

Go to page (83 pages in total): [First page] << < 42 43 44 45 46 47 48 49 50 51 52 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4601 |
PF15085 (NPFF) |
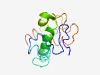 Estimated TM-score=0.51; very hard target. |
>Neuropeptide FF (Length=109) SQALHIQGGLDKNDILPGSSEENMPDRLLGLESENRDSDTDDRVLTSLLRALLHGAQREARESVLHQPQRFGRSSRGQVV LEDQMHSRDWEAAPGQIWSMAVPQRFGKK |
| 4602 |
PF15061 (DUF4538) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4538) (Length=57) LKGWRYGAFIGSIVGLIGLACYPIIIYPMMHVDEYKKIQKINRAGINQEDIQPGNMR |
| 4603 |
PF15031 (DUF4528) |
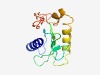 Estimated TM-score=0.51; very hard target. |
>Domain of unknown function (DUF4528) (Length=126) KPKASEVLSAHLRQRGLPHWTSYFVKYSSVRNDQFAKSHFNWSVDGQNYHILRTGCFPYIKYHCTRRPYQDLSFEDKFYT GLKIINFGIPCLAYGIGAWFLVTTTEDVKMPQGTVKVYFWYKEDHD |
| 4604 |
PF15017 (WRNPLPNID) |
 Estimated TM-score=0.51; hard target. |
>Putative WW-binding domain and destruction box (Length=73) DQFSSFLFWRDPLLTFSDELLGLLKEGNESPQTEERAEQRASAGQSDDEDKENEPDDEVDEEDDDDGGGWITP |
| 4605 |
PF15013 (CCSMST1) |
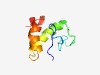 Estimated TM-score=0.51; hard target. |
>CCSMST1 family (Length=75) DKPLQYFNSPASRWRAEHTRSGQDNEDMPWYQPYVVIASMAVFMLYFCVLREENDIDRSLEKSLYEHIPGLEEKQ |
| 4606 |
PF14990 (DUF4516) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4516) (Length=46) MPAGVSWPRYLRMFTASVLSMFAGAQVVHQYYRPDLTIPDVPPKPG |
| 4607 |
PF14971 (DUF4510) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4510) (Length=161) CAPEVPAAASQPTFLPWVPEHGGGELDLVVRELQALEEELRAAAERRRAAWEAKAGGCGRGPEWSAARRASREAVEKELG ALQQCWEQDGGPAQPHGPHRLVRREDGAAGDRDLRAAVVIRTLRSQEACLEAVLRRLQGQCRQELARLVGALPGLIWIPP P |
| 4608 |
PF14929 (TAF1_subA) |
 Estimated TM-score=0.51; hard target. |
>TAF RNA Polymerase I subunit A (Length=360) EALLKHQWQRAAEYMHSYLQILEDSDSNKRQAAPEIIWKLGSEILYYHPKSSVETFNTFADRMKNIGVLNYLTISLQHAL YLLHHGMFDDASRNLSQAETWRYGEKSSSQEALINLVQAYKGLLQYYTWSKKKKELSELDEDDYAYSTASQNMFKHSWKT STNFSALIQTPGVWDPFVKSYVEMLEFYGDQDGAREVLTNYAYDEKFPSNPNAHVYLYNFLKREKAPREKLISVLKILYQ IVPSHTLMLEFHRLLRKSDKEEHHKLGLEVLFGVLDFAGCTKNMTAWQYLAKYLRQILMGSHLAWVQEEWNSRKNWWPGF HFSSFWAKSDWKEDKALAYEKAFVAGILLGKGCRYFRYIS |
| 4609 |
PF14632 (SPT6_acidic) |
 Estimated TM-score=0.51; very hard target. |
>Acidic N-terminal SPT6 (Length=90) SSEEEEDDDDEEEARKVREGFIVDEDEDEEEGGESDGDVRPVHKRKRKHRDREEEAQLDEEDLDLIGEQFGERPKPQTQS KFKRLKRGYR |
| 4610 |
PF14449 (PT-TG) |
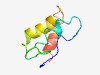 Estimated TM-score=0.51; hard target. |
>Pre-toxin TG (Length=66) QTFSAVLDFLPFIGNAKAGQEASTGVDLITGQELDEWDRGIAAASIFLGGIGKVGGRTLKNIVKGS |
| 4611 |
PF14380 (WAK_assoc) |
 Estimated TM-score=0.51; very hard target. |
>Wall-associated receptor kinase C-terminal (Length=106) NCKTFSNGPDGGDSFVFTSGELEAPVESELARRCSQVIVVPVNGSILNSSNQSALPSGGYGEVLNKGFDLAWTSRKDGQC YQCEKFQGQCAYSQNRIFLSCLCSDG |
| 4612 |
PF14331 (ImcF-related_N) |
 Estimated TM-score=0.51; hard target. |
>ImcF-related N-terminal domain (Length=258) ADKAAWTGFLQLLKKHRRRRPINGVIVTVPVSDLLQGTEAGRQAQAQAIRDRLKELHDQLGVRFPVYVLVTKCDLLGGFV EFFDALGREERAQVWGMTFPLQDGMDGDAALAVFPAEFEQLEQRLQERVLARMQQERDPHKRALVYSFPQQFAGIEDVLK RFLDDVFASNRYERQALLRGVYFSSGTQEGSPLDRVMSAMAASFGLERQALAPNAASGRSYFITNLLQQVVFPEADLAGV NHALEKRRRMIQWGTVAA |
| 4613 |
PF14267 (DUF4357) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4357) (Length=54) REQLLNQGVLEQQKNVYVFTSDYFFSSPSAAAAAVLGRRANGWAEWKDEDGKTL |
| 4614 |
