

Go to page (83 pages in total): [First page] << < 43 44 45 46 47 48 49 50 51 52 53 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4701 |
PF07530 (PRE_C2HC) |
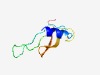 Estimated TM-score=0.51; easy target. |
>Associated with zinc fingers (Length=70) CSWIVEELLKLGFQARFVRNMTNPATGGPMRMFEVEIVMAKDGSHDKILSLKQIGGQRVDIERKNRTREP |
| 4702 |
PF07458 (SPAN-X) |
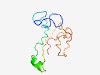 Estimated TM-score=0.51; very hard target. |
>Sperm protein associated with nucleus, mapped to X chromosome (Length=94) MEQPTSSTNGEKRKSPCESNNNKNDEMQETPNRDLAPEPSLKKMKTSEYPRVLVFCYRKVKKINSNQLENDQSRENSINP VQEEEDEDLDSAEG |
| 4703 |
PF07432 (Hc1) |
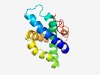 Estimated TM-score=0.51; hard target. |
>Histone H1-like protein Hc1 (Length=123) ALKETTKNMKHLLVQIAEDLEKADGGNKAASQRVRTGTVKLEKIAKLYRKESIQSEKGTKGRAKKPAAKAAKPKAAAKHA APKHAAAKAPKAKATKAHAKPASKAKKTTAKARPMSFKRPTAK |
| 4704 |
PF07384 (DUF1497) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF1497) (Length=59) MGYYDTRNEARRISKLASQNISSEQTKKEFELDRQSKFNQEMQAEFHEKIKKLGEKNGS |
| 4705 |
PF07277 (SapC) |
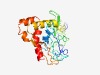 Estimated TM-score=0.51; very hard target. |
>SapC (Length=220) LQTALHSNLKLKADNTFAFASGIHVAAIMANEFYRAASTYPIVFIEDAGKDGFMPVVLLGLTENENLFVNAEGRWEATYV PAILRRYPFALAKTGEEGQYTVCIDEESGFFTEEEGQALFNEDGQPGEVLEKVKTYLAQLQQMEQLTRQFCEVLKEHNLL APLNMRVQKDNAVQNITGAYAVNDERLSHLSDETFLELRKKNFLPLIYSHLVSVTQIERL |
| 4706 |
PF07215 (DUF1419) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1419) (Length=110) TLSPIRKVFQGVADRRQMFRMFDRHAQRPNRWEGDDSALYCGEWFEIDEREHNYMFEILPPLWMRGDMFAMREFLTGNIT SIFFTLKIDDRMRYFHAYCDLSDKGSPERM |
| 4707 |
PF07203 (DUF1412) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1412) (Length=50) GLIRQRRQSYWNSNGVVHNVVTDNMAGGPTSLGWAQVPHRLSPMFSPIFG |
| 4708 |
PF07174 (FAP) |
 Estimated TM-score=0.51; easy target. |
>Fibronectin-attachment protein (FAP) (Length=293) MDQQDAVRVHRKGLSKSLAIAALTGATAVALALPSIAQAQPAPTPAPPPPPPSGNTFLQGPPPPPGAPAPPPPPADPNAP PPPPPADPNAPPPPPADPNAPPAPPPVEAGRVDNAAGGLSFVVPAGWKVSDSTQLSYGQALLTKIAPEGAEPPNDTSVLL GRLDLKLFAGAETDNTKAAQRLASDMGEFFMPFPGTRINQKTVPLDAAGMPGVASYYEVKFTDANKPTGQIWAGVVGEPV EPGTPRGQRTPQRWFVVWLGTATNPIPQEEAVTLANSIRPYTAPAAPPPADPN |
| 4709 |
PF07029 (CryBP1) |
 Estimated TM-score=0.51; very hard target. |
>CryBP1 protein (Length=164) QTDTEVEFTCVTCVPESFAVDIESLEEIRFIFNTDKLYCCLDKTTEECNIPGIGDIEVDVCVVKIVGAIEYILNAYPAVR GDKYGRGEGDIEVIPDREANVCCEGTTFVDNVVGAFAVDDPELIEDPCEELDFCDIEGVLTDISIITDEESDKQCIKFEG RFEL |
| 4710 |
PF06834 (TraU) |
 Estimated TM-score=0.51; very hard target. |
>TraU protein (Length=289) XCIFPLSLGSIKVSQGKVPDTANPSMPIQICPAPPPLFRRIGLAIGYWEPMALTDVTRSPGCMVNLGFSLPAFGKTAQGT AKKDEKQVNGAFYHVHWYKYPLTYWLNIITSLGCLEGGDLDIAYLSEIDPTWTDSSLTTILNPEAVIFANPIAQGACAAD AIASAFNMPLDVLFWCAGSQGSMYPFNGWVSNESSPLQSSLLVSERMAFKLHRQGMIMETIGKNNAVCNEYPSPILPKER WRYQMVNMYPDSGQCHPFGRSVTRWETGKNPPNTKKNFGYLMWRKRNCV |
| 4711 |
PF06755 (CbtA_toxin) |
 Estimated TM-score=0.51; hard target. |
>CbtA_toxin of type IV toxin-antitoxin system (Length=112) MKTLSDTHVREVSRCPSPVTIWQTLLIRLLDQHYGLTLNDTPFADERVIEQHIEAGISLCDAVNFLVEKYALVRTDQPGF STCPRSQLINSIDILRARRATGLMTRDNYRTV |
| 4712 |
