

Go to page (83 pages in total): [First page] << < 39 40 41 42 43 44 45 46 47 48 49 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4301 |
PF04136 (Sec34) |
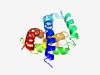 Estimated TM-score=0.53; hard target. |
>Sec34-like family (Length=146) DVLSVCQEQCDSILSDVNAALERLDSLQKQYLFVSTKTGTLHEACEQLLKEQSELVDLAESIQQKLSYFNELENINTKLN SPTLSVNSEGFIPMLSKLDECIEYVSSHPHFKDYPVYLTKFRQCLSRAMLLIKSHTVSSLQNLTAQ |
| 4302 |
PF04001 (Vhr1) |
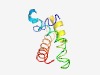 Estimated TM-score=0.53; hard target. |
>Transcription factor Vhr1 (Length=88) SGTTHRIREQLNFSDDKKWKQFSSRRLELIDKFNLSERKASEQDENIRQIANILRTEFGYPTTTSSEFEKLVTAAVQSVR RNRKRSTK |
| 4303 |
PF03929 (PepSY_TM) |
 Estimated TM-score=0.53; hard target. |
>PepSY-associated TM region (Length=301) NIHLWLSLPLGILISIICLSGAALVFEQDITQALQRSIYRVTPPQKGAEALSPEALIECIQKQASDSLHLTALQLSHNPE ETAFASFRETGKKRLSINPYTGKALGWTETYPFFQTMRKLHRWLMDPPPSKGEKSVGKVVVGITTLLMVFILISGLVIWI PRTRKALKNRLCVSCTKGWRRFWYDTHVSIGFYSTIFLLVMALTGLTWSFGWYRTAAYSLFGENPSASVHRVDKGKEKPR KDEVRITWKDAQPSAPQSMKGWFYAFHTGSWGGMFTKIVYFLAALSGGVLPLSGYYLWWKR |
| 4304 |
PF03824 (NicO) |
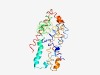 Estimated TM-score=0.53; easy target. |
>High-affinity nickel-transport protein (Length=300) LWLLVGLSFAYGIFHAAGPGHGKAVISSYMVANEVALKRGILLSFVSALLQGLTAVAVMLLAYFVLRGTAISMTDAAWFM EIMSFVFVTLFGAWLLWRKLGPSILRLFGAGPAYSLSAAHAGHSHAGHSHAAHSAHAHSHAHGDHDHAHDHHAHDHDHGV HDHDHGAHAHHDHAAGEVCETCGHSHAPDPALLSGDRFDWRTAWSAVAAVGIRPCSGALIVLSFALLNGLWLGGLLSVLA MSLGTAITVSALATIAVTAKNWAVYFAGDGRMGNRIHSIVEIGGAAFVFLFGLLLLSASL |
| 4305 |
PF03766 (Remorin_N) |
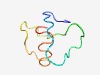 Estimated TM-score=0.53; hard target. |
>Remorin, N-terminal region (Length=52) KDVAEEKASVPPPAEEKPDDSKALAIVEKVEDPPAEKSSGGSTERDAVLARL |
| 4306 |
PF03672 (UPF0154) |
 Estimated TM-score=0.53; hard target. |
>Uncharacterised protein family (UPF0154) (Length=60) IVVALLGGAVAGFFIARNYFKKYLEKNPPINEQMIRAMMSQMGRKPSEKQVRQVMASMKQ |
| 4307 |
PF03608 (EII-GUT) |
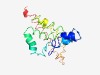 Estimated TM-score=0.53; hard target. |
>PTS system enzyme II sorbitol-specific factor (Length=167) LAKLAEGFIGLFEEGGKTFVGLVTGILPTLIVLITFVNAIIKIIGEDKVQRVAQKATKNAITRYTILPLISVFFLTNPMC YTFGKFLDEKYKPAYYDAAVSFVHPITGLFPHANPAELFVYLGVAEGIKKLGLPLGQLAVRYFIVGLVIIFIRGLVTEFM TIRMMKK |
| 4308 |
PF03369 (Herpes_UL3) |
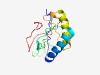 Estimated TM-score=0.53; hard target. |
>Herpesvirus UL3 protein (Length=135) TFDTLFMVSSIDELGRRQLTDTIRKDLRLSLAKFSIACTKTSSFSGTAARQRKRGAPPQRTCVPRSNKSLQMFVLCKRAN AAQVREQLRAVIRSRKPRKYYTRSSDGRLCPAVPVFVHEFVSSEPMRLHRDNVML |
| 4309 |
PF03312 (DUF272) |
 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF272) (Length=125) YLWNLDSKTEGLFVSKNHSLAQGHHFEGIFKKNPDGRWTCQKYENPIEGLLTGGINPNNKIYFTVKIDKYQPAKGNTKYG HATAKYIGDVLEGDSENTKLSAECNGKMVNIQRRGIGDKDFVWMS |
| 4310 |
PF03248 (Rer1) |
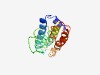 Estimated TM-score=0.53; hard target. |
>Rer1 family (Length=165) TQQRWQHYLDEVTPYTTNRWIATGVLVVIYAIRVLVAQGWYIVTYALGIYLLNLFLAFLQPKFDPSLEMDIQESEVEEGP ALPTKNDEEFRPFIRRLPEFKFWYSATRAIVIAFFCTLFSIFDVPVFWPILLVYFCILFYITMKRQIKHMIKYRYVPFNV GKKSY |
| 4311 |
PF03086 (DUF240) |
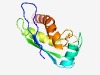 Estimated TM-score=0.53; hard target. |
>Domain of unknown function (DUF240) (Length=121) MKFHTVYVGKNTKIDLDFALQAQTNNFSSLEELRESFTNSGQTLSTQLFWKPVIDKLITDEGNDLTTIARTAIGENLFDL KVNLTDSVIDGTVLTKARKSFEERILNPFIEQRKEAKRIHD |
| 4312 |
PF03050 (DDE_Tnp_IS66) |
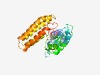 Estimated TM-score=0.53; hard target. |