PF14217 (DUF4327) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4327) (Length=67) YSLDVIQDEARQLVEKGVISRQQPIYTLCQYIPPREWVCVECELEKCDFLLRDRIADLIGREEWDND |
| 4615 |
PF14209 (DUF4321) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4321) (Length=48) LWWLSYEQEFGINAPIVLDLSVLKLTFGLMFKINIASIIGMVLAIFIY |
| 4616 |
PF14194 (Cys_rich_VLP) |
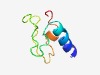 Estimated TM-score=0.51; hard target. |
>Cysteine-rich VLP (Length=56) TPAQRRRANALVRKTCCNYDNGNCLLLDDGEECVCPQTISYSVCCKWFRWAVLPQD |
| 4617 |
PF14162 (YozD) |
 Estimated TM-score=0.51; hard target. |
>YozD-like protein (Length=55) MKEIEVVIDTEEIAEFFYRELIRRGFVPTQTELEELADITFDYLLEKCIIDEELD |
| 4618 |
PF14138 (COX16) |
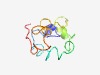 Estimated TM-score=0.51; hard target. |
>Cytochrome c oxidase assembly protein COX16 (Length=78) LLFGLPFISIIVAGSFVLTPATAIRYEKHDRRVRQMTKEEELGLGKDRRQVDIREEYYRLAAKDLDNWEQKRVERFKG |
| 4619 |
PF14058 (PcfK) |
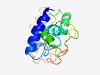 Estimated TM-score=0.51; very hard target. |
>PcfK-like protein (Length=135) SSNFETTIQAYLENRAKTDSLFAETYRKANKSIEECIKYIYSKARKLAKGGNAVGVDDATVYGWAVHYYDEDDIKVKDVK ERVEVVAPTTVQEPVVQEPVKEEKPEPVKQKSARKKTKQELQKIFDSRQLSLFDM |
| 4620 |
PF14047 (DCR) |
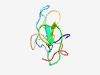 Estimated TM-score=0.51; very hard target. |
>Dppa2/4 conserved region (Length=65) DKWCVVHGCSLPADAGGWVQLQFHAGQAWVPEKKGRVNALFLLPSGTFPPPHLEDNLLCPTCVRR |
| 4621 |
PF14035 (YlzJ) |
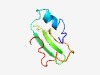 Estimated TM-score=0.51; hard target. |
>YlzJ-like protein (Length=66) LYTPMQLELVLEGFDRTEYPEYREINYDGVPLLVERTEHGKNRIVKLLSTNPYDYLRAELAPGNLI |
| 4622 |
PF14022 (DUF4238) |
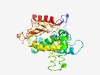 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF4238) (Length=284) NQHYVPRGYLRGFTIPGEKSLIWQYDKNTGEIFKRPKSVNRICSEFQYYAQKNEDGSTDKETMENAFHEIEDKTPRIIKK INSLNNAEKVIISEDEKAVLSFFTAMQLTRVPNFRNGIEEMYRKVVEIGLSHQVDKARKDGTLPVSIDSLYKQGKINIDI ESFVSLKAMLATAQEGAINLMGKVWHFAMPAKGMSFVTSDNPVFFQAPEKYRELNSQIGPFHPVSEVTLPLSSDLLLIFS PSANYSKSEYDLINMTAVQLDEKDTDNLNKRSILAAKQYVYSSE |
| 4623 |
PF13978 (DUF4223) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF4223) (Length=54) MKKFIKVALVGAVLATLTACTGHIENRDKNCSYDYLLHPAISISKIIGGCGPTA |
| 4624 |
PF13974 (YebO) |
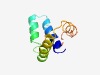 Estimated TM-score=0.51; hard target. |
>YebO-like protein (Length=80) ISLVVLLVGLVIWFFVNRASSRANEQIELLEALLDQQKRQNALLRRLCEANEPEEKDAPKDAVVEENKDEDDFIRLVAER |
| 4625 |
PF13922 (PHD_3) |
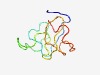 Estimated TM-score=0.51; easy target. |
>PHD domain of transcriptional enhancer, Asx (Length=68) QQPQQQAVHPLLQLRQSGENTPPGSEAATAPANNCACSLNAMVICQQCGAFCHDDCIGAAKLCVSCGI |
| 4626 |
PF13890 (Rab3-GTPase_cat) |
 Estimated TM-score=0.51; very hard target. |
>Rab3 GTPase-activating protein catalytic subunit (Length=155) SKAEGRLHPYNNMTLLNSMEPLYVPVTQEPAPMTEDLLEEQSEVLAKLGTSAEGTHLRARMQSACLLSDMESFKAANPGC VLEDFVRWYSPRDYVKEEVADERGNTVVKGELSARMKIPGNMWVEAWETARVTPARRQRRLFDDTKEAEKVLHYL |
| 4627 |
PF13763 (DUF4167) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4167) (Length=78) RGRNNNNRSNNNKGPNPLTKSYESNGPDVKIRGTAQHIAEKYSQLARDAQSSGDPVAAENYFQHAEHYFRIIAAAQEQ |
| 4628 |
PF13747 (DUF4164) |
 Estimated TM-score=0.51; easy target. |
>Domain of unknown function (DUF4164) (Length=86) KTIEAALKELKQAISSLENAVDMRIERQREQGEIEGEVRRVHADRSRLAQELDQAEFRANRLEEVNREVSRRLVTAMETI RAVLDR |
| 4629 |
PF13726 (Na_H_antiport_2) |
 Estimated TM-score=0.51; easy target. |