PF06727 (DUF1207) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF1207) (Length=337) QFKVDVPALIVRNGTITIPADKISARDRAKVVQILSEIPGVSRVEMGEHAVQRSVATASVPDKEILPKKSTPAIGSTMLA TGLLPEGHLFKPLLADPRWAHFSAAYRNYVGNNIDGNNNGSVSFGETIPFYRANIGQSIVQWETGIQAAVFSDFNLGASS SDLINTDFIASAYSSMRANNASAFARIYHQSSHLGDEFLLRKLTNLERVNLSYEGIDLKLSYELPYGIRVYGGGGGIFHK EPSTIKPWSVQYGLEFRSPWRIESAAMRPIMAVDVKNHEQNNWNADISARAGVQFDNFQAFNRKFQVLAEYFNGNSPTGQ FFREKVEYIGLGLHYHF |
| 4713 |
PF06587 (DUF1137) |
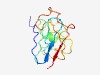 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1137) (Length=117) EKAPPFLKIKKLGVHKQITSPDRQFFNCHVDKSCMELHLSDANYACKEVLSKLSGHIHTQDLDKLMTFQGNGGLLNYQDC SLNIYDCRFHVDPIHPDPQAPEERAVGGMKTLSLSLL |
| 4714 |
PF06547 (DUF1117) |
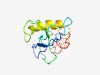 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1117) (Length=112) VGLTIWRLPGGGFAVGRFSGGRRAGERELPVVYTEMDGGFNNSGAPRRISWASRRSRSRESGGSLGRAFRNIFSFLGRFR SSSSSSSRSTSEPGSITRSHSHSSSVFSRYVR |
| 4715 |
PF06465 (DUF1087) |
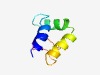 Estimated TM-score=0.51; hard target. |
>Domain of Unknown Function (DUF1087) (Length=61) EDAEEEPQREIIKQEENVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRKQVNYNDTSQED |
| 4716 |
PF06277 (EutA) |
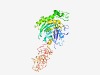 Estimated TM-score=0.51; easy target. |
>Ethanolamine utilisation protein EutA (Length=475) EELLSVGIDIGTSTTQLIFSKLIVENEASAFTVPRISIVDKEIIYRSEVYFTPLISNTEIDGRKIKEIIEEEYKKAGINK DDIQTGAVIITGETARKENANDVLHTLSGFAGDFVVATAGPDLESIIAGKGAGAHIYSKEHSTSVVNIDIGGGTSNLALF LRGEVKDTGCLDVGGRLIKINPNTREIIYISPKIEKIIKEKNLGLEIGTKATVENIKPVVNMMVAWLEESVGLRPKDEYF DYIVTNKSIALENEIKCISFSGGVADYIYYEGEIQDYFKYGDIGIILGQAIKNSDLCKKLKVVKSIETIRATVVGAGSHT TEISGSTITYTKDSFPIKNLPILKLSLEDESQGAYELETALKKKIEWFRLENDFQKIAIAINGKKNPSFKEIQEYAKGLV NGMKDLIEKEGQLIVVVENDMAKVLGQAIYSLLNFQKEIICIDGIKVENGDYIDLGTPIADGKVLPVVIKTLVFN |
| 4717 |
PF06266 (HrpF) |
 Estimated TM-score=0.51; easy target. |
>HrpF protein (Length=75) MSSSSPIQRRLDGAFVDAQSRLDEMALNVSEGEVSAADSYAFYEASMDYSNANWAAGQMLSVKHGLAKAIINDFN |
| 4718 |
PF05883 (Baculo_RING) |
 Estimated TM-score=0.51; hard target. |
>Baculovirus U-box/Ring-like domain (Length=134) MFCTFCLKDRNYYMFKLFKDYWPTCATECQICLEKIDDNGGIVAMPDTGLLNLEKMFHEQCIQRWRREHTRDPFNRVIKY YFNFPPKTLEECNVMLRETKGFIGDHEIDRVYKRVYQRVTQEDALDIELDFRHF |
| 4719 |
PF05824 (Pro-MCH) |
 Estimated TM-score=0.51; very hard target. |
>Pro-melanin-concentrating hormone (Pro-MCH) (Length=84) NIGSKHNFISHGSPLNLGIKQLPYLALKGSMAFPADTEIQNIESIQERRETGDEENSAKFPIGRRDFDMLRCMLGRVYRP CWQV |
| 4720 |
PF05766 (NinG) |
 Estimated TM-score=0.51; hard target. |
>Bacteriophage Lambda NinG protein (Length=196) MAKPARRRCKNDECREWFHPAFANQWWCSPECGTKIALERRSKERKKAEKAAEKKRRREEQKQKDKLKIRKLALKPRSYW IKQAQQAVNAFIRERDRDLPCISCGTLTSAQWDAGHYRTTAAAPQLRFNERNIHKQCVVCNQHKSGNLVPYRVELISRIG QEAVDEIESNHNRHRWTIEECKAIKAEYQQKLKDLR |
| 4721 |
PF05650 (DUF802) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF802) (Length=53) LSARLEQTSAAVAGNWSHALAEQQQTNQILNQDLRATLEGFSAGFDTRAGALL |
| 4722 |
PF05561 (DUF764) |
 Estimated TM-score=0.51; hard target. |
>Borrelia burgdorferi protein of unknown function (DUF764) (Length=180) MIFTLDMVLNHLTQIFKGFKAYATENNFECDIINTYNHPYLSKITAASSNIIALKFDGTENLFDHNSRAGAFYENALEFS LNFQIYIIAIVLNAQDFDANSRMLMLYGMLSNFLHNKAHKYTLESQSQPEYISKVNFYIYPISNMQTVGLINLGTKYSNH AYSASIAFNASVKAIEILKE |
| 4723 |
PF05533 (Peptidase_C42) |
 Estimated TM-score=0.51; hard target. |
