

Go to page (83 pages in total): [First page] << < 47 48 49 50 51 52 53 54 55 56 57 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 5101 |
PF12498 (bZIP_C) |
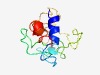 Estimated TM-score=0.49; very hard target. |
>Basic leucine-zipper C terminal (Length=118) TLRAKVKMAEDSVKRVTGMNALFPAASDMSSLSMPFNSSPSEATSDAAVPIQDDPNNYFATNNDIGGNNNYMPDIPSSAQ EDEDFVNGALAAGKIGRTASLQRVASLEHLQKRMCGGP |
| 5102 |
PF12438 (DUF3679) |
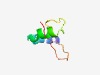 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3679) (Length=56) KCFLLVTIMFLGVLFGMQQANNGMLSMKGYHDPSLKGAFTLTDGKNNEKEASILGE |
| 5103 |
PF12425 (DUF3673) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3673) (Length=53) RFTPEDIITTARRMGYVEDDKGYVEEEPAHAKPVAVYANKKMVYAAKSQLPRL |
| 5104 |
PF12398 (DUF3660) |
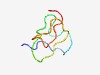 Estimated TM-score=0.49; very hard target. |
>Receptor serine/threonine kinase (Length=42) ETPFVDQVRSQDLLMNEVVISSRRHISRENKTEDLELPLMEF |
| 5105 |
PF12378 (CytadhesinP1) |
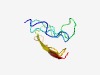 Estimated TM-score=0.49; hard target. |
>Trypsin-sensitive surface-exposed protein (Length=67) NPQTPTRDQTGQITFNPFGGFGLSGAAPQQWNEVKNKVPVEVAQDPSNPYRFAVLLVPRSVVYYEQL |
| 5106 |
PF12232 (Myf5) |
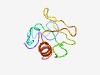 Estimated TM-score=0.49; hard target. |
>Myogenic determination factor 5 (Length=73) ENYYTLPGQSCSEPTSPTSNCSDGMAECNSPVWSRRNSSFDSVYCPDMPNVYSTDKNSTLSSLDCLSSIVDRI |
| 5107 |
PF12045 (DUF3528) |
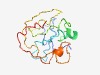 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF3528) (Length=135) ADFSSLPSFLPQTPSSRPMTYSYSSNLPQVQPVREVTFRDYAIDASSKWHHRSNLSHCYSAEEIMHRDCLPAPTTMGEMF AKNNSTVYHSSSNSTSNFYSSVGRNGVLPQAFDQFFETAYGNTENPPSDYSGDKN |
| 5108 |
PF12020 (TAFA) |
 Estimated TM-score=0.49; hard target. |
>TAFA family (Length=90) AGTCEIVTLDRDSSQPRRTIARQTARCACKKGQIAGTTRARPACVDARIVKTKQWCDMVPCLEDEGCDLLVNKSGWTCTQ PSGRVKTTTV |
| 5109 |
PF12005 (DUF3499) |
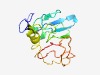 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF3499) (Length=123) MRRCCRPGCKNPAVATLTYVYSDSTAVVGPLATVAEPHSWDLCETHASRITAPKGWELVRHEGGFSSSTPDDDDLTALAE AVREAGMRRRTPEPEQRRYPDQPPAPPRPSRTGRRGHLRVLPD |
| 5110 |
PF11985 (DUF3486) |
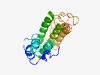 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3486) (Length=182) RGRASKVDLLPPNIKTQLAMMLRDKQYSQTQILEEINDLIRDCGLDESYCLSKTGLNRYANRMEQLGAKIRQAREVAEVW TKQFGEMPQTDIGKALMEMVKQIAFETSLKLGEQEGGIEPKQLALLSSAIQRLEQAESLSYKREQAIRKEIAQQAAETAE KVVTQAGLSADTVKMIKEQILG |
| 5111 |
PF11983 (DUF3484) |
 Estimated TM-score=0.49; very hard target. |
>Membrane-attachment and polymerisation-promoting switch (Length=71) VDVIAQKAVTGEELLRRKPIDTGRTTVVSESVAYSETNNIPEIKKPQDNVEPKQKENLGDKVRNIFGSMFD |
| 5112 |
PF11891 (RETICULATA-like) |
 Estimated TM-score=0.49; hard target. |
>Protein RETICULATA-related (Length=168) RVAADPQFAFKVLMEEVVGVSSAVLGDMASRPNFGLNELDFVFSTLVVGSILNFTLMYLLAPTLGSTSTTLPFIFANCPT SHMFEPGPFSVMNRLGTLVYKGTLFAAVGLGAGLAGTALSNGLIKMRKKMDPNFESPNKPPPMLLNAVTWAIHMGVSSNL RYQTLNGI |