>Na+-H+ antiporter family (Length=87) NPVVISVIVMSVLCLFKLNVLLSILLAAMTAGVAAGMPIKETMNTLIDGMGGNSQTALSYILLGTFAMAISKTGLATILA KKISQVV |
| 4630 |
PF13598 (DUF4139) |
 Estimated TM-score=0.51; hard target. |
>Domain of unknown function (DUF4139) (Length=285) IKLSYVTNNASWEPAYDVQVESINQPVKLSYKATIFQNSGINWDNVKLTLSTINLSDRAQTPHLSPWSIELYNANSKRSL SYYEISGANSLYTRFRLPMPCNIATGGKGQVVLIKQSEVLGDYRYVVAPRIDTTAFLQVQVKNWQKLNLLPGKASIFFAG SYVGQQRIDTTDDQDTWDLSLVRDDKIMVSRKKEKHLTSKAGFWGNRIRQQYSYVTQVKNTHNEPISLVVFDQIPLSSDE AITIAGLDYGGGIYDRNSGDVHWKVNLAAGDSQQFTLGYTVNSPK |
| 4631 |
PF13552 (DUF4127) |
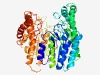 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF4127) (Length=504) IMYIPLDERPCNYEFPKMIIDISDIDMIRPPKEILGCKKKPADVDKLWEWMRQNIEGITHLVVSIDMLVYGGIVPSRIHQ KQVEECLSKLLRLRELKKANNNLKIFAFNLIMRTPSYNSSEEEPDYYKEYGERIFRFGAFSDKIDLNIATEEEIKEFNAL KREIPEEILEDYLRRREVNHKINLSVVDLVKESIIDFLIIPMDDSSKYGFSPMERRKIMKYIDELDLTDRIYSYPGADEV GCTLTARAFNDIKGVKPAVFTRYSSENGKNVIPLLEDRRLGETVKYQIIAAGGIVTEDSEKADFILMINPPSEITQQIGD SWNSYLTRKDIGDPERNLNEFVEAIKDYIEKGKNCSVADVAFLNGADNSLMKLIKKYGLLKKLLSYGGWNTSANTLGTVI AHSMASSFYINSDNFTEDRKERSYRFLFLRYLEDWGYQYEVRTKVAKEIDKYGVSYFDLGEKEKIISEVIKGELEKFKKE NLNEFDYEFNVYMPWNRLFEVGIE |
| 4632 |
PF13400 (Tad) |
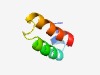 Estimated TM-score=0.51; easy target. |
>Putative Flp pilus-assembly TadE/G-like (Length=47) GATAITFGLLLVPLLGFAGLAVDYSRASNDRSRLQNAADAAALAAAS |
| 4633 |
PF13062 (DUF3924) |
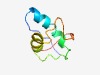 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF3924) (Length=62) MNTLTIELPKETAEKLDLLKQAYEKKTGASISESTLVQTLISKEFIQAITPFDLQQFVNGKE |
| 4634 |
PF12992 (DUF3876) |
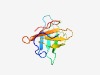 Estimated TM-score=0.51; easy target. |
>Domain of unknown function, B. Theta Gene description (DUF3876) (Length=92) SCSESRFKDCDRLCGSWSSVEGKPDVLIYKEGDAYKVTVFARSGKTRKLKPETYLLVEENGNLFINTGYRIDIAYNEAAD VLTFSPNGDYVR |
| 4635 |
PF12926 (MOZART2) |
 Estimated TM-score=0.51; hard target. |
>Mitotic-spindle organizing gamma-tubulin ring associated (Length=89) SAPDSPALVVTSNVQKYAIKKKKVLNAEEMELFELTQAAGITVDQEVFKIIVDLLKMNVAPQAVFQTLKAMCAGQRVADS NGGESSTIS |
| 4636 |
PF12856 (ANAPC9) |
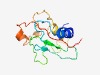 Estimated TM-score=0.51; very hard target. |
>Anaphase-promoting complex subunit 9 (Length=128) RLIKSKLLNLHQSSKQKTTWKYHPNKSVPSFIIRKPINMNGYDYSVFTNYPTEIDLQHGEHLPPAIKESQIKSWESAERI STNVIFGNSSESEDDIDELDEDEHTEASRSSKVNSRKNNVIQAIPGYT |
| 4637 |
PF12805 (FUSC-like) |
 Estimated TM-score=0.51; hard target. |
>FUSC-like inner membrane protein yccS (Length=285) ALLVTLLCFSVAAFAVELLFPYPWLFVSGMALSAFGMILLGALGERYAAIASATLILSIYTMIGVDQRGGEVADVWHEPL LLVAGAAWYGLLSVLWNALFSNQPVQQSLARLYFELGEYLRIKSSLFEPLRQLDVEGQRLALARQNGKVVVALNQAKETI LNRLGHGRPGPKVSRYLKLYFIAQDVHERASSSHYPYNRLAEAFFHSDVLFRCQRLLNQQGKACQALARAIRLRQPFDYD DSEQALEDLQASLEHLRQQSNPAWKGLLRSLGALAANLATLDRKL |
| 4638 |
PF12749 (Metallothio_Euk) |
 Estimated TM-score=0.51; hard target. |
>Eukaryotic metallothionein (Length=65) MDTQTQTKVTVGCSCNPCKCQPLCKCGTTAACNCQPCENCDPCSCNPCKCGVTESCGCNPCKCAE |
| 4639 |
PF12601 (Rubi_NSP_C) |
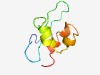 Estimated TM-score=0.51; very hard target. |
>Rubivirus non-structural protein (Length=66) VCAVGGGPRRVSDRPHLWLAVPLSRGGGTCAATDEGLAQAYYDDLEVRRLGDDAMARAALASVQRP |
| 4640 |
PF12551 (PHBC_N) |
 Estimated TM-score=0.51; hard target. |
>Poly-beta-hydroxybutyrate polymerase N terminal (Length=40) DTADRALHAAQARLTGGISPISIASAAFNWTLQLANAPGR |
| 4641 |
PF12505 (DUF3712) |