>Beet yellows virus-type papain-like endopeptidase C42 (Length=86) YPDGRCYLAHMRYLCAFYCRPFRESDYALGMWPTVARLRACVEKNFGVEACGIALRGYYTSRNVYHCDYDSAYVKYFRNL SGRIGG |
| 4724 |
PF05384 (DegS) |
 Estimated TM-score=0.51; easy target. |
>Sensor protein DegS (Length=159) DSILEKMVHTVDHSKEEIFRIGEQSRNEFVTLTDELKDIKERVGTIIDEVDRLENHSRFARKRLSEVSSHFKKYSEEEIR NAYEKAHDLQMQLTVIRQEEKQLRERRDELERRLVGLNETIERADHLVGQISVVLNYLNSDFKMVGEMIEDAKQKQAFG |
| 4725 |
PF05377 (FlaC_arch) |
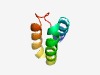 Estimated TM-score=0.51; easy target. |
>Flagella accessory protein C (FlaC) (Length=55) RINSLEKKLSKFEMSFNMMERENKDIKDTVQKIDQSVIELLSLYEIVSNQVNPFV |
| 4726 |
PF05288 (Pox_A3L) |
 Estimated TM-score=0.51; hard target. |
>Poxvirus A3L Protein (Length=70) KYHINLNPPRRCSRCLANLFEFLSEDSETIKMVLESQPNKLRILKEFLSACRNKEFIYKILDDEIKRVLT |
| 4727 |
PF05226 (CHASE2) |
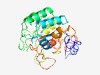 Estimated TM-score=0.51; very hard target. |
>CHASE2 domain (Length=288) LLVGWLALTPTADRADNLFYDGLIRVQDGPADPSIVIIAIDDRSLNALGRWPWPRTVHAALVDRLNQAQPRAVAYDVLFT EPEAGDPALAAALARARDVHLPLLVEAPGQNGAAWQVSEPASDLARAAASLGHVNMIVDGDGVIRRVPLYMQAGARSWPQ LVLPLAETAGAAPPAPRPDADSGEALIAAAPEAIAYRGPPGRFRTLSFVDVLNGEVPADFLKDRLVLVGATAPGMGDRYA TPAAPHGELSPGVEIQAALLQTLLEGGGPRTLSPPWVLGLSLLPLALL |
| 4728 |
PF05083 (LST1) |
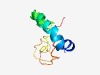 Estimated TM-score=0.51; hard target. |
>LST-1 protein (Length=79) LGGLLLLAVVILSACLCRFRQRVTRLERNWAQFRDQELHYASLQRLPGPSGEGPDLGDREEGTKADPSSDYACIAKNKP |
| 4729 |
PF05077 (DUF678) |
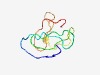 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF678) (Length=74) MEDTGGAKRIRKRKPKTTIQDNDDCVTCSSCYSKLIKVSDITKVSLDLYKVSGKGNTLSCAACGSELRLLNDFV |
| 4730 |
PF04932 (Wzy_C) |
 Estimated TM-score=0.51; very hard target. |
>O-Antigen ligase (Length=159) ASIIFAGLVASIARSNWISFVVYSAVLFIYCLKKKKYIKRYMMCLGIFVVVFLLLNLLTDSSIASRLNSIGLDAKKITMK DNDTAGSSRLYIWKRGITFIPEHPLLGSGPDTFGVVFLEKYFDELKYMQDITGGIVDKAHNEYLQLAVTSGIPALLIYL |
| 4731 |
PF04757 (Pex2_Pex12) |
 Estimated TM-score=0.51; hard target. |
>Pex2 / Pex12 amino terminal region (Length=237) QDSLMGAVRPALQHAVKVLAESNPSRYGPLWRWFDEIYTVLDLLLQHHFLSRTSASFSENFYGLKRVVTSGLQRSAHLGL HSRQRWNSLLLLVLVPYLHAKLEKVLARQREEDDFSIQLPQTRFQRMYRAFLAAYPFVSTVWEGWTFCQQLLYVFGRTWR HSPLLWLAGVRLVHLTAHDILNMDLKPTTTSLATSQRFGEKLQQMLSTVVGGIAVSLSTSLSLGVFFLQFLEWWYSS |
| 4732 |
PF04713 (Pox_I5) |
 Estimated TM-score=0.51; hard target. |
>Poxvirus protein I5 (Length=74) SAKEIFSAISLTLLALLMIISGGALAFKSLAPHRIVSMRSATFNRVVSILEYLAILIFIPGTIALYSAYIKKLF |
| 4733 |
PF04668 (Tsg) |
 Estimated TM-score=0.51; very hard target. |
>Twisted gastrulation (Tsg) protein conserved region (Length=131) LSKKSHVEDFSEPVPGLFQALTAEPDPHERWLTFTFPVDFDVSLFTPKHEKEMKYHMQTVEQEERTLKPNVMTVNCTVAY MAQCNSWNKCRASCQSMGATSYRWFHDGCCECIGDTCINYGINESRCVHCP |
| 4734 |
PF04623 (Adeno_E1B_55K_N) |
 Estimated TM-score=0.51; very hard target. |
>Adenovirus E1B protein N-terminus (Length=68) MDPPNSLQQGIRFGFHSSSFVENMEGSQDEDNLRLLASAASGSSRDTETPTDHASGSAGGAAGGQSES |
| 4735 |
PF04610 (TrbL) |
 Estimated TM-score=0.51; hard target. |
>TrbL/VirB6 plasmid conjugal transfer protein (Length=218) IVIDITLAGLFWAWGADEDILQRLVKKTLYIGFFAFIITNFTKLSGIIFNSFAGLGLKAGGSSISTADFLRPGRLAQVGL DAGQPLLDSASQMMGFTSFFSNFVQIAVLMVSWLLVLIAFFILAVQLFVTLIEFKLTTLAGFILIPFALFNKTAFLAEKV LGNVVASGVKVMVLAVIVGIGTGLFSQFTTAYAGGQPTIEQALSVVLAALAMLGLGIF |
| 4736 |
PF04579 (Keratin_matx) |