| 5113 |
PF11878 (DUF3398) |
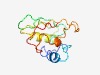 Estimated TM-score=0.49; very hard target. |
>Domain of unknown function (DUF3398) (Length=110) KVIEPLDYENVITQRKTQIYSDPLRDLLMFPMEDISISVIGRQCRTVQSTVPEDAEKKAQSLFVKECIKTYSTDWHLVNY KYEDFSGDFRMLPCQSLRPEKIPNHVFEID |
| 5114 |
PF11867 (DUF3387) |
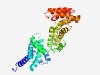 Estimated TM-score=0.49; hard target. |
>Domain of unknown function (DUF3387) (Length=327) SGGRGRTALDQEEAVAVMQEKYEICCDLFHGFDWSAWKTGAPEERLALLPAAQEHILAQPDGKERFVKAVLELSKAFALA VPHEEALRIRDDVAFFQAVRSALVKRSPSDARPEEELDHALRQLVDRAVAPEGVVDIFAAAGLKKPDISILSEEFLAEVR DMPQKNLAVELLRKLLQGEIRTRRRKNVVQARRFSEMLERAVRRYQNRAIEAAQVIEELIALAREMREADRRGEELGLSE EEVAFYDALAANESAVEVLGDKTLRQIAQELVRLVRENVTVDWAQRENVRAYLRVLVKRTLRKYGYPPDKQEEATQTVLK QAEVLSE |
| 5115 |
PF11808 (PhoR) |
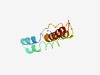 Estimated TM-score=0.49; hard target. |
>Phosphate regulon sensor protein PhoR (Length=86) SWKKLAWELAFFYLPWFIIGWLFGGLPWLLLGATVAMLLWHLHQQIKLSDWLWKDKSLTPPSGTGSWEPLFNGIYRLQKR NRRRRK |
| 5116 |
PF11805 (DUF3326) |
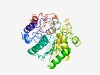 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3326) (Length=336) TVVLIVPTGVGASIGGYAGDALPVARAIANCCDRLITHPNVLNGASLYWNLPNGFYVEGYALDQFAANSWGLRPVRNNRV GLILDQGIEPDLRVRHLQAAEAARATLGLNLTDYVVTDQPLGVELRIADSGASWGTIQNPDSLLRAADVLIKQEKADAIA VVARFPDDCSSEVLHNYRHGQGVDPLAGAEAVISHLVVRTFQIPCAHAPALLPLPLDPDISPRSAAEELGYTFLPCVLVG LSRAPQFITNKSQQPRPEDIWADQVDAVVIPASACGGSAILSFSQLPVKIIAVEENQTQMQVPPEKLGIKALQVNSYLEA LGVLVADRAGININAL |
| 5117 |
PF11786 (Aft1_HRA) |
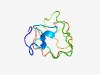 Estimated TM-score=0.49; very hard target. |
>Aft1 HRA domain (Length=72) NTGFNWGINSLRSGPLSPAMLQGPAQSAALAFVDSHLRSGLTPNESGIRTGLTPGGSGSIFPAPSPTTAAIF |
| 5118 |
PF11770 (GAPT) |
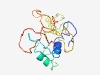 Estimated TM-score=0.49; very hard target. |
>GRB2-binding adapter (GAPT) (Length=156) MLKICGNTSVAVSIGIFLLLLLVICGIGCVWHWKHQNTMRFTLPKFLQRRRSRKKDYTKTFSLSPQFIGPRHKTSVQTQD RHSAGKDTNIHDNYENVKVCPPKAKGETDKKLYENTWQTNPEEHIYGNETPCDYYNFEKPSTSEAPQEEDIYILPD |
| 5119 |
PF11759 (KRTAP) |
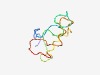 Estimated TM-score=0.49; hard target. |
>Keratin-associated matrix (Length=66) MSYYSGYYGGLGYGHGGSRGLGCGYGCGCNSFRRLGYGCGYGYGSGYGNGSGYGYGCCRPSCCGGY |
| 5120 |
PF11694 (DUF3290) |
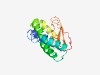 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3290) (Length=144) NFYGIDYLESQSNLNDYIKYFFIFSALIVLIVAFSLYMRHRIQTKYRDLSIITFLLLLFLLGVQYSDYTQNQNKNSQSSQ MVNFVKQIAKEKGVDQEEILVNSTQLVDGTLVKIEDSYYKTTLSGDQTSYVLEEAHLMNPEITI |
| 5121 |
PF11690 (DUF3287) |
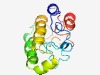 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF3287) (Length=134) SELESNIKEHESNLLLLDQVHEEDFSEDPLFLSGDDFEPAGVLFPHTAFHFPPKDKKEAYDYMDKVVDKHRRLCDERQRL GLKRQTLEARIARVERKVREMESDPFQWELRNFDCVAKIPTMLWMYLRARGQVL |
| 5122 |
PF11660 (DUF3262) |
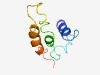 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3262) (Length=78) MNPSQQAAFEAAAGPGMSPSVLNLLCIGALLAVLFLWAAWGLVDVYRGWANDSVRAAKLGQFAVRAVILLVVCIWMFA |