>Transposase IS66 family (Length=288) GRAGPKLLAGILCDKFLQHLPLNRQSAAFAREGIDLDTSTLADWVGACTATLAPLTALIRAHVLAARRLHADDTTVPVLA RGRTVTGRLWNYVRDDAPFGGLAPPAVWFRYSRDRKGEHPADHLTGWTGILQSDAYGGYTHLAKPGRQPAPVVPAGCWAH GRRGLFKIAEKDKAPLAIEAVRRIDAIFDAERAINGSSAAHRLEIRKESIAPLVEELLDWMRDTCRRMSSKNPIAQAMNY ILRRGDTFTVFLEDGRICLTNNAAERAIRGIALGRKAWLFAGSDRGGE |
| 4313 |
PF02711 (Pap_E4) |
 Estimated TM-score=0.53; very hard target. |
>E4 protein (Length=89) LCAVPVTDRYPLLNLLPNYQTPPRPIPPQQPHAPKKQSRRRLESDLDSVQSQSPLSPTECPWTILTTHSTVTVQATTPDG TSVVVTLRL |
| 4314 |
PF02554 (CstA) |
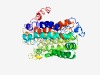 Estimated TM-score=0.53; easy target. |
>Carbon starvation protein CstA (Length=377) VNAIWLIVAAVCVYAIAYRFYSKFIAKKVFQLDDNRKTPAEIFNDGKDYVPTNKWVLFGHHFAAIAGAGPLVGPILAAQM GYLPGTLWIIVGVVLAGAVQDFIILFASMRRNGKSLGEMIKEEIGPVTGFIAMLGILGIMIILLAVLALVVVKALVGSPW GMFTIAATIPIAIFMGVYMRYMRPGRVGEASFIGFVLLMLSIVAGQYVAENPTLANMFTFKGETIALLMIGYGFVASVLP VWLLLAPRDYLSTFLKIGTIVGLALGILFVAPDLQMPAITKFVDGTGPVFAGNLFPFLFITIACGAVSGFHSLVSSGTTP KMIERESHARPIGYGAMLMESFVAVMALIAACVLTPGTYFAINSPAAAIGQDVVQAA |
| 4315 |
PF02349 (MSG) |
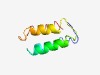 Estimated TM-score=0.53; hard target. |
>Major surface glycoprotein (Length=75) NLTDEKCRKYEEKCLLLEEADPSNLEEKCVKLRDRCYGRRRQGVTKEILFRALEGKVNDTDECKKRMKEICQGLS |
| 4316 |
PF02124 (Marek_A) |
 Estimated TM-score=0.53; easy target. |
>Marek's disease glycoprotein A (Length=204) RAEFVWFEDGRRVFDPAQIHTQTQENPDGFSTVSTVTSAAVGXQGPPRTFTCQLTWHRDSVSFSRRNASGTASVLPRPTI TMEFTGDHAVCTAGCVPEGVTFAWFLGDDSSPAEKVAVASQTSCGRPGTATIRSTLPVSYEQTEYICRLAGYPDGIPVLE HHGSHQPPPRDPTERQVIRAVEGAGIGVAVLVAVVLAGTAVVYL |
| 4317 |
PF02010 (REJ) |
 Estimated TM-score=0.53; hard target. |
>REJ domain (Length=463) ADMRVNCPGPKSTKVSWNIYKVSNLSDKPDWSKPFNPTGIEGRHFIRLKIPCSSLDTGLYLFNFTVQLISPTFPQMIERS DAVLIEVGSDLWGAVIMGGHFRSVGFSDRWILKGSVLSNGEMVSLSQGLSYTWYCTKQERDYTSMALSTVGKCYPDQVDL KWTTSSNSIQMVLPETLQANNTYHFLLLVQNGSRITQAQQTVHVRAHGALVLNIACTENCGTSVIPTERFSLSGKCGNCR EIKSWYYWSLRSAQSHEIKFDWSSRTTTGRSSSYLHFKAFALVSMAEEFYILSLKVVTKNGQSSSCEYFFYINAPPQIGN CVLHPKEGIAFLTKFIIQCHGFEDRNQPLAYKVIAASDQMTISSISSIQNNTLGIIIYVGHESKTPWSFLPPGRPSQTFA LPIYVQVFDALGASSQVTLQATVLDQRQPPTNAVLHQQLYDLMNGSSALMTKLLITKDYLSIG |
| 4318 |
PF01508 (Paramecium_SA) |
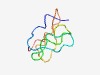 Estimated TM-score=0.53; easy target. |
>Paramecium surface antigen domain (Length=59) TLDSECQAISNRCITDGTHCVEIDACSTYKKQLPCVKNTAGSLCYWDTTNNTCVDANTC |
| 4319 |
PF01107 (MP) |
 Estimated TM-score=0.53; hard target. |
>Viral movement protein (MP) (Length=199) LKNISKTQLTLEKEKIFKMPNVLSQVMKKAFSRKNEILYCVSTKELSVDIHDATGKVYLPLITKEEINKRLSSLKPEVRK TMSMVHLGAVKILLKAQFRNGIDTPIKIALIDDRINSRRDCLLGAAKGNLAYGKFMFTVYPKFGISLNTQRLNQTLSLIH DFENKNLMNKGDKVMTITYVVGYALTNSHHSIDYQSNAT |
| 4320 |
PF18850 (LPD30) |
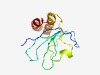 Estimated TM-score=0.52; hard target. |
>Large polyvalent protein associated domain 30 (Length=119) DMEISIGTRINCVLHWAGPGTVYAIHGEQRPGSVRVTCGIAHSGGSASFDVVFDNGRMSAKVPECIIRGVQWSVLPGIAS AEQIAEALAHAACVKAEKANAAEQAAQRFAAEVERLRIV |
| 4321 |
PF18169 (SLATT_6) |
 Estimated TM-score=0.52; hard target. |
>SMODS and SLOG-associating 2TM effector domain 6 (Length=177) MNKAELLRYIAETGYNVGFGAKKNFATYDLIEKMPGWIGFISTAVGILGLIIDALSTKLVSAIFVIIGICGLYISFYDKD KNRYENAGKELTKLFNELKSLYFDVKCLSENSDVSAYQVRIQDIETRYFSLCISKQILFSNWYAHYKFFWEHQISWIDEQ LKFRFIRDKIPLSLIFI |
| 4322 |
PF17748 (VID27_N) |
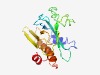 Estimated TM-score=0.52; very hard target. |