 Estimated TM-score=0.51; very hard target. |
>Keratin, high-sulphur matrix protein (Length=96) CCVARCCSVPTGPATTICSSDKSCRCGVCLPSTCPHEISLLQPTCCDPCPPPCCQPEVYVPTCWLLNSCHPTPGLSGINL TTFVQPGCESPWKEEC |
| 4737 |
PF04571 (Lipin_N) |
 Estimated TM-score=0.51; hard target. |
>lipin, N-terminal conserved region (Length=102) MNYVYYIFNNVKYLYNSMNSATLSGAIDVIVVEQPDGTMLSTPFHVRFGKYGVFNYDDKYVDIQINNEEIDLKMKLGENG VAFFVEKPSDETELPSHLETSP |
| 4738 |
PF04570 (zf-FLZ) |
 Estimated TM-score=0.51; easy target. |
>zinc-finger of the FCS-type, C2-C2 (Length=51) FAAAVETAPFLRSCGLCKHSLNLGRDIFMYRGEIAFCSLECRQQLMNQEDR |
| 4739 |
PF04569 (DUF591) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (Length=51) GRTRGSEPPDRIRRKEARGGGLRRRTLATRKRGNGGGATRIKFKVVGVSPG |
| 4740 |
PF04528 (Adeno_E4_34) |
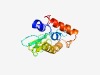 Estimated TM-score=0.51; very hard target. |
>Adenovirus early E4 34 kDa protein conserved region (Length=145) LASWFRMVVDGAMFNQRFIWYREVVNYNMPKEVMFMSSVFMRGRHLIYLRLWYDGHVGSVVPAMSFGYSALHCGILNNIV VLCCSYCADLSEIRVRCCARRTRRLMLRAVRIIAEETTAMLYSCRTERRRQQFIRALLQHHRPIL |
| 4741 |
PF04517 (Microvir_lysis) |
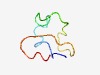 Estimated TM-score=0.51; hard target. |
>Microvirus lysis protein (E), C terminus (Length=42) RPVSSWKALNLRKTLLMASSVRLKPLNCSRLPCVYAQETLTF |
| 4742 |
PF04431 (Pec_lyase_N) |
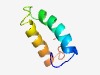 Estimated TM-score=0.51; hard target. |
>Pectate lyase, N terminus (Length=53) HIADFDEEWQKRAEEARKVALEAYQPNPEDVIDHFNNQVQGALNGTNSTRRNL |
| 4743 |
PF04393 (DUF535) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF535) (Length=278) WENPAYRRKFMLRSLATPFATRRWLSVLARQPMLDALLHAQPGLPCRLHRPWLSVNVSRKAALAALEYHYQHIERLLPDT LMQGHLTRQGSTLATLSGKNDAQYRLDLAAIADLDKEGETTVIFRDANGVVLAEMTFTLCEVDGQSALFIGGLQGAKAWV EHDQIQLATKACHGLFPKRLLLEAVCLLAQHFGVSQILAVSNETHIYRSWRYAKKKKDKLHADYDSFWESLGGEKDARGL YHMPLQITRKSLEEIASKKRAEYRRRYELLENMTAQIA |
| 4744 |
PF04380 (BMFP) |
 Estimated TM-score=0.51; hard target. |
>Membrane fusogenic activity (Length=75) MNKPNFFDDIQAKINQAIENSPAKDIEKNVKAMLGQGFSKLDLVTREEFDVQAQVLANTREKLEALEARVTELEA |
| 4745 |
PF04363 (DUF496) |
 Estimated TM-score=0.51; easy target. |
>Protein of unknown function (DUF496) (Length=92) FQDVLEFVRLFRRKNKLQREIVDNEKKIRDNQKRVLLLDNLSDYIKPGMSIEEIQAIIANMRGDYEDRVDDYIIKNADLS KERREISKKLKA |
| 4746 |
PF04347 (FliO) |
 Estimated TM-score=0.51; hard target. |
>Flagellar biosynthesis protein, FliO (Length=84) WLVRRLGLQNRQGGRLLKVVSSLMVGQRERVVLVEIGDTWLLLGVGAGQVNTLHTMAAQHAPDASPKVDDTDPDTFANRL RQSL |
| 4747 |
PF04284 (DUF441) |
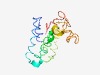 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF441) (Length=140) LFLLLLLIIGYIANNQSLMIAVSFLLIVKLIGLDTKIFPSLQKNGINWGVTVITIAVLVPIATGEIGFKQLLDSLRSPYA WIALVSGISVAIIAKQGLSLLANDPHITTALVFGTILAVSFFKGVAVGPLIGAGIAYLLM |
| 4748 |
PF04276 (DUF443) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF443) (Length=205) NLRYRILEINGEEYVLDFGNSIWKIIFPFLHWVLPNPVYKIDNHEIIEKLEVPQENHPKKISTGTGALGGLVGGVFLANL LRPLANVFNIPSTSLVNLIITIIVLIIALSVFVYVNRRHEKNLTAIVDYTHYPRKKIWIKPPTLKFFFNICFAYLFFMAL TVVPLMGFIETGDMFIFVPGVVFLFVSFMITYMTIPPGNTTVRFK |
| 4749 |
PF04194 (PDCD2_C) |
 Estimated TM-score=0.51; very hard target. |