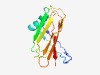 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF3712) (Length=127) LGTFSLQSLSANPKIRRAYINQTTELTVADEDAFGRFTAYMVTSQNFTWRLKSDNLEARALKLPISKGLKFTKDITLNGI NNFQNNAVITDFQLPSDSPAGGINYVATTELYNPSPFKLGLGVVSFA |
| 4642 |
PF12442 (DUF3681) |
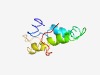 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF3681) (Length=91) NAEKLPAAMISCGVVEAAAAIFLAAFKAPGGIFLHHGKAPFYLYYGIIGGVAIFGFAEAWAGFWVSGDLNGRRIVGKTIL WVGMLPLVLFM |
| 4643 |
PF12395 (DUF3658) |
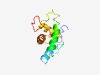 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (Length=112) KNEPPLSQHERENFEKEWLALANTREVLRIWQNRTIHSVSEDYYDLFIINRAKKLHNEMKTKDFIKSVRLIGEVLGHLDQ YVGDEFLEYRLRKLIEKRAFEMEGSLEAMAFY |
| 4644 |
PF12313 (NPR1_like_C) |
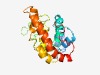 Estimated TM-score=0.51; very hard target. |
>NPR1/NIM1 like defence protein C terminal (Length=205) RRLTRPKDYHAKTEQGKETNKDRICIDVLEREMRRNPMAGDACMSSHTMADDLHMKLLYLENRVAFARLFFPSEAKLAMD IAHAETTSEFAGLSASNSKGSNGNLREVDLNETPIVQNKRLLSRMEALTKTVEMGRRYFPHCSEVLDKFMEDDLPDLFYL EKGTHEEQRIKRTRFMELKDDVHKAFNKDKAEFSRSGISSSSSSS |
| 4645 |
PF12271 (Chs7) |
 Estimated TM-score=0.51; hard target. |
>Chitin synthase export chaperone (Length=298) GFGSFVYLCRNTGSYPFCNLFWRQLSKANFTLPVVTRAPVGILPRCGIPLAGNGRFGNVADIIFCAISFIFIVYLSQRCL GNAPSITGRIEIRAMFVLYAVLMLLQTVTAGSIFEQGSLPLLILTCLHAGAVAAFFWCLLANAFVATQYVEDGTPSSLIP FYGFAGIFFATTAYISADTAFSYTNLFKSTPPRDLRNIALFILLVIWPAVSVLLYAGIMSYIVVNILGEKRPLMYYLGAL VVFIGAQLAYLFLGTAICQGTNRFIDSAFIATLLETTAMGLLFIGWRTITESKWEDQV |
| 4646 |
PF12251 (zf-SNAP50_C) |
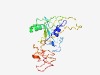 Estimated TM-score=0.51; very hard target. |
>snRNA-activating protein of 50kDa MW C terminal (Length=204) QTMLVLGSQKLTELRDSICCVSDLQIGGEFSNTPDQAPEHISKDLYKSAFFYFEGTFYNDKRYPECRDLSRTIIEWSESH DRGYGKFQTARMEDFTFNDLNIKLGFPYLYCHQGDCEHVIVITDIRLVHHDDCLDRKLYPLLTKKHWLWTRKCFVCKMYT ARWVTNNDSFAPEDPCFFCDICFRMLHYDSEGNKLGEFLAYPYV |
| 4647 |
PF12233 (p12I) |
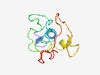 Estimated TM-score=0.51; very hard target. |
>Human adult T cell leukemia/lymphoma virus protein (Length=99) MLFRLLSPLSPLALTALLLFLLSPGEVSGLLLRPLPAPCLLLFLPFQILSNLLFLLFLPLFFSLPLLLSPSLPITMRFPA RWRFPPWRAPSQPAAAFLF |
| 4648 |
PF12141 (DUF3589) |
 Estimated TM-score=0.51; hard target. |
>Beta-mannosyltransferases (Length=463) CEDVNIPETMSINKYYQFQEDLYQVVEILQDQLDNDPAFKDLAPFFEGKIPEMIKNQTVTDHFYKFMGTSVWLKQYGVHL MISRVLFTRTGIRFKPQLSLMYGQLFNENWQELHNVTLRIPDSFSEQPGSSSDMAFTFPTFLPIPFFHDSNNVKYRYYGP EDGRILLVQNQNGFEEPIVVFNSFTKTKENDNSKKVKLHRSMFVGWLFQSQTGKENTDAFKDTKYDNVTYTKVRQFTIAN ETSTKRQEKNWTPFIDPQERTTHDGNIYLIYEWNNLLVLKCELTNFTSNAFSYCHRIYNETSQDKIGPLRGGTELIPVNH IKTDNQIWIGFIRSHIGKCGCGTTMYHPHLVILQRQGDLFRVSHMSSSISFNIPAKSLTNNNPNKCGVGVANVLMANGIS NWEIDETSGIDYLTLTFSSGDEDTSIVHIKDFSKMLERLDLDEPWNETTDKMRDCVLIESKQF |
| 4649 |
PF12133 (Sars6) |
 Estimated TM-score=0.51; hard target. |
>Open reading frame 6 from SARS coronavirus (Length=62) MFHLVDFQVTIAEILIIIMRTFRIAIWNLDVIISSIVRQLFKPLTKKNYSELDDEEPMELDY |
| 4650 |
PF12084 (DUF3561) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF3561) (Length=107) MRNSHNITLTNNDSLTEDEETTWSLPGAVVGFVSWLCALAIPMLIYGSNTLFFFIYTWPFFLALMPVAVVVGIALHSLMD GKLRYSIVFTLVTVGIMFGALFMWLLG |
| 4651 |
PF12025 (Phage_C) |
 Estimated TM-score=0.51; hard target. |
>Phage protein C (Length=68) DLSLRSSRSSYFATFRHQLTILSKTDALDEEKWLNMLGTFVKDWFRYESHFVHGRDSLVDILKERGLL |
| 4652 |
PF11879 (DUF3399) |
 Estimated TM-score=0.51; very hard target. |