>VID27 N-terminal region (Length=176) MFMLRNVSKYLFGDSSKESIVEVPQGQLYLVRPLSVKGYSELIFKDAAASIRRTGQDFQYQLVIQRAYEEGERELLGEDE EEDGATEGLGAEKDERTFLLDEALRFRTDVRQTGEKVFAWKDLSGDEGDLWEFVCDSSVQPPTVATFEVVAVQCQYERKY RRSHQQATEQDLQQFS |
| 4323 |
PF17697 (DUF5543) |
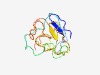 Estimated TM-score=0.52; very hard target. |
>Family of unknown function (DUF5543) (Length=118) MYCQDSNICAVFAVQGGKVGRKHGIKRGRRPSIRSPAQRARGPWIHESKHPAFAKQQINLEMPNSRATTELAWVCRSLTL STAPLSPPPSLVHCEDCSCLPGCHSGDLYNLAPAERTC |
| 4324 |
PF17646 (Zemlya) |
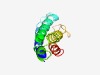 Estimated TM-score=0.52; hard target. |
>Closterovirus 1a polyprotein central region (Length=105) LPFEVAVSVIQKFIFDIPSVLTGKISPMQCVLRCATNLVFELNLNLAIAKIIGPSDTLKKDMFNRTVSSLISSAFLDRFT FSPETFIRLSTVVPMVVRKLLVSFF |
| 4325 |
PF17577 (ETM) |
 Estimated TM-score=0.52; hard target. |
>ECORI-T site (Length=109) MDVLHGICVNINKFYKCRRIVIEYDNCSTSFTFDCAHDNRKVNCLEIVGLRRNEYVCMGKIINGDKIICVHENSVNGKII VPVEDTFDFGLFTLKNKITDAVIKLNVYI |
| 4326 |
PF17498 (DUF5433) |
 Estimated TM-score=0.52; hard target. |
>Family of unknown function (DUF5433) (Length=67) MFASCFNVNGTNFILALCIIFVPTIKLYIQIIMRCVYANGINKRSTPFSHNSNPKFFKVYLNERSCV |
| 4327 |
PF17467 (E7R) |
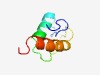 Estimated TM-score=0.52; hard target. |
>Viral Protein E7 (Length=60) MISMGDIITYNGCKDNKWMLEQLSTLNFNNLHVWNSCSIGNVTRIFYTFFSYLMKDKLDI |
| 4328 |
PF17442 (U62_UL91) |
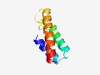 Estimated TM-score=0.52; hard target. |
>Functional domain of U62 and UL91 proteins (Length=65) MNSALNEIKDDFENCETKNDLFKIIDKISKNCNFIVEQVESLPRRVDSAAILFDNLAVEIFNDVI |
| 4329 |
PF17412 (VraX) |
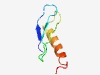 Estimated TM-score=0.52; hard target. |
>Family of unknown function (Length=55) MIIYRRNIENGTPVYEIITKTFKTITIKCDETFNKYEIYQLLSLLENDVDNMPTS |
| 4330 |
PF17411 (SmaI) |
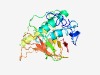 Estimated TM-score=0.52; hard target. |
>Type II site-specific deoxyribonuclease (Length=239) IVKAIPLLTEGQLYWLDRVIQVFNSSQQFKILHSDLFDQNTLENFGDALRIHHSFSMEPFSKDKFEYILEKVLKMSGHQA SLAPKGNRGHDISINGCPISLKTQANKGIREDKIWISKFMELGKGKWGDDEKDLIGLRKMFLGHLKNYKRILILRALEKG PQWHYELVEIPINLMKSAQKGELELKKKSKQFPKPGYCHVRTTKGENIYQLYFDAGSERKLQVKNLLKSYCKVHATWEF |
| 4331 |
PF17388 (GP24_25) |
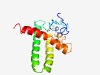 Estimated TM-score=0.52; hard target. |
>Tail assembly gene products 24 & 25 (Length=130) MTNVFTIDAFREEVKKKYAPVLIGLSDDVTVELKPLLKLGQKAREAVVEVFKEFADIPDLEEDDDDELVDEYSLQVCDII AKAFRLIATKPKKLIAALDEEPDPRIRAELYAAVLNTWKRETQLGEAAPS |
| 4332 |
PF17362 (pXO2-34) |
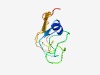 Estimated TM-score=0.52; very hard target. |
>Family of unknown function (Length=78) RICSKKAIRSKALTIRLFDGSTWANLGNGQFINQETKKELPDYKIYPQIKTAVSSGGLIFAKRRNTKQNLQYVERPMK |
| 4333 |
PF17336 (DUF5368) |
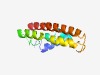 Estimated TM-score=0.52; hard target. |
>Family of unknown function (DUF5368) (Length=111) MKELTFETLIAVFEEIFGRGLFWVMVAVAVVVTLAYLYVLIRDRSVSWRKFLWAQLSMPVGAVLAVLFVLRMTRSGVSDM GGPIDVIVLLGVAAAGAVGLAILVYTVESLI |
| 4334 |
PF17325 (SPG4) |
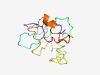 Estimated TM-score=0.52; very hard target. |
>Stationary phase protein 4 (Length=112) SFWDAFAVYDKKKHADPSVYGGNHNNTGDSKTQVMFSKEYRQPRTHQQENLQSMRRSSIGSQDSSDVEDVKEGRLPAEVE IPKNVDISNMSQGEFLRLYESLRKGEPDNKVN |
| 4335 |
PF17299 (DUF5350) |
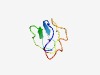 Estimated TM-score=0.52; very hard target. |
>Family of unknown function (DUF5350) (Length=56) MGKTGSIEWVKIKGRKGRVIKVPKSNAQKAHPGPAQRFTSSGHKRRFIRRSPKALV |
| 4336 |
PF17246 (CDC24_OB1) |