>Programmed cell death protein 2, C-terminal putative domain (Length=172) FKSYYLSVIEEPPEVTSVDVHVEDLIRKYEQDEGKQLDELFKESCTPGKPKHGEHYEKTELKHGDKMFHKFLKRLQRCPE QCIRYDRNGDPLLVSVMDSSLTIPKCPYCGCQRIFELQLLPSLIPWLKTENGSDVEIDYGTVMIYTCQNNCWPPSSDPNC TLTVMEEFIMVQ |
| 4750 |
PF03856 (SUN) |
 Estimated TM-score=0.51; hard target. |
>Beta-glucosidase (SUN family) (Length=243) DGTISCSDFPSGQGAVSLDWLGLGGWASIMDMNGNTATSCQDGYYCSYACSPGYAKTQWPSEQPSDGRSVGGLYCKNGKL YRSNTDTNSLCVEGQGSAQAVNKVSGSIAICGTDYPGSENMVVPTVVGAGSSQPINVIKEDSYYQWQGKKTSAQYYVNNA GVSVEDGCIWGTEGSGVGNWAPVVLGAGYTDGITYLSIIPNPNNKEAPNFNIKIVATDGSTVNGACSYENGVYSGSGSDG CTV |
| 4751 |
PF03529 (TF_Otx) |
 Estimated TM-score=0.51; very hard target. |
>Otx1 transcription factor (Length=90) AVAPLPEPGPPSNASCMQRSVSGSAAAATSYPMSYNQTPGYGQGYPAPSSSYFSGVDCGSYLAPMHSHHHHPHQLSPMTA STMSGHPHHH |
| 4752 |
PF03516 (Filaggrin) |
 Estimated TM-score=0.51; very hard target. |
>Filaggrin (Length=55) GRSRSSTKSRQQSHQHNSADHPRHSGTGRGRSTIESGAGSHRGSSVGQASDSEGQ |
| 4753 |
PF03511 (Fanconi_A) |
 Estimated TM-score=0.51; hard target. |
>Fanconi anaemia group A protein (Length=62) EELLVVLFFFSLMGLLSSHLTPHAVDSLKAVDICAEILRCLDKRKVSWLVLFQLTDTDGGLG |
| 4754 |
PF03478 (DUF295) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF295) (Length=56) RRMKSLEGQALFVGTHCSKSLHAAECVGAQDDCIYFMCDYSQTTADPLHDAGVYNM |
| 4755 |
PF03112 (DUF244) |
 Estimated TM-score=0.51; very hard target. |
>Uncharacterized protein family (ORF7) DUF (Length=161) LEIINLKKDIYSNYREEYLMAHNFNEDTFIKLVEDLVERSDFYSSGVEFDWAREFIEYVDCADLEIKDNQSAENLAYDLM EIDSLQKELNRIQNENKKREKPIKDRLKMLIYNITNTYPLIEQLNYKFGEFVFTLDPKKRAISDRLKGLLPTSGTVFFPS N |
| 4756 |
PF03092 (BT1) |
 Estimated TM-score=0.51; easy target. |
>BT1 family (Length=576) RGVADRILTGQTYAMMIDRYGIDVARYQRLSTIATMGWSIKAFTAMLCDGFAFLGYTKRWYMFISCVGGGAFALIYGLLP AKEASADVAAAFIFLSTWGKANVDILSEGHYSRLMRQNPKPGPSMVSWIWFWIMVGAIIATVMNGPLADAGKPQISIFVS AALQLITCVFYLFNWYGEKKNRVLRSEDALFILEETRKERDRFRLPAHDEPVTGVPQHGGAAKGKRSPQRSHSDEDVEGA VRDALNDGQRDNGELVQDVYDDAYDDGEEVAEGEVYYGKPPVPCLFGLFEANTEVISKNWKIFVYSVVMTCAVIAMLCCN ILADTLGLLIACVVVSTICCATSFWALPLVIAKANVFCYLQMAVYIRATSPLYAFYLNSNECQGDYPNFTYTFYNTVAGV IGNLAGLVGVTLFNFLFAKHNYQVTFIVTTIMQVLAALFDIIMVKRWNKRIGIPDHAMYIWGDAVVAQIVYMLGFMPLVV MLSRLCPRGSESVVYALMAGFANLGQTTSSSLAAIIMEYGWPIFSDNDPCNYDNLPLLLFVTSVCTPLLVIPLSYLLPMA RIIDDVDIDGKVVRQK |
| 4757 |
PF03004 (Transposase_24) |
 Estimated TM-score=0.51; hard target. |
>Plant transposase (Ptta/En/Spm family) (Length=149) DSHIWKAMCEYWDTEEAIARSKIYSKARLSDRNGLGPHIHLSGPKSYQNIKHELEKELGRRVTVGEVFIKTHTKPDGTYV DKKAEQIVKNYEKKLQEKLSEMEAETSNVSDGESRPRELTIEDHTVIFLEVTETDTRGNYYGVGSLKDN |
| 4758 |
PF02713 (DUF220) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function DUF220 (Length=75) SGSIPVHLTFNESRKDFSTLYMIPKKNVMFMKTFYGKWQIEPWYVDNMRFCKPRLPKNREEYRQCTGGKGLIGSR |
| 4759 |
PF02701 (zf-Dof) |
 Estimated TM-score=0.51; hard target. |
>Dof domain, zinc finger (Length=57) QALKCPRCDSLNTKFCYYNNYNLSQPRHFCKNCRRYWTKGGVLRNVPVGGGCRKSKR |
| 4760 |
PF02496 (ABA_WDS) |
 Estimated TM-score=0.51; hard target. |
>ABA/WDS induced protein (Length=78) DYKEDEKHRKHLERFGKLGVDVVGAYAMHDEYGEQKDLDHAHIHKVKAEIGATPTLGVEGFTFHEYHENKQAKQEDEE |
| 4761 |
PF02468 (PsbN) |
 Estimated TM-score=0.51; easy target. |
>Photosystem II reaction centre N protein (psbN) (Length=43) MEPATVLSISIAVMVVAITGFSVYTAFGPPSKELADPFEDHED |
| 4762 |
PF02444 (HEV_ORF1) |
 Estimated TM-score=0.51; very hard target. |