>Domain of unknown function (DUF3399) (Length=102) MQSKRNGLLSNQLQSSEDEQAFVSKSGSSFETQHHHLLHCLEKTTNHEFVDEQVFEESCMEVATVNRPSSHSPSLSSQQG VTSTCCSRRHKKTFRIPNANIS |
| 4653 |
PF11703 (UPF0506) |
 Estimated TM-score=0.51; easy target. |
>UPF0506 (Length=58) CKEYGESCMKTVFNKCCGSNVCQLTGPFNGKCVRCIQTGYSCLHNGECCSRNCWLFRC |
| 4654 |
PF11661 (DUF2986) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2986) (Length=43) MNRKKKINQTLQKRLKKQHAKLHPSNKPRYISKAERAKLAAEQ |
| 4655 |
PF11279 (DUF3080) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF3080) (Length=319) DEYLERLARVLDTAPVPRAELPAASIPPRRRERILALPELDLGMLDFLSLYGCELQYVVGERNSVMGKVMQPINQLRYEI RFIRAAEACLPEVDDEELTEALESAIESKRDSLPLAVWNATWGTEEVERQFTLSKGYYPVAEAGNPASDLVRDLQQLNRQ VEAILAQKLEISLKNLGQVHQRWQADVLAGQTINSARLLISTLNAGTELLGSRLEGRPLCLNGQPNNESEIVQNFFFSIY IEKIQPYMSDVSRARDSLIAGFAELARQQQAVMPESFTPWYQRHLAADTPDSLWQELDQAMMRHTRHWQDLLGQCGLRP |
| 4656 |
PF11262 (Tho2) |
 Estimated TM-score=0.51; hard target. |
>Transcription factor/nuclear export subunit protein 2 (Length=334) WQKITPELYANFWALQLGDLYCPDKIYRQEKDRLRAEEVAISRDRSDMSRRGQERKVEKRKELMQLQIGLSEENSEHLLR QAKWKIYLNKQFQTSFPEPRAKPDTISDILLEQCFIPRMLLSKADAEYTYKFIKALHEGNAPGFQLMSLYDRLFNANRLR ALIFTSTVMEAEYLGRFLKLILEDLSRWHKNATVPNEKDKSRDQPRLGVYEKEAKGPPDQPRLGFALTFDDNDKPLTFVE HEQFRDLLFRWHKNLNSALRSCLEGTEWMHIRNAITVLKAVLDFFPAIDFMATKFSSLLQTITKQESAAKPSPDSEVGGR VDLSVAAQGAMSEL |
| 4657 |
PF11157 (DUF2937) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2937) (Length=159) LRSYLRLTLFAFGLLLGVQVPGFIDDYAKRVEAHRLESQQSLKGFQETAQKFFKGDMDALVAHYRVSDDPVMRSDAKSVG HLVQRSALLEREWQAMQGPWYAQAWHLATGADHELFQETLRAYRYQVLFTPDAILWGVISALLLAWLAELLVLIFGWLF |
| 4658 |
PF11146 (DUF2905) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2905) (Length=62) KLLITIGVVLIVLGVLWPLLQKSGLGRLPGDIAVEKENFKFYFPITTSIIVSIVLSLILWFF |
| 4659 |
PF11084 (DUF2621) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2621) (Length=139) EGWFSWFIVLWTVILLGLMSIGGYFMFRKFLKRLPKEDGMSILDWEEHYINKTRHLWNDEQKQLLEELVSPVPELFRDVA KSKIAGKIGELALKEKASQITQDLIIKGYIIATPKRDHKFLVKKLQEKEIDYSHYESLL |
| 4660 |
PF11072 (DUF2859) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2859) (Length=146) VEDRGGASALPYYQSLNPQPDQATPPAPMPAPRVGNAADAEAAMLPVRSTQLSPGDVQRRVIRAPGLTPLFLIGDDERSR AWLRQRQAALRELQAVGLVVNVESMAALTALRRLAPGLTLSPASGDDLAQRLGLRHYPVLITSTGI |
| 4661 |
PF11023 (DUF2614) |
 Estimated TM-score=0.51; hard target. |
>Zinc-ribbon containing domain (Length=111) KYSSKINKIRTFALSLIFIGFIVMYGGIFFRNSPIIMTIFMILGLLCIVASTVVYFWIGMLSTKTVQVKCPNCGKATKIL GRVDICMHCNEPLTLDKNLEGKEFDEKYNRK |
| 4662 |
PF10866 (DUF2704) |
 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF2704) (Length=167) VKTEQAEASGDNNRKVVDANVDEYTVDGLKLKSKYVAYYKCLKILVDFLVMYVSKEVSMKEYEQVYTLGRQLYEVLRSIF VDEPFKLWLERNTQEFDNNKDKILETLQSELKFALADKNKLKTCTFKDIITNLLNTKLDCKYDCVDEYIKPNCIVDTYNC CNLVFKK |
| 4663 |
PF10809 (DUF2732) |
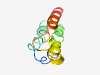 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2732) (Length=74) MRNIVTRSFNTDSDALAILLTDAKKEERKDRALAVSIRLEALAIHIIKEGMSCTEAAELLRREATRFENESQEL |
| 4664 |
PF10795 (DUF2607) |
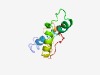 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2607) (Length=94) TRLRCPHYRAQSIALCAMLLMLWLNVAYVDHHLDTNPAHHAQHHCQLFSGFQNGLSSALPILPVISGSEIRSSVEKTQRR PSVHYAYLARSPPH |
| 4665 |
PF10784 (Plasmid_stab_B) |
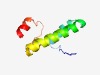 Estimated TM-score=0.51; trivial target. |
>Plasmid stability protein (Length=70) QYSFYLHSDDRTDIAAMATISTISQPLRGEFIRTAATAGAVVYQVDARLPALIPVFFAGQLSAVRLCAVM |
| 4666 |