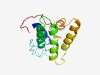 Estimated TM-score=0.52; very hard target. |
>Cell division control protein 24, OB domain 1 (Length=133) FLRFIDYARSMLSPEDENDDDEDFDLNGRIEADIKRPSWNWIASRILKTCTAYSSGVTAAILLSDLSQAWNEQHRDGAPR KRPECINQLKKKRGRAKLPNTVTIDSIYEKNFLSMNSVLEAVIVDAFVLPGTN |
| 4337 |
PF17079 (SOTI) |
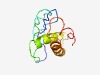 Estimated TM-score=0.52; very hard target. |
>Male-specific protein scotti (Length=100) ENPQVAMLLDAPHEPPIELHHMLEPVNVPQRPRKKRSFLTISKPFHVQPERCALISNGWRAVQCVQPEKRGEYFANYLIK HMNSRNYPNGEGLPNRWGQF |
| 4338 |
PF17027 (Bromo_TP_like) |
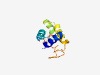 Estimated TM-score=0.52; easy target. |
>Histone-fold protein (Length=119) KILRLAVVEILSQSGFDKTADQALNVLTDILRYYIEHLGCRMRRKGENGIVPELICRFLVDEEYGECEYQIPELLSFLRY QVTVKNYLNDRYSVGSEESILHILRVLPKNAQLRMVMRN |
| 4339 |
PF16954 (HRG) |
 Estimated TM-score=0.52; easy target. |
>Haem-transporter, endosomal/lysosomal, haem-responsive gene (Length=52) LKFFIAVGVLFSILAVTALITFLTLAITQKQSPTDPRSLYLSCVWSFLTLKW |
| 4340 |
PF16931 (Phage_holin_8) |
 Estimated TM-score=0.52; hard target. |
>Putative phage holin (Length=121) MSDPVSGTVAAGAALTGASIYGLLTGTDYGVIFGAFAGAVFYVATAADLTLIRRAAYFVVSYIAGVYGAGLVGSKLASWT EYSDKPLDALGAVILSALTIKILTFASQQDPTQWFQRWRGG |
| 4341 |
PF16663 (MAGI_u1) |
 Estimated TM-score=0.52; very hard target. |
>Unstructured region on MAGI (Length=59) TGDALQDSLPGSQQSTPRRTKSYNEMQNAGIVPTETEDDDEVPEMNSSFTGDSSEPDDH |
| 4342 |
PF16637 (zf-C2H2_assoc3) |
 Estimated TM-score=0.52; hard target. |
>Putative zinc-finger between two C2H2 zinc-fingers on Patz (Length=72) GPSNFCSICNRDGMSDGEKCPHQEQHDNSDSFGEFSDTSDLHSPTKQNENGPFSCDPVVNPKSDRDLEAERK |
| 4343 |
PF16587 (DUF5061) |
 Estimated TM-score=0.52; hard target. |
>17 kDa common-antigen outer membrane protein (Length=104) LLALVACLLLAACGGKQPEMAASPIDDPVVEKPLLDYLSTHDGGSSATLDDPEFGKSIRVFVEGSFLSASGQECRRATLL TRDGQAEVVVACKGDDGTWTMAPR |
| 4344 |
PF16501 (SCAPER_N) |
 Estimated TM-score=0.52; hard target. |
>S phase cyclin A-associated protein in the endoplasmic reticulum (Length=90) GKKNDLRARYWSYLFDNLKRAVDEIYGTCEADESIVECQEVIMMLDQCRRDFSALIERINVQDAFEKADYKDRPTSLAWE VRKSSPGKTL |
| 4345 |
PF16494 (Na_Ca_ex_C) |
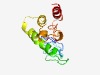 Estimated TM-score=0.52; easy target. |
>C-terminal extension of sodium/calcium exchanger domain (Length=129) RRLLFYKYMGKRYRADKRHGIVVETEGDLTPNKGGMEMIIDGKFPVRGGAVVPAENKELDESRKEVIRILKELKQKYPDK ELDQLVELANYYALLHQQKSRAFYRIQATRMMIGAGNVLKKHAADHARR |
| 4346 |
PF16429 (DUF5026) |
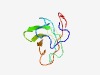 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF5026) (Length=82) LIVDSITKVFDTAQIKKGYLMYAKHRSWPDGRGGFVTAVTDKQITVQYHPGIANVTNHFFLPADEVAAGEWEVRWSADMI EV |
| 4347 |
PF16339 (DUF4969) |
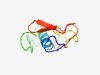 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4969) (Length=79) MKKRGLLLVSVLMLFIFLTVACGSSEDNEESGEISTKEIELTVDDPIIPTDENGEATIEGTVDPKSKLTIDGKAVKKDD |
| 4348 |
PF16306 (DUF4948) |
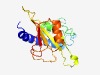 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4948) (Length=171) PPSDEEMIRHFTTHEVAFRKVYEIMAESSEGSFHYPPLSPEEVIILDSMEQSDTSHETNDEQDIPVYGLLKPERILLDSL LSEIGCGFILVDRREWGTADSVYVSLVMPYYSHGIVDAGTSKSFVYDPGLRSRRNIRITEHGDLNEIYRRTYNDTTLYKP IKGNWYIELDH |
| 4349 |
PF16101 (PRIMA1) |
 Estimated TM-score=0.52; very hard target. |
>Proline-rich membrane anchor 1 (Length=122) LQKSCSKTIAEKGTDSCQEICQCRRPPPLPPPPPPPPPPRLLSLTPPNSTSCPAEEPWWSGLVIIIAVCCATLVFLVVIV IICYKAIKRKPLRKEENGTSGTEYAMTSSQNNKTVDVNNAVV |
| 4350 |
PF16085 (Phage_holin_3_5) |
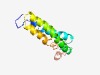 Estimated TM-score=0.52; hard target. |