>Hepatitis E virus ORF-2 (Putative capsid protein) (Length=114) MGSRPCALGLFCCCSSCFCLCCPRHRPVSRLAAAVGGAAAVPAVVSGVTGLILSPSQSPIFIQPTPSPPMSPLRPGLDLV FANPPDHSAPLGVTRPSAPPLPHVVDLPQLGPRR |
| 4763 |
PF02194 (PXA) |
 Estimated TM-score=0.51; very hard target. |
>PXA domain (Length=184) SAKVSAALNQVLSYIIRDFVNSWYLSISKNPAFSNEVDKALRHALLGIKDRLTSLDLAETLTTRLVPILTAHFRDFSDAE RSVRGKKLNRSVTESEELDLAIASKYRDGKLHPAATLSFSDTKTAQQDYLRKMVFKILPKVLPENLLASRAVATIIREIV ACAVLFPVMQMLSDPDTWNQLMEN |
| 4764 |
PF01524 (Gemini_V2) |
 Estimated TM-score=0.51; hard target. |
>Geminivirus V2 protein (Length=78) MWDPLLNEFPETVHGFRCMLAIKYLQLVENTYSPDTLGYDLIRDLISVIRARNYVEATSRYNHFHARLEGTPPSELRQ |
| 4765 |
PF18871 (HEPN_Toprim_N) |
 Estimated TM-score=0.50; very hard target. |
>HEPN/Toprim N-terminal domain 1 (Length=230) MGTEIQLKLGGLGLTYAKNHRGPDHGALFQPADRQRIRSDQVDYEYCEEEGEDPSPMEWAFARKLKHVVPRLQLLGFTID LAREAYSNWVVDWKEERESLQDDDDPPIPEPLSFDEFCAFATRQSILDLDDTFLSGDEEWRDRRKPKGRFASDMAIIERI PIGFDHDDMAWSERSYFGSRVNILHPYLMLQILAGNAANLETDVTWQYGPLVENGYTAAAAFTPSARRTE |
| 4766 |
PF18857 (LPD38) |
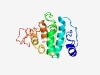 Estimated TM-score=0.50; very hard target. |
>Large polyvalent protein associated domain 38 (Length=176) DPRYQELPQWQKDLFWIIPAKDTLIRIPKPFEVGILFGTSVERMLDWMYKKDPNAFKGYGKTVWDAMVPGWMPTALLPIV EWTSNYSYFMERNIVPPSQEKLPPKLQYGPNTSAIGKWIGEHAITPWTPQGVSPAKVDNTIRGYTGGLGGLAMTLGDLVA GEFDKRPSLKWTEYPG |
| 4767 |
PF18839 (LPD24) |
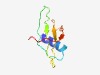 Estimated TM-score=0.50; hard target. |
>Large polyvalent protein associated domain 24 (Length=70) EVMEQVRIYFKENLPKYTVLKIRKKSYHPDDSHLYMAAAKKDDGTYAVWTCWNQKLKSLNHGHYGLQSKE |
| 4768 |
PF18812 (PBECR3) |
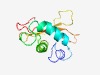 Estimated TM-score=0.50; hard target. |
>phage-Barnase-EndoU-ColicinE5/D-RelE like nuclease3 (Length=118) ASNKELELLKDANFKHPENIRASLDHDAITHILKRHGVNSVNVKNGESPITYEDIANYRYIVNNADAILRTLDNEDKEVI SAFKQINGYAVVVEQALNKKNELVLKTMYKSNGDYKDN |
| 4769 |
PF18303 (Saf_2TM) |
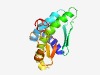 Estimated TM-score=0.50; very hard target. |
>SAVED-fused 2TM effector domain (Length=153) WNTLDFVVRSLVSWFVRARHAESWIFRGAVAVLVAIHGANWVFKYSGTIGGESGSIEFGTAGAVPDSILWIITVVCIALM LGSAMWAWIRYANEQKRLARKKVFVIEGRGLRDDDGSSLKSAVPESITGTRIDYMLDLRQRKDGVIVEPEDLL |
| 4770 |
PF18296 (MID_MedPIWI) |
 Estimated TM-score=0.50; hard target. |
>MID domain of medPIWI (Length=250) RDVAYIVVCPENEALLEGAKTFFRDLSAVYEMCRLGQHKPICKVLRDGIMRVGKTVAQKLAEELVSEWFNQPWGSEESDN HSRLKLYAQVCRHHLGFSGSVGAQNPSAGASSAERTQGNIGCGGDTEPGQSSSQPSQDGQESVTERERIGIPTEPDSADS QAYPPAVVIYMVDPFTYTAEEDSTSGNFWLLSLMRCYTEMLDNLPEHMRNSFILQIVPCQYMLQTMKDEQVFYIQYLKSM AFSVYCQCRR |
| 4771 |
PF18175 (HU-CCDC81_bac_2) |
 Estimated TM-score=0.50; easy target. |
>CCDC81-like prokaryotic HU domain 2 (Length=70) SLLAQSYIEAYDISYPEALQRIEDEVNELTQTLQNEGRYELNNIGILELNEDGNYVFTPCEAGILTPSLY |
| 4772 |
PF17694 (DUF5540) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5540) (Length=78) TASFQRTKEREEDRHSSDLSTKNNDTPRFCSAGVGKRMQPSPSAGKGGKCCTVHGLTRRIHNAQANLHSPILSAACAD |
| 4773 |
PF17690 (DUF5537) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5537) (Length=160) MSVLSPEIKCETSKFTRSSFGSCLIFESSWKKAVLETQKIKKEYTTAFGLEELKECIKMPYLPGLQSCQKSVSSTPLEVP KRLPRADAEVSAVRLKKTKETCSVAPLWEKSKGSGFSDPLTGAPSQYLERLSKIAILEYDTIRQETTTKSKKSKKRDLRD |
| 4774 |
PF17640 (UL17) |
 Estimated TM-score=0.50; very hard target. |