PF10768 (FliX) |
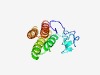 Estimated TM-score=0.51; hard target. |
>Class II flagellar assembly regulator (Length=134) MRIYGPNGTTLGTPASQARRTGSGTFVLPDTSSAQETRSAAAPKAAANIDALLALQGVEEDPVERRKRSVARGKTALDVL DELKMGLLSGNLDASTVMRLRDAAANLKSSSGDPGLDSVLSEIELRVEVELAKA |
| 4667 |
PF10742 (DUF2555) |
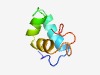 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2555) (Length=55) KKDISAMTTADVQELANRLEEDDYSNAFEGLNDWHLLRAIAFQRPELVEPYIYLL |
| 4668 |
PF10737 (GerPC) |
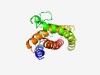 Estimated TM-score=0.51; hard target. |
>Spore germination protein GerPC (Length=173) IQQLERQLKEMQTEMNTMKQRPATTIERVEYKFDQLKIERLDGTLNIGLNPTDPNSVQNFEVSQTTPDIGMMQQEESAQL MQQIRQNVDMYLTEEIPAVLEQLENQYGSRLDDTNRHHVIEDIRKQIDSRIHYYMSHIKKEENTPPKQYAEHIAEHVKRD VIRAVEHFLEHIP |
| 4669 |
PF10696 (DUF2501) |
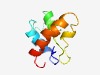 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2501) (Length=79) QKLVSATNTENVKNQLLDKLGLSSQQEKQQQTDYTQGLAGLLNTSDGKQLNLNSIGNTPLAEKVKTKACDVVLKQGANF |
| 4670 |
PF10506 (MCC-bdg_PDZ) |
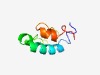 Estimated TM-score=0.51; easy target. |
>PDZ domain of MCC-2 bdg protein for Usher syndrome (Length=64) RLNSRIEHLKSQNDLLTITLEECKSNAERMSMLVGKYESNATALRLALQYSEQCIEAYELLLAL |
| 4671 |
PF10496 (Syntaxin-18_N) |
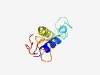 Estimated TM-score=0.51; hard target. |
>SNARE-complex protein Syntaxin-18 N-terminus (Length=89) SDITPLWKASLKTVKIRSKTLGINQENAKNSILPHSRQKSEFVTKSKEVVNSITKLRDFLVEHRKAYINASSHLSLGVSR MTDSERDQI |
| 4672 |
PF10457 (MENTAL) |
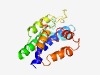 Estimated TM-score=0.51; hard target. |
>Cholesterol-capturing domain (Length=167) SDVRRTFCLFVTFDLLFISLLWIIELNTNTGIRKNLEQEIVHYRFNTSFFDIFVLALFRFSGLLLGYAVLRLRHWWVIAV TTLVSSAFLIVKVILSELLSKGAFGYLLPIVSFVLAWLETWFLDFKVLPQEAEEERWYLAAQAAVARGPPLFSGARSEGQ FYSPPES |
| 4673 |
PF10364 (NKWYS) |
 Estimated TM-score=0.51; very hard target. |
>Putative capsular polysaccharide synthesis protein (Length=137) IEGKKNDAIHRDMERTYQHLEFCFDHYINLNYFANWFDNELNKNFGIDVIKSDIDNNEPFYLFKNKKTEVMLIKCEHLNK LDSDIAKFLNVDGFILKNSNEAKNKWYSNIYSHFKDTYDFSKLFYMYDLPLYRHLYS |
| 4674 |
PF10332 (DUF2418) |
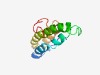 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF2418) (Length=105) RDVWELAVWDPLPVSLQLFSLFSPGHVLVYMLFLPLENLDPRPSVTVFNCLALQVILSVQMLFLQSRFSQQIKDTAIIHK EVLHEYDTKFVHPRLHPLVRDVGTQ |
| 4675 |
PF10172 (DDA1) |
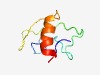 Estimated TM-score=0.51; hard target. |
>Det1 complexing ubiquitin ligase (Length=63) DFLKGLPSYNQTNFTRFHSDSGVKTTSRRPQVYLPTTDHPSEQVITTEKTNILLRYLHQQWDR |
| 4676 |
PF10164 (DUF2367) |
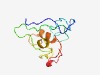 Estimated TM-score=0.51; very hard target. |
>Uncharacterized conserved protein (DUF2367) (Length=107) PSYGTIPAAQVGFQPPPPYPYPAAPGFSGGPPPASVQPPYSSTYTIIQQPATTSVVVVGGCPACRVGVLEDDFTCLGILC AIVFFPIGILFCLALRQRRCPNCGATF |
| 4677 |
PF10011 (DUF2254) |
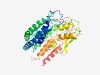 Estimated TM-score=0.51; hard target. |
>Predicted membrane protein (DUF2254) (Length=376) SSYWFVPTLMAASAIALAFTMLTLDRAGKSGPIEKLAWIYTGGPDGARTLLSTIAGSMITVAGTAFSITIVALTLASSQF GPRLLRNFMQDTGNQVVLGTFIATFIYCLLVLRTVRGDDYNVFVPQISVTVGIVLAIASIGVLIYFIHHASTSIQAWHVI TEVGSDLDKAIDRLFPQKIGYGGSGEKRRWVEEIPVGFDREASPILATSSGYLQAIDNDKLMKIAQSKDLLVRLKHRPGK FVVQGSELVRVWPGERVNKTLSQQLNEAFILGKQRTEQQDVEFCVNQLVEVAIRAISPAVNDPFTAIRCIDQLSAGLCRL AEREFPSPYRYDDDNNLRVIVDPVTFAKLTDDAFNQIRQYSKPDVAVRIRMLEAIA |
| 4678 |
PF10001 (DUF2242) |
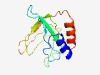 Estimated TM-score=0.51; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2242) (Length=121) CEAARRTLLSQGYIIKQAKADLVDGHKHFQPDQDTHVEIEFHVVCAPNDKDGKSSTLFVSATQDRYALKKTNNSASVGLS VLGSVSLPIGSTSDSLVKVASETIPAGRFYDRFFGLIEHYL |
| 4679 |
PF09999 (DUF2240) |
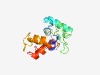 Estimated TM-score=0.51; hard target. |
>Uncharacterized protein conserved in archaea (DUF2240) (Length=135) XVAAPFKARAQDRLGQGEFVVALSLEREWFSPDQAKRLVDVALGRGLLASEDGDLVPQFDPAEVTVPEGFTPDESVLREQ STFESALDAVVAAGVEKQRAVAAINERQRALAVTIEAAAVLYAREQGVAVDRLAV |
| 4680 |
PF09991 (DUF2232) |
 Estimated TM-score=0.51; hard target. |
>Predicted membrane protein (DUF2232) (Length=287) AIYAILLLISMYVPILGTVVTFALPLPFILLTLRHRLSNVIVIFVAALLITVIVGQPLSLIKTVMFGLIGVVLGYMYKKE KKPVEILIAGTLAYLIGILLIYVASIKFFNIDLMKQMQNMFSESMAQSEKIVNAAGMPVSKEQKELFAQMSDILQTLFPS ILVLVSVCFSWITLILSGSVLRKLKYDVTPWPKFKDIQLPKSIIWYYVICILLATFIKVEPTSYLHMVFSNLNVIFALLL VLQGLTFITFLAHRKGFTKGVPIISFIACMFIPMLFPLVTILGIIDL |
| 4681 |
PF09875 (DUF2102) |
 Estimated TM-score=0.51; hard target. |
>Uncharacterized protein conserved in archaea (DUF2102) (Length=103) YIVISSDKVLPSDAAMKIYESESPVTVKETCFGLIVSGAKNDVQTVVEEIRQLDKNHIFVKDRGFPPGDERRCRASRGGG PRPGFHFLREEVEMLPAIGAALD |
| 4682 |
PF09777 (OSTMP1) |
 Estimated TM-score=0.51; hard target. |
>Osteopetrosis-associated transmembrane protein 1 precursor (Length=244) CRRLLTAFAEGSATLSGCLARRARPVRLCQACGGLYRSLLTQYGDIARAVGNSSESHNCAKSILTSDRLQVVVTLSSFFN DTWEAANCANCIKNNSEGLSNSTQEFMDLFNKSLACFEHNLQGQAMDLSPNNYTEVCKNCNGTYKKLNDLYNKLQRLNRH GDESGHSAHLCIDVEDAMNITRKLWSRTFNCSVPCSDTVPVIAVSSFILFLPVVFYLSSFLHSKQKKRILILPKRIQSNA SLVN |
| 4683 |
PF09767 (DUF2053) |
 Estimated TM-score=0.51; easy target. |
>Predicted membrane protein (DUF2053) (Length=149) TLFHFGNCFALAYFPYFITYKCSGLSEYNAFWRCVQAGATYLFVQLCKMLFLATFFPTWEGGAGVYDFVGEFMKSTVDLA DLLGLHLVMSRNAGKGEYKIMVAAMGWATAELVMSRFLPLWVGARGIEFDWKYIQMSFDSNISLVHYIX |
| 4684 |
PF09740 (DUF2043) |
 Estimated TM-score=0.51; hard target. |
>Uncharacterized conserved protein (DUF2043) (Length=105) EIPILSFGLDLKYWGEDDVVPAEVPRNNADCHRFWRPSDESCCTSRDSAEVYRARVITFVGPGLKATRRCRAPLKNGELC SRMDFRRCPLHGNIIDRDEMGYPID |
| 4685 |
PF09538 (FYDLN_acid) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (FYDLN_acid) (Length=108) MAKPELGTKRLCPSCGAKYYDLNRNPITCPKCGTLFDIVSSSRAAKASRVVQQEDEDEDEEDLVAPELVSLEEADAEVEG DIVPDLGDDDEAAIDDDADDDVFIEDEE |
| 4686 |
PF09072 (TMA7) |
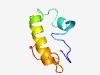 Estimated TM-score=0.51; hard target. |
>Translation machinery associated TMA7 (Length=62) GREGGKKKPLKQPKKQAKEMDEEDKAFKQKQKEEQKKLEELRAKAAGKGPLATGGIKKSGKK |
| 4687 |
PF08983 (DUF1856) |
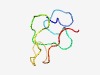 Estimated TM-score=0.51; very hard target. |
>Domain of unknown function (DUF1856) (Length=47) KFNKEDSDSVSRRQTSYTHNRSPTNSMGIWKDSPKSSKSIKFIPVST |
| 4688 |
PF08845 (SymE_toxin) |
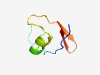 Estimated TM-score=0.51; easy target. |
>Toxin SymE, type I toxin-antitoxin system (Length=51) ERFTRVGYRPIKGNHNTPAINIAGQWLKDAGFTTGQPLKLRIMPGCIVITT |
| 4689 |
PF08811 (DUF1800) |
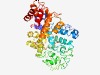 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF1800) (Length=440) TLRRITFGPLPNEVARARAIGLEAFILEQLNPESIPEPDILAPFDFETLEMNAGELLRVDPPGRPIGELMGATLARAVFS PRQLYEKMVVFWSDHFNIYIRKGIERALKPVDDRLVIRPHALGRFHDLLSASAHSPAMLYYLDNASSTKEKPNENYAREL LELHTLGVDGGYTHQDIVEVARALTGWSITRPRRFFGGGGEFVFRPRLHDTGPKTILGQSFPAGQGVEDGEQLLDLLSHH PSTARFISTKLVRRFVADEPPEDLVTRATQTFQETDGDIAEVMRTILTSDELRASLGGKLKRPFEYLVSALRQTQAEIEP NRALFRYLTMMGQPLFHWPTPDGYPDVAAAWMGTGSLLARWNFALALAFNTIPGVRVDWDVLAGEASAPEGALDTLSDRF LGAPLPEEARSLILDFLNTLDAGFVLPALGALLLASPYFQ |