>Bacteriophage holin Hol, superfamily III (Length=113) MNSEQQALVEMPIWLVIFLSLVGGVSGEMWRADMAGVSGWFIFRQVLLRSGACVVCGLSTIMLLYSAGMSMWSASAIGCL TATAGADVAIGLYKRWVAKRLGVCDVTSRSGEP |
| 4351 |
PF16040 (DUF4792) |
 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4792) (Length=70) NATFNAFVVPGPPEMAEELVPVSMTRELELEDDMKEYWGFYLLKGSSVTVSACVSWPGASLIMIKGYKHL |
| 4352 |
PF16020 (Deltameth_res) |
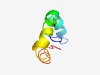 Estimated TM-score=0.52; hard target. |
>Deltamethrin resistance (Length=48) DDLPVPSGSWQARYDANQRRYNGILLFGIAFSVGSIIAVRYSSTLHCH |
| 4353 |
PF16018 (Anillin_N) |
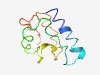 Estimated TM-score=0.52; hard target. |
>Anillin N-terminus (Length=88) VKTRMQKLAEQRRHWDNNSMTDDIPKSSLLPPMPAEEKAASPAKIPSAAASATPLGRRGRLANLAATICSWEDDVNYSSA KQNSVQEQ |
| 4354 |
PF15947 (DUF4755) |
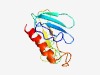 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4755) (Length=128) NYLNENNDGIYLHKFNDCEIRLDTNKQEVFLKNSEGANTYSFEEIKEWSYNITYGGEVAGTGLSSIRENGKTLSQNIADT GLFIRVKDVRKPEWHIRFYPRNDSFNSRKGINDLKAQLNQWMEIFDQN |
| 4355 |
PF15904 (LIP1) |
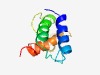 Estimated TM-score=0.52; hard target. |
>LKB1 serine/threonine kinase interacting protein 1 (Length=90) SGQCPLVHSLAKLLRNDGDLVLDGSSTLTLTASSLQQLTRMFEQYLLPRTKQHGFLALPSHPADTASLLQLQFLFDVLQK TVSLKLINPP |
| 4356 |
PF15848 (ODAPH) |
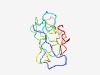 Estimated TM-score=0.52; very hard target. |
>Odontogenesis associated phosphoprotein (Length=108) EEVVNPPGDSQNNANPTDCEIFTLTPPPATRNPVTRIQPITRTPRCPLHFFPPRWPRVYIRFPYRLYPPPKWNHHFQFHP YFCPHSRLPPYYNYFPRRRLWRGSSSEE |
| 4357 |
PF15810 (CCDC117) |
 Estimated TM-score=0.52; very hard target. |
>Coiled-coil domain-containing protein 117 (Length=143) EIEAAQRKLQEIEDRITLEDDEDEDLDVEPAPRRPVLVMSDSLKKGLQQGISDILPHTVAQSVSRSCMELVLWRPPEDPF SQRLKDSLQKQRKQQTACRQTPTPCPSPSPHSPLNPSSPPADTHSPLYSFSVAHNSGEEEMEM |
| 4358 |
PF15751 (FANCA_interact) |
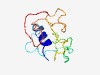 Estimated TM-score=0.52; very hard target. |
>FAAP20 FANCA interaction domain (Length=109) ECARLWAELLRAAREDLNLDGEPPPLPAFPGQEPGRSPERTTPSEVFTVGPETFSWTPFPPALRRGRGRSYRVLRPAGGR PRSPDRAPQGCPAPEPRGTPSAEEQPSVE |
| 4359 |
PF15747 (DUF4687) |
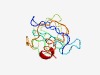 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF4687) (Length=120) MVLNYVSMSAGDQGNRVAYRSSRGDLSPSALALVMVSGDSFLVTRPEAIHPGYTLRQAVRPGIRTENRRATGGAWGPTRF IKGRDLDGRNRSRQSRFSPYPTPGVKLDLLRSVLQQRLIA |
| 4360 |
PF15701 (DUF4668) |
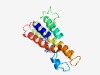 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4668) (Length=162) KKLSRVADIVYIVDTFLIPPLHPLKKQHPKVAKFLKVQLVFDLISLFIFATHQLLLLEDGSFGKHYFKRKTKRCSKFSCS RCNANAHHPKWFKFKHSLLCLGTFCFGVYSLVKINKFFKTDQTVDLNRLLELFFWQLNAILNMKLFAFYGDHLESHSAPL DV |
| 4361 |
PF15613 (WSD) |
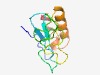 Estimated TM-score=0.52; hard target. |
>Williams-Beuren syndrome DDT (WSD), D-TOX E motif (Length=99) NIFPLGRDRIYRRYWIFPSIPGLFIEEDYSGLTEDMLLPRPSSFQNNVQSQDPQLPKPVHKPNRWCFYSSCEQLDQLIEA LNSRGHRESALKETLLQEK |
| 4362 |
PF15604 (Ntox15) |
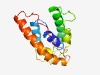 Estimated TM-score=0.52; hard target. |
>Novel toxin 15 (Length=146) QHPAEFARQLSRQYEALSYLTVDEYLKGRNLYNTQKRAPTPPSLELQREDLEKKIVNRLVDDYRFDPDKAEKKAVEAVQA LAALHEPDQSLQGTHQLAAGEIVWGNSSVNSSIGGQHKAGLARLDEKARDAQVAGKGHRYLNVRVV |
| 4363 |
PF15543 (Ntox5) |
 Estimated TM-score=0.52; very hard target. |
>Bacterial toxin 5 (Length=135) MRRRLLAQRASGKESLLDFLLDADGQWQKGSFIARSGRPMRGRYALSDPDRQIVQAGHMLSDWYVKAMSRRDHLMLEDAD LNWMSGFTEASGSVTSKSAVLIEGYPVDIPTARLWEAHGKLPAGTVSAAPVVEPP |