| 5123 |
PF11394 (DUF2875) |
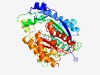 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2875) (Length=438) AQSGGKFVLEVRGLGLVTGKETNEEIWKAIETKADNHSTYMSQNSADYPANEDERMTEVGLSTRISFKYGARHSVEYWPV PVFIWEPPKASHVSRPANELSGIRQEASLGVTLLLWQEDANTNDGSTIIEKLFAFFDAHPDIPEAIIVTFDGAATRDLNQ TPGYVDIFKQSNIPTMPDSMVTLLVSRSDRVDRLIRPYAVEQTENVDKNTTEYDITKLWNFFWEKNNGEGPGSFEAYYQE QQKAAGIQPRAFLGFMSAQWWQTQLPDFWKTISNKGPGEFKPTPYIPVRWTTWQVRQFDNAPLLGYLHRPIDVKLADAHG KPLKTAQQAQALKAGWQQAVDTLPTGETPKRIFYDTTGDRAWVAPINQALAQSGPSAPSLDDVKEGYDIGRRIGNTGISS PLVQIGLGLIASYHEGGASATIHRRPNGTATIVMVSPP |
| 5124 |
PF11365 (SOGA) |
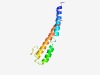 Estimated TM-score=0.49; easy target. |
>Protein SOGA (Length=89) SADLKCQLHFAKEESALMCKKLTKLAKENDGMKEELVKYRSLYGDLDSSLSVEELADSPHSREAELKVHLKLVEEEANIL SRRIVELEE |
| 5125 |
PF11273 (DUF3073) |
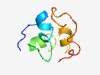 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3073) (Length=63) GRGRAKAKQTKVARELKYFSPDTDLNALERELRSSSSTHAAENAAADVDQGDDDDYDDYADWA |
| 5126 |
PF11241 (DUF3043) |
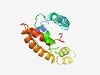 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3043) (Length=157) GKGAPTPKRSVQQAARKRPLVPNDRKAAREQGKSAARDDRARMRQALDTDDERYLPLRDKGPHRRYIRQFVDARWNIGEF LMIAALVFVVLSFFQSLTVQSAVLFAFWVLIMAVILDCFLLRRKLKSKLTAKFGGPLSGDLWYGMTRALQLRRLRLP |
| 5127 |
PF11233 (DUF3035) |
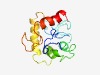 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF3035) (Length=139) RRSIGLDRQSPDEFTVVSRAPLTMPPSISNLPKPRPGAPRPQDVTPTAAAAGAVFGGASRAVNKAGGSAGESALVAQAGT KGIDPDVRAKVDQETTALIIADKSWVDTLLFWQKQEQPFSVVDPAKEQQRLREAQAQGK |
| 5128 |
PF11223 (DUF3020) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF3020) (Length=49) NKERKRKWRNANIVKNWENDVRARLKKRSKTDFIDLTQDEKDEWVEVEF |
| 5129 |
PF11135 (DUF2888) |
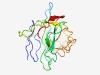 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF2888) (Length=144) GEPYTCKGDLCEIPFNRNFTIDLVNLSVSTEFQVKITMTPHHDLGTFVVEPKNVFSIKRAVKGDAAFKVERAAGWLPDTP QVLTLFVYERLNPVEWHSECMYENLETDGGTVIVPGEATGQRFGTATEVPTMFLFKRMFVVKGV |
| 5130 |
PF11125 (DUF2830) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2830) (Length=54) FKHEDYPCRRQQRSSTLYVLIFLAIFLSKFTNQLLLSLLEAVIRTVTTLQQLLT |
| 5131 |
PF11041 (DUF2612) |
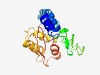 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2612) (Length=180) YTGLITSEHADKPKFSAMVAAVAQCFVDQQNALGGFIPAFDLDQAVGDQLDIIGLWVGVSRRVNTPLTGVYFSFDITGVG FDQGVWQGPFDPSTEVTLLDDDTYRTLIRAKIGANHWDGTLGSSAAILNQIFESGTHVFIQDNQDMTMTIGVSGTIPSAI LLALLKGGYIPLKPEGVLVN |
| 5132 |
PF11021 (DUF2613) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2613) (Length=55) KFLVPGAVSAVIGIALGTVATLGITAAAQQNTRPEVDRSGNADSSLLNQVEYGSR |
| 5133 |
PF10972 (CsiV) |
 Estimated TM-score=0.49; very hard target. |
>Peptidoglycan-binding protein, CsiV (Length=247) LFTFLSCLIFGSLSAEDRTYDVEMIIFENSRAPISNTNYWKPEILVPNLSNTLAFPGDEAAEGHPSAPSAASKQQFIKLK SDTYFLNTQKQALSRSSRYNVLTHLRWRQPGLAKANALPLRIKAGTPFPLYLLSQPGHSