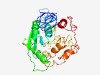 Estimated TM-score=0.50; hard target. |
>Glycosyltransferase family 17 (Length=332) DLRALLVLLIGLPILIFVIYLHGQKVTYFLRPIWEKPPKPFKVLPHYYNENVSMGHLCKLHGWKVRETPRRVFDAVLFSN ELDILDIRWHELSPYVSEFVLLESNSTFTGLKKDLHFKENRQRFEFAESRLTYGMIGGRFVKGENPFVEESYQRVALDQL IKIAGITDDDLLIMSDVDEIPSGHTINLLRWCDDIPEVLHLQLRNYLYSFEFFLDDKSWRASIHRYRAGKTRYAHFRQTD DLLADSGWHCSFCFRYISDFVFKMQAYSHVDRIRFKYFLNPKRIQHVICQGADLFDMLPEEYTFQEIIAKLGPIPSTYSA VHLPAYLLETCS |
| 4985 |
PF04708 (Pox_F16) |
 Estimated TM-score=0.50; easy target. |
>Poxvirus F16 protein (Length=215) SAAIITSLISLLDTSIEYQIKACRDYCKLLSVVVELEFKEFGYIDKHTLETKRWNDLARSNINILVFYQVRQLSISAEYL YDVFIKNREVPIRMYFVKDHLAFDGCPPLFRTINIDVKFTYRKKIRDLITISNMTTCNDKIMQDFINNNFGNVSSLLRII DSDSIWLRKVIMSGKYKHGSKIRSYRHFMFTIRRIKKKEASSIDDICNNIQSITI |
| 4986 |
PF04667 (Endosulfine) |
 Estimated TM-score=0.50; very hard target. |
>cAMP-regulated phosphoprotein/endosulfine conserved region (Length=80) DLSKLTPQELKLYKMYGKLPSKKDLFKHKMQERRYFDSGDYALSKAGVIQSEGASHNNLPVTNPSGLRESIIRRRMSSSA |
| 4987 |
PF04660 (Nanovirus_coat) |
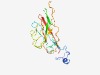 Estimated TM-score=0.50; hard target. |
>Nanovirus coat protein (Length=170) MARYPKKSIKKRRVGRRKYGSKAATSHDYSSSGSILVPENTVKVFRIEPTDKTLPRYFIWKMFMLLVCKVKPGRILHWAM IKSSWEINQPTTCLEAPGLFIKPEHSHLVKLVCSGELEAGVATGTSDVECLLRKTTVLRKNVTEVDYLYLAFYCSSGVSI NYQNRITYHV |
| 4988 |
PF04534 (Herpes_UL56) |
 Estimated TM-score=0.50; very hard target. |
>Herpesvirus UL56 protein (Length=197) MALGAGHAHACRDDGDDSVIDAPPPYESVAGASAGQFVVIDIDTPTDSPPPYSAGTSPVGLVSPASSGDGEVCERGRSRR AAWRAARRARRRAERRARRRSFGPGGLFVETPLFLPETMIGAHPGVGGDLPSGLPTYAEATSDRPPTYAMVMAACPTEPP GGSVGPADQPRVQSSRTWRPPLVNSRELYRAQRAARC |
| 4989 |
PF04382 (SAB) |
 Estimated TM-score=0.50; hard target. |
>SAB domain (Length=49) MLEDLDKTQEDLMKHQTNISELKRTFLETSTETAVSNEWEKRLSTSPVR |
| 4990 |
PF04285 (DUF444) |
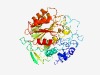 Estimated TM-score=0.50; hard target. |
>Protein of unknown function (DUF444) (Length=419) HFIDRRLNGKNKSAVNRQRFIRRYKKQIKQAVSDAVSKRSIQDVDKGESISIPTKDIGEPHFHQGQGGKREMVHPGNDQF VSGDKIDRPKGGGSGQGGGEGQASDQGEGADDFVFQISKDEYLDLLFEDLELPRLQKTQLNQLMEMKTYRAGYTSNGVPA NINVVRSLQNSLARRMALQAGKKRQLRELEERLAQLTSAPNDHFAEIAQLEKEIRELKRRIEAVPFIDTFDLKFNNFVKR PVPSSQAVMFCLMDVSGSMDQATKEMAKRFYILLYLFLSRTYKNVEVVYIRHHTQAKEVDEHEFFYSQETGGTIVSSALK MMNDIINERYPANQWNIYAAQASDGDNWADDSPQCREILMRDIMPRVRYYSYIEITTRAHQTLWHEYESVAGQFPHFAME HIRKVEDIYPVFRELFKKQ |
| 4991 |
PF04236 (Transp_Tc5_C) |
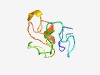 Estimated TM-score=0.50; easy target. |
>Tc5 transposase C-terminal domain (Length=65) WKKSGYMDRDANEPEFTTPAEYCMGKATPEDCYISGCGELGCLKCARCQNWVCFDHLVVSQIHLC |
| 4992 |
PF04061 (ORMDL) |
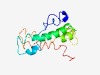 Estimated TM-score=0.50; hard target. |
>ORMDL family (Length=135) NLNANWVNAKGAWTIHIVLICLLKVLYDIIPGVSQETSWTLTNISYMFGSYIMFHWVRGVPFEFNGGAYDNLNMWEQIDD GAQYTPAKKFLLSVPIVLFLLSTHYTHYDLTYFTINFLAVLAVVIPKLPFSHRMR |
| 4993 |
PF03908 (Sec20) |
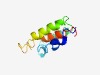 Estimated TM-score=0.50; easy target. |
>Sec20 (Length=92) SQNKQITKSLQASRQLLSAGILQSELNIDNIDQQTKDLYKLNEGFIQFNDLLNRSKKIVKFIEKQDKADRQRIYLSMGFF ILCCSWVVYRRI |
| 4994 |
PF03384 (DUF287) |
 Estimated TM-score=0.50; hard target. |
>Drosophila protein of unknown function, DUF287 (Length=55) ISSVLEPDYEEKDLLRRIVDGDHVDDGIGFVDPVVDSWRNRLIKERKRIFWKDMF |
| 4995 |
PF03383 (Serpentine_r_xa) |
 Estimated TM-score=0.50; easy target. |
>Caenorhabditis serpentine receptor-like protein, class xa (Length=156) GPFVYIIFLTGFGIMEKLNAFLMVDGWPVSEWFDPEHGYERYRNFFGAYITLIFLICYLTPLLLDCIMTIHRICIFISPL KSTKWFSDFKVVCYCITTCLLVLIWLLVQQISNCTLNFNALTSFLESACAPDRHPVTWFQNKYLIYVPILSMVINA |
| 4996 |
PF03298 (Stanniocalcin) |
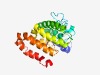 Estimated TM-score=0.50; hard target. |
>Stanniocalcin family (Length=198) LLLLLLLSASASHETEQNDSVSPRKSRAAAQNSAEVVRCLNSALQVGCGAFACLENSTCDTDGMFDICKSFLYSAAKFDT QGKAFVKESLKCIANGVTSKVFLAIRRCSTFQRMISEVQEECYSKLDMCGIARRNPEAITEVVQLPNHFSNRYYNKLVRS LLECDEDTVSTIKDSLMEKLGPNMASLFHLLQSDHCAQ |
| 4997 |
PF03233 (Cauli_AT) |
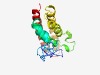 Estimated TM-score=0.50; hard target. |
>Aphid transmission protein (Length=167) MSNASANPHIYKKDKYVRLKSLEPVSSKQRQYYFATDDKKSNIKSIAAHCNNLNVVVAKSWLKLSKLLSYLGLEKDESEA YSKNKLPFGQFFKDLTKSFQEGGDNKEEKTNLLPTLQEMSNRIGKISEETKKLAEQDTVTNKQLEAMVKDFDKRLKEIQD SVKAIVG |
| 4998 |
PF03202 (Lipoprotein_10) |
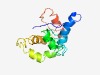 Estimated TM-score=0.50; hard target. |
>Putative mycoplasma lipoprotein, C-terminal region (Length=134) ITQGPNIIGIHANEKENAETQKFVNWFLNNTESWEVKNGKDSSTKQQTAAEFFAESASYILPLKEIFDKDNKKATENKDN NTSNKKQANTYAEKALDLFQQISKGDIVSYSDPSDFRSGKFRDGIGSNFNAAVS |
| 4999 |
PF03158 (DUF249) |
 Estimated TM-score=0.50; easy target. |
>Multigene family 530 protein (Length=193) GALEGDRYDLIHKYYEQIGDCHKILPLIQDPQIFEKCHELSNSCNIRCLLEHAVKHNMLSILQKHKDQIRLHMALTQILF ELACHERKNDIIRWIGYSLHIYHLETIFDVAFAHKNLSLYVLGYELLMHKVNTEAANIDLPNLLSYHLRTAAAGGLLNFM LETIKHGGCVDKTVLSAAIRYKHRKIVAHFIHQ |
| 5000 |
PF03121 (Herpes_UL52) |
 Estimated TM-score=0.50; very hard target. |
>Herpesviridae UL52/UL70 DNA primase (Length=73) FSPTNQCIIQVRPERGSNFRCLRYNHRGGSKTVRVFLTLHLNRESKLVVTFMSQCFANKCNNNKPVAHFSVTV |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417