| 4364 |
PF15475 (UPF0444) |
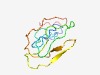 Estimated TM-score=0.52; very hard target. |
>Transmembrane protein C12orf23, UPF0444 (Length=91) KDHPQQQPGMLSRVTGGLFSVTKGAVGATIGGVAWIGGKSLEITKTAVTSVPSMGVGLVKGSVSAVAGGVTAVGSAVASK VPLTGKKKDKS |
| 4365 |
PF15464 (DUF4633) |
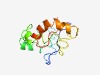 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF4633) (Length=115) MGTGLRSQSLRGPRPSYGKLQEPWGRPQEGQLRRALSLRQGREKSRSQGLERGTEGPDATAQEWVPGSLGDTEQLIQAQR GGSRRWLRRYQQQVRRRWESFVAIFPRVTLSQPAS |
| 4366 |
PF15459 (RRP14) |
 Estimated TM-score=0.52; hard target. |
>60S ribosome biogenesis protein Rrp14 (Length=61) IHQHSLFFDKLVELIPAKFYLPTDDKDKPWFQGLSKAQKASAKKESKENIKKARRDRLDPD |
| 4367 |
PF15257 (DUF4590) |
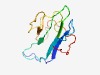 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4590) (Length=107) KDIDLRHEVKVFQQHCEGENICVYKGKLREGEFFQFISRRSRGFPFGLTFYLNGLQVEHLNSCCEFRHQKGSEPKGRDRH FSFVSMDGASPCYKCVLAFGLYKKSIR |
| 4368 |
PF15255 (CAP-ZIP_m) |
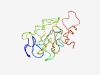 Estimated TM-score=0.52; very hard target. |
>WASH complex subunit CAP-Z interacting, central region (Length=129) PFKTKEPSPRIWKIQANLAINPAALLPTAAPQISGMKSVLPELGVPSSEPERIHSLESVPTLPGSGEAGVSFDLPAQADT LHSANKTRVKVRGKRRPQTRAARQLAAQESSEAEDMSIPRGFVAQLADG |
| 4369 |
PF15224 (SCRG1) |
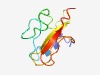 Estimated TM-score=0.52; very hard target. |
>Scrapie-responsive protein 1 (Length=76) PSRRPSCYKRALKDHDCHNIPEGMENLRQIDDGLQDHFWEGKGCEVICYCNLNELLCCPKDIFFGPKISFVIPCNS |
| 4370 |
PF15221 (LEP503) |
 Estimated TM-score=0.52; hard target. |
>Lens epithelial cell protein LEP503 (Length=63) MQPQRPLPQAMPSSPGRNLGDAALGTGTGKRLLGGNMAYGVIQSLKECLYFLLCCWCIKEILD |
| 4371 |
PF15167 (DUF4581) |
 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF4581) (Length=130) KELVDAAEKWCTGNPFDLIFAEDVDERRLDFYAEPGISFYVLCPDNLTGGTDNFHVWSESEDCLPFLQLAQDYISSCGKK TLLEVLDKVFRSFRPLLGLPDIDDDTFDQYHADVEEEPEPDHQQMGVSQQ |
| 4372 |
PF15143 (DUF4575) |
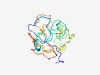 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF4575) (Length=129) RGRGEGLALSPLLPGVPPPPAMGVPRDRGGRGAGSQSTPRGGCLLPPRLGSQQPAGEEGLVLSPRLAGDASPCCDGSPKS QGVKRGWLSVPTSRGVPPPPAIGVLIARGGRGAGSQSLPRGWSFTPLRW |
| 4373 |
PF15108 (TMEM37) |
 Estimated TM-score=0.52; easy target. |
>Voltage-dependent calcium channel gamma-like subunit protein family (Length=183) QAQRLLAQRRPRRPFFESFIRTLIIVCAALAVVLSSVSICDGHWLLAEDRLFGLWHFCTTSNQTRPHCLRDLSQAHVPGL ALGMVLARSVSALAVVAAIFGLELLMVSQVCEDLYSRRKWALGSFLLLISFILSFGGLLSFVILLRNQVTLIGFTLMFWC EFTASFLFFLNAISGLHINSITH |
| 4374 |
PF15084 (DUF4550) |
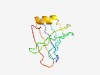 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4550) (Length=94) RFYHMEYFLLPDDIEPRKLDLVLFGPVAKLFLDTESKVVKPWLENDQIWVSWNHSIEINITNEFLIKLREHSIKLRLWDL KEKVCSKARFCKLK |
| 4375 |
PF15062 (ARL6IP6) |
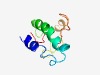 Estimated TM-score=0.52; hard target. |
>Haemopoietic lineage transmembrane helix (Length=86) QQLTGYVQGQWSEALGWSERHQLVSKLSPLLCGLIVATFAYGMVYLDSAVPGVTPPSPFSPRSKQRFRQQKSASLHLGYL CALFSG |
| 4376 |
PF15045 (Clathrin_bdg) |
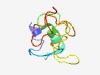 Estimated TM-score=0.52; very hard target. |
>Clathrin-binding box of Aftiphilin, vesicle trafficking (Length=79) SRKLWRALQNTNSTSASRCLWSESRCQENLFLVLGIDAARKNLSGDPGHILECSDLKEPEELGVPTFRLQPCRALIQTK |
| 4377 |
PF14936 (p53-inducible11) |
 Estimated TM-score=0.52; easy target. |