>Uncharacterized UL17 (Length=102) MDHALFTHFVGRPRHCRLEMLILDEQVSKRSWDTTVYHRRRKHLPRRRAPCGPQRPAEIPKRRKKAAVLLFWHDLCWLFR RLFFPREDSEPLMSDPARSPEE |
| 4775 |
PF17600 (DUF5496) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5496) (Length=87) MILRFKDTSGVVLFTLPNPSELEVPGPEQPITIYGKKYYTHKMTREYFDNKISTVKTSSDCYYDITVLTEKQYDELFQRG PSMPGSE |
| 4776 |
PF17588 (DUF5486) |
 Estimated TM-score=0.50; hard target. |
>Family of unknown function (DUF5486) (Length=53) TSEIDYNKIRSSREEMMRRFKEAHDKAKAEGTITYKRIKFKSSNEPLYGVLCG |
| 4777 |
PF17584 (ComS) |
 Estimated TM-score=0.50; hard target. |
>Bacillus competence protein S (Length=44) MNRSGKHLISSIILYPRPSGECISSISLDKQTQATTSPLYFCWR |
| 4778 |
PF17522 (DUF5446) |
 Estimated TM-score=0.50; easy target. |
>Family of unknown function (DUF5446) (Length=72) MSGYVCKTDILRMLSDIEDELTRAEQSLNHSVNALEAEMRETENDRLSVLEEEIIDLHICINELYQSPEHRY |
| 4779 |
PF17508 (MccV) |
 Estimated TM-score=0.50; very hard target. |
>Microcin V bacteriocin (Length=103) MRELNEHEIKCAAGGQDYGREIAQGAGAAAGTFVAGGGGAIVGPIVAGMVYDWASNKTPDSALSPSGLGSTLKKKPEGLP SEAWNYAEGRMCNWSPNNLNDVC |
| 4780 |
PF17504 (DUF5436) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5436) (Length=79) MRRCIHIKEREIHMTDIVDRNVTFILIVVHKYVRYKPHTIANDAHNLVHLAHLIHFIIYFFIIRNVQKKKKTKTEQNIF |
| 4781 |
PF17471 (GP63) |
 Estimated TM-score=0.50; hard target. |
>Gene product 63 (Length=73) MPRKVSPLRQQVLAALIKTRPTAWTQKKVDLRSENPRKPELIEVVHDGSELKYPLARNVSEASVEQAAKRWLP |
| 4782 |
PF17453 (Sigma_M_inh) |
 Estimated TM-score=0.50; hard target. |
>Sigma-M inhibitor (Length=96) MELVRIFKEHNVFGWISVGTAILSLLLLNLAIISNVTFYSYQMLPFAMAAVPFGVIELFIKRGRTGPGLLGVILNLFVII CVYTIVSVDTNLQFGF |
| 4783 |
PF17397 (DUF5404) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5404) (Length=144) MSEEQEPKTSEAEYGTMDFPEFENEEEWLFKVLGIKPRPSSDLDDADKQEDEPLGHTEFLRLQDILQEDKVSSTNDSDTC QAGYTEENDEASHSDSDIDDNVNVIIGDIKANSSMYMEMFTNMNSQADQDLKLTESDNAMYPTD |
| 4784 |
PF17394 (KleE) |
 Estimated TM-score=0.50; hard target. |
>Uncharacterized KleE stable inheritance protein (Length=108) MSNIIKFPKEIEQPAAPAPVEPAAAPAAPNKAPGAGLLAGLVKFVWVATVLVWPVLKWVLSIITFFQFVRMLYHWNTPGV YAGWSFLAYFAVLTAITYFVSIYKPKGL |
| 4785 |
PF17357 (FIT1_2) |
 Estimated TM-score=0.50; hard target. |
>Facilitor Of iron transport 1 and 2 (Length=86) STIKTIVATNGDHVYTKVVTDSAAPVYTESTTVTTTITNNGASYVKTLTIAPQTTIGSSTTSKITITNSDNGATYTKTNV VAISSS |
| 4786 |
PF17328 (DUF5366) |
 Estimated TM-score=0.50; hard target. |
>Family of unknown function (DUF5366) (Length=157) TYIVNQLKELGLYEGMREFFSDSGIKLTLLFLAMLFFFMVFSALKLIADTINELSMLFFSKDEEGNELKKIRGGSWIFLV ASLTSLAFISYPLGIVLLLCLSCFVYFVYFVYKVNESLSGAGLVGFIFFQVIFWFSFLLAVIYACLKLYNSFMASLP |
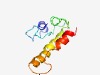
| 4787 |
PF17318 (DUF5361) |
 Estimated TM-score=0.50; hard target. |
>Family of unknown function (DUF5361) (Length=87) IICDLAETYNIYDYRQLPVSKVAVFVCGLRSDSRIMMKISDSKLPLDTLLLAGVADRLSLMLWQNTKDGQKGKNQPVSIV DQLTKQQ |
| 4788 |
PF17311 (DUF5358) |
 Estimated TM-score=0.50; very hard target. |
>Family of unknown function (DUF5358) (Length=162) PAEFAGADYQLSDKNAKQWAIASKQAEQCIYPNLTRIQQQHFAKEDSYIHSQYIFFYPLEKIIGEDYVKIIQNDEKSMNY ATYQFKKFRTEIANVEPLENKNCLILRTQARDDLDVVKGQYKNGMVDNSKNEDGTQKNTDGVATNQNKFFFDIIKWGSAL LL |
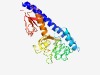
| 4789 |
PF17155 (GAPES1) |
 Estimated TM-score=0.50; easy target. |