| 4690 |
PF08469 (NPHI_C) |
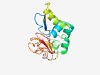 Estimated TM-score=0.51; very hard target. |
>Nucleoside triphosphatase I C-terminal (Length=148) HIFTPEERRYVNVHFIIARLSNGDSTVDEDLLDIIRTKSKEFTQLFKVLKESSIEWIYEHHKNFSPIDDESGWTSLISRA IDMNSKPREIIHVAEGQNIWYSHSNRLITIYRGFKTEDGKIYDTDGNFIQNMPDNPIIKIHNDKLVYL |
| 4691 |
PF08188 (Protamine_3) |
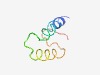 Estimated TM-score=0.51; hard target. |
>Spermatozal protamine family (Length=48) ARRRHSMKKKRKSVRRRKTRKNQRKRKNSLGRSFKAHGFLKQPPRFRP |
| 4692 |
PF08175 (SspO) |
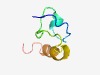 Estimated TM-score=0.51; hard target. |
>Small acid-soluble spore protein O family (Length=48) AKRKANHVIPGMNAAKAQGMGAGYNEEFSNEPLTEAQRQNNKKRKKNQ |
| 4693 |
PF08157 (NUC129) |
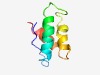 Estimated TM-score=0.51; hard target. |
>NUC129 domain (Length=63) YTAMHLKDQSLTGLHQQTAKDFVYSHLYGPGTNRTSANEFFSLANKKDPVKKAAVQFVDKSWG |
| 4694 |
PF08101 (DUF1708) |
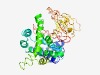 Estimated TM-score=0.51; easy target. |
>Domain of unknown function (DUF1708) (Length=397) EVAPDEVQELIHVCTQELKSRALDTPFLLLPFRPGSDATSARSFIKSFFRDAYEGGGSYRGEGLRAELRLTEPMVLCSIM KWCWSRLPGGVVTWDVYELFRVGEQDSGMAKHAFDTFVPISVDSEARKQIIIDFFDLLAAISARGKTNGLGGRKLSRMAG WWAFEHPDSGKGFDGGYKSWTGAADATSHLFFAYLRSLAPEDMSLTGINTLPRSLQALLNQTEYPPEPPTLMRTTTTKVV MIVDTVSPTPFALLRRAKHFEYRDDDRVLQKFSHYDDPIRALTEECQRPNVLPTLRGQDGTTNHSVMGGFPFVWVLPPIG EVTRVQSSQSHLRLGEDDLEPGELASVSHVDLDETFWWVWMTSLAIEEPNERKAVFGRCALLETNISGGKWLILEEQ |
| 4695 |
PF07956 (DUF1690) |
 Estimated TM-score=0.51; hard target. |
>Protein of Unknown function (DUF1690) (Length=134) WKGSAPTSVSQDMLDSLQSSTETDASRAQTLELQIQARVAEELKRLQAKEDAKLKEAEEKLSELTDKDEKDSPSRQTVSK EVEALRAKLEGRKKVRELPDTVESARSEVIRCLRENDRRPLDCWKEVEKFKDEV |
| 4696 |
PF07937 (DUF1686) |
 Estimated TM-score=0.51; hard target. |
>Protein of unknown function (DUF1686) (Length=179) VGVSWNTALYSVAVAGIIAYQLWVLSRSYRTEGVSGMLRQDWSAIGCMVPMMVGLPHAGMIRSGVYSLTMAASGAVMLYV QDAMRSGMSLKQVCAIGGGNLVLAVACLAGSKKLFMPGACGLSQRICVGLVVFVALLVVVSYAEKGRSGGKKYSSTVGSV VFVMTLAAVTLGSWVCGRD |
| 4697 |
PF07898 (DUF1676) |
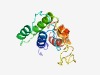 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1676) (Length=156) NCDDSYSLTCLKLDVVSWVDKLSENEDISVLPGVSIVRENSSAKVTTSDIVTELAREFPDDPNARLDAFMLKKVSGFLNN HSIKLNLWNAAQDESAGTARKKDKKGGDGMGMILAAGAMMKGMLMALALGGIAALAGKALMTGLISLLLSAIIGLK |
| 4698 |
PF07777 (MFMR) |
 Estimated TM-score=0.51; very hard target. |
>G-box binding protein MFMR (Length=93) MGSSDDVKSPKSEKTSPPATEHNGVHMYPDWAGIQAYYGPRVALPPYFNSAIAPRHPPPPYMWGPPQPMVPPYGAPYGAI YAPGGVYAHPAIP |
| 4699 |
PF07600 (DUF1564) |
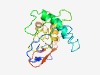 Estimated TM-score=0.51; very hard target. |
>Protein of unknown function (DUF1564) (Length=166) MDILLFDSNQKIQSTLIENAVGTDSLLVPNVYWDRLNLKEKKALRGKLPFLLRKYSKQIASMKRLHNRAGKIKYNRSVGK MKKLSIRVHTGVWATLGVLAAAHGVSRCYLFNYMLWLEDLGVGDSTVETLNRGVPSFHWTYKMIWKINRRQNLISRELEF EPNPMT |
| 4700 |
PF07577 (DUF1547) |
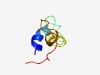 Estimated TM-score=0.51; hard target. |
>Domain of Unknown Function (DUF1547) (Length=56) DILAAVRKHLDKVYPGENGGSTEGPLPANQNLGKVISDMEQTGTAEETIVSPGNGS |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218