>Tumour protein p53-inducible protein 11 (Length=179) MKKHSQTDLVSRLKTRKILGVGGEDDDGEVHRSKISQVLGNEIKFAVREPLGLRVWQLVSAVMFSGVAIMALAFPDQLYD TVFDEESASSKTPIRLYGGALLSIALIMWNALYTAEKVIIRWTLLTEACYFTVQFLVTTVSLVESSRIATGAVLLLVSRA LFILISIYYYYQVGRRPKK |
| 4378 |
PF14855 (PapJ) |
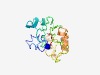 Estimated TM-score=0.52; hard target. |
>Pilus-assembly fibrillin subunit, chaperone (Length=187) MNRATIGFYLAVVLGSCSMNGVSQADELLTRDDFFVADESRHQWVTEHNGRTGALNVKGALVSSPCILDTPEVNFPLQKD KGRYVLNLKVSRCGDGHSDIPERQSTEHGNIVVKQSAVLKTGKDQILLSAWRNGGLSRKNLQHGDNQITYYINEAQYQKI AKTQMPVTAHKTSDSNRGVMHLNIMYE |
| 4379 |
PF14837 (INTS5_N) |
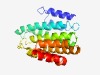 Estimated TM-score=0.52; hard target. |
>Integrator complex subunit 5 N-terminus (Length=214) QELAQEIKAFLSGVDPVHGTKLTLKEHARCAILLLRSLPPARHAVLEHLRAVFDEHVCTHLLERESGEAGLGAAAGRAPG PSLDDVVQEVQHVLSDFVRANPKAWAPVVSAWSIDLMGQLSSKYAGRHGVPHASSLNELLQLWMSCKATRTLMDIYTQCL SSMISTCPDACVDALLDTSVQHSPHFDWVVAHIGSSFPNTIISRVLSCGLKDFC |
| 4380 |
PF14820 (SPRR2) |
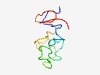 Estimated TM-score=0.52; hard target. |
>Small proline-rich 2 (Length=68) SYQQQQCKQPCQPPPVCPTPKCPEPCPPPKCPEPCPPPKCPQPCPPQQCQQKYPPVTPSPPCQQKYPP |
| 4381 |
PF14774 (FAM177) |
 Estimated TM-score=0.52; very hard target. |
>FAM177 family (Length=117) FETVEIAECKARKVKVPRRIIHFANGETMEEYSTDEEEEEEEEEHVKQVVPVDTSKLTWGPYFWFHMWRVATSTISVCDY MGEKLASLFGITSPKYQYAIDEYYRMKKEEEEEEEEN |
| 4382 |
PF14773 (VIGSSK) |
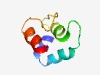 Estimated TM-score=0.52; hard target. |
>Helicase-associated putative binding domain, C-terminal (Length=62) SQLAALLTSENQEKMLESKKAKMPKSDAIQAILTSAGVEYTHDNSEVIGTSKVEEQLSRRAA |
| 4383 |
PF14610 (DUF4448) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF4448) (Length=174) TNKDGTGSFLVCNNADGANAPFCQPSRNSSLYPGTTYYVTWDTSVFNSTNTTVILEGSYINATTGEVASQAFSSDKITAG WSYYAWAVDSKLLQGKSAANITLFLNALDADQNTTAKSYTGPTVLVTKPPTYHQPPPKMPTGAALYIGLPTVLGFCVIMI FGVCLWNRQARKIS |
| 4384 |
PF14491 (DUF4435) |
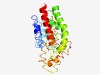 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF4435) (Length=230) AVFCFVEGEDSKYYAAKIESIEPSYKYHFINSKGKEGVLKLFRLITGISEYDLIKTLYFVDRDFDELIDNPHIYETPCYS IENLYTTKNTICRVLQREFQLDETDDDYYTALHAFSTHQSEFHDAMDQINAWIACHRDLSKQGITTRLNLSDLNPSNLVT VSLTGVTQKYSFDSLIEMFPHAAEIDQETLENKLRSFRSIDRQKWYRGKFELFFFKKFIEILSEDSNRRH |
| 4385 |
PF14463 (E1-N) |
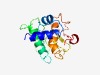 Estimated TM-score=0.52; very hard target. |
>E1 N-terminal domain (Length=150) MNKATQQNAMMLASLLGVGEAEAGERLARTVLITAAPGWKSGWAVEVGELIGRTVQVSHQQEPTDPDLELVIGDVTPRTS ARRVYADLGSEGAAASLEPVAKLAGEPHGLYAAAAACAVSAVVVHAVIDAADLPQARLPMRLDYAQLGVP |
| 4386 |
PF14448 (Nuc_N) |
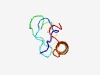 Estimated TM-score=0.52; hard target. |
>Nuclease N terminal (Length=57) ADSVSPIFKGVEILPDGSVVRSGTNYSGKFQEAHDASKASIQSRISNLESGGVKGTG |
| 4387 |
PF14409 (Herpeto_peptide) |
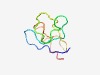 Estimated TM-score=0.52; very hard target. |
>Ribosomally synthesized peptide in Herpetosiphon (Length=56) MEFETTQINETPMIFGLTYLEEEAAEIDDVVGCFVVDGYSGTGCDDSDMPPGTQIP |
| 4388 |
PF14388 (DUF4419) |
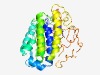 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF4419) (Length=296) NGFVNTIIRAYNTHQHLVLRPDDVWIAILGQFNFYVNAHAEQLRKQFVAHEGKKRLTVHAVGTRFTVDFGSMARQMSDLI DQNVVDKELKDWILPDFSTTSQNDTVICSVMMMATLKAYFEYTFCLTCGIPSVTLEGTKADWEKLFKKVDKFESFGEEPK AWARLLRPVLKGFVSAFEGKPDTAFWAKICHYEDFGSGSPFISGWITAFCVWSSEGKWLGPPLSGPYTDWRPAVQLDDVR YMTVDETDIPPAFCEVDVLLNDNGEEFNCMMVAGHLATKVLDKERNKVAPLSAWFM |
| 4389 |
PF14222 (MOR2-PAG1_N) |
 Estimated TM-score=0.52; hard target. |