>Gammaproteobacterial periplasmic sensor domain (Length=274) SDRAMNAYMRYIMERADSSFLYDKYQNQSIAAHLMRTFEAPGDPVTAEKRRAFCDAFEAINGTHGVNLTRHNYPALHGTL QTAATQCTDNLDDALLLPAFDQAVSINRSQDDHSHGLGTLELKFRYYVDLNKHYVYFYDLINSRRFAMHRWTFLQKGTMG INRKDIDKLFTGRTVISSIYMDDITQENVMSFLTPVYLAGSLKGIVMVDVNQDNLKNIFYTQDRPLVWRYLNVTLKDMDS GKEIIINQSKNNLFQYVNYSHDIPGGLRVSLSLD |
| 4790 |
PF17153 (CHASE9) |
 Estimated TM-score=0.50; easy target. |
>Periplasmic sensor domain, extracellular (Length=116) HKKIAENSAQINLILINNNLNNFFSKVEKEADALKDAIYLLHDEDEIKRAVLHRMKKIDAVNLVGIMLNNGKYLSFIRTE NGEVKFLGQSEPGKPFIGDDGDISDNYFNPESRPWN |
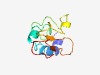
| 4791 |
PF17051 (COA2) |
 Estimated TM-score=0.50; hard target. |
>Cytochrome C oxidase assembly factor 2 (Length=86) MFRTLKASQKRSLVNLMFGTTALFATATVIFPSLLPCPAMKNPYLDTQSDPGYEELPENSRVIVIDQQSPKSSLVASKQN NPPSKS |
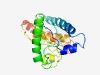
| 4792 |
PF17018 (MICSWaP) |
 Estimated TM-score=0.50; hard target. |
>Spore wall protein (Length=193) VVRFHFYISPYARARYINENRKVEDLVQFVAAELERALNLYLEKSDRPVNITFVPIFVREIPPEIELDKCDTNMLELATT LNNFNLETSDTSIIAVLSCDSRPYEGVFITRGQEVPYITHSLSSQCTTRTLIFLEIEEHKFLAALSTAMIRAAGVDVLNP LLFEEVINGDAGVRYDIRVNNETMDKIKENMCF |
| 4793 |
PF16936 (Holin_9) |
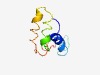 Estimated TM-score=0.50; hard target. |
>Putative holin (Length=77) MLVGNAVGLLAGVACSVLVHARIRPDIVIAMVVGIPSAIGLLVILFSGRRWVTMLGAFILALAPGWFGVLVAIQVAS |
| 4794 |
PF16720 (Albumin_I_a) |
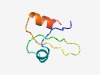 Estimated TM-score=0.50; hard target. |
>Albumin I chain a (Length=48) EHPKLCQSHDECMKKGSGKFCARYPNPESEYGWCIDSDSEALKGFLKM |
| 4795 |
PF16606 (zf-C2H2_assoc) |
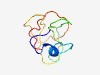 Estimated TM-score=0.50; very hard target. |
>Unstructured conserved, between two C2H2-type zinc-fingers (Length=77) RRGMDEALHQSVDERRGSASDPESQSISRSTTPGSSNVTEESGAGGGQSQTGSIQDDSPHPSSPSSDVGEDSGKVPV |
| 4796 |
PF16498 (SWIRM-assoc_3) |
 Estimated TM-score=0.50; hard target. |
>SWIRM-associated domain at the C-terminal (Length=66) VDPRVASAAAKSALEEFSKMKEEVPTALVEAHVRKVEEAAKVTGKADPAFGLESSGIAGTTSDEPE |
| 4797 |
PF16481 (DUF5058) |
 Estimated TM-score=0.50; hard target. |
>Domain of unknown function (DUF5058) (Length=218) NHPFMWVAAGLAVAVVVVQAVIFFTKSYKAGKEIGLTDKQMKSAVKSSAISSIGPSLVILIGMVSLIVVMGGPISWMRLS FIGSVTYELMAAGFGADAMGVTLGKAGMNNVVYTNAVWTMVLGSLGWLIFTFLFTHKMDKLNNVLSNGKKAMVPIISAGA MLGSFAYFNADRILKFDNGSVACISGLIIMAVLTYLNNEKGVKWLKDWALSIAMFGGM |
| 4798 |
PF16311 (TMEM100) |
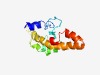 Estimated TM-score=0.50; very hard target. |
>Transmembrane protein 100 (Length=133) MTDEPIKEILGAPKHPEPVTMEKRNNNDCVVTTVPLVSECQLTAATGGAELSCYRCTIPFGVVILIAGVVVTAVAYSFNS HGSIISVFGLVLLSSGLLLLASSALCWKIRQQHKKAKRRESQTALVANQRSLF |
| 4799 |
PF16208 (Keratin_2_head) |
 Estimated TM-score=0.50; very hard target. |
>Keratin type II head (Length=154) GFSSGSAIVGGSSGARSSFSSVSVSRVGGGRAGGGGGFGAGGGFGSRSLYNLGGSKRISVGGGLRSGASAGYGFGGGSGF GLGYGGGAGAALGLGAGGGGGYGLGGFGMGGPGFGGRGGPGFPVCPPGGIHEVTVNQSLLAPLKLDIDPEIQKV |
| 4800 |
PF16166 (TIC20) |
 Estimated TM-score=0.50; hard target. |
>Chloroplast import apparatus Tic20-like (Length=175) RRHRRPIEPARVAKDDFFKIKLPKIAERPEWWWRTLACVPYLISLQISDVGFYVQPFLEKHDAIGDMIYFIPGAINRWPT WFFMVYCYLGYMWVVKNKELPHYLRFHMMMGMLLETALQVIWCTSNFFPLIHFKGRFGMYYWMAIGFTYICLLLECIRCA LAGVYAQIPFMTDAA |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218