>Cell morphogenesis N-terminal (Length=533) ERRDLAVEFLFCLALIEVLKQLPFHPGHEDLITYIEQISFKHFKYREGIQSGPNAANIHIIADLYAEVVGVLAQSRFMSV RKHFMNELKELRTKEASPTTTQSIISLLMGMKFFRVKMVPIEEFEASFQFMQECAQYFLEVKDKDVKHALAGLFVEILVP VAAAVKNEVNVPCLKNFVDMLYSQTLDASTKSKHRLALFPLVTCLLCVSQKTFFLANWHYFLAMCLSNLKNRDQKMCRVA LESLYRLLWVYMIRIKCESNQATQSRLQSIVNSLFPKGSKAVVPRDTPLNIFVKIIQFIAQERLDFAMKEIVFDLLSVGR TIKIILTPERMSIGLRSFLVVADSLQQKEGEPPMPRTMGVLPSGNTLRVKKTFLNKMLTEDTARSIGMNSYFSHVRRVFV DILRALDVQYGRPLMMTNTQNVNKEPDEMITGERKPRIDLFRTCVAAVPRLIPDGMTGAELVDLLARLTVHLDEELRVLA YQSLQTLVLDFPDWRQDVICGFTQFLARDVLDTFPQLVDNGLRMLLQLLTSWK |
| 4390 |
PF14221 (DUF4330) |
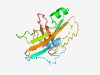 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4330) (Length=173) MAILDSKGRLFGKVNILDLGALLVIVMVIFGIFVFPGTSGSVAQVGAKTVPIEVDLVVRGLNVRDPQQLAGNGLKKGGKT KVIIRNQPYGEIEIKSVEQLPRTVVATQPDGSVKELPDPKANNFSTDMLLTLNGKAQVTKDGPVLGNSKVKIGMPFELEG FNYNFNATVIDVR |
| 4391 |
PF14177 (YkyB) |
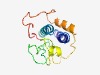 Estimated TM-score=0.52; hard target. |
>YkyB-like protein (Length=138) TVENLSKAVYTVNRHAKTAPNPKYLYLLKKRALQKLVKEGKGKKIGLHFSKNPRFSQQQSDVLISIGDYYFHMPPTKEDF EHLPHLGTLNQSYRNPKAQMSLTKAKHLLQEYVGMKEKPLVPNRRQPAYHKPVFKKLG |
| 4392 |
PF14161 (FAM110_N) |
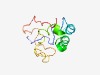 Estimated TM-score=0.52; very hard target. |
>Centrosome-associated N terminus (Length=105) GVSSTAPFTSAMPFRILNKGPEYFRRQAETGARKPSAVERLEADKAKYVKSQQVASTKQEPVKPLVLKQPLFAPGVRKAM LTPSRRTPPGARRVEACGSKPSLNL |
| 4393 |
PF14006 (YqzL) |
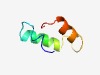 Estimated TM-score=0.52; hard target. |
>YqzL-like protein (Length=44) DFTWKVFSQTGNIDTYLLFKELEKDTQEMPGNQEEELAEIDFPI |
| 4394 |
PF13966 (zf-RVT) |
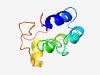 Estimated TM-score=0.52; hard target. |
>zinc-binding in reverse transcriptase (Length=77) FSSARTWNHLHPAGPTVPWYSQIWFKERILKLAFMAWLTVKDRLTTSDRLISWNILVPASCLLCLNGDESRDHLFFH |
| 4395 |
PF13947 (GUB_WAK_bind) |
 Estimated TM-score=0.52; very hard target. |
>Wall-associated receptor kinase galacturonan-binding (Length=99) HCPSSCGDVDDIAYPFGIGPGCFREGFELKCNTSTKTPKLYMKDGTTQILYVGDDDLWAPMHFNITMKPGTDTYNISWVS PRKGVTISQRNTFYIIGCN |
| 4396 |
PF13901 (zf-RING_9) |
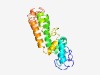 Estimated TM-score=0.52; very hard target. |
>Putative zinc-RING and/or ribbon (Length=188) RRCEYTGRYYCSLCHWNSSSVVPARVLHNWDFEPRKVCRASLQFLRLMVKKPVLNIDKLNPMLFTFVEDLRHVKKLREDI LRMKQYLTLCHAAQQQKLLLLLHKRQHFVEGSELYSLQDLIDLHDGNLLGYLTQVHQVFLDHITKTCEGCRGRGYLCKLC DSDEVIFPLGGDTYTCTDCGSVYHRECR |
| 4397 |
PF13865 (FoP_duplication) |
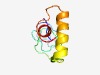 Estimated TM-score=0.52; very hard target. |
>C-terminal duplication domain of Friend of PRMT1 (Length=78) GLGRGAMGRGGISGRGRGIIGRGRGGFGGRGRGRGRGRGALSRPVLTKEQLDNQLDAYMSKTKGHLDAELDAYMAQTD |
| 4398 |
PF13572 (DUF4134) |
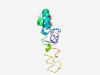 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF4134) (Length=91) TALVVLATATASLAQGNGAAGIQEATTQVTSYFDPATTLIYAIGAVVGLVGAVKVFSKWNSGDQDTQKTAMSWFGSCIFL VVVATVLRAFF |
| 4399 |
PF13309 (HTH_22) |
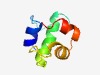 Estimated TM-score=0.52; easy target. |
>HTH domain (Length=62) LEAMIEQVINRIGIPPARMQKEDKVRVVRELDLKGVFLIKGAVEKIASVLGVSRYTIYNYLD |
| 4400 |
PF13162 (DUF3997) |
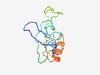 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3997) (Length=105) NDEYELIRTSGNAFEIFPSQDAVYATQYIPAKITDIAWDDKYIIAKQTEEKSDPNNPDAAIANKKSEHYWIIDVKHNKRF GPYNEKQFNEQKDAFKIKVPFQNVD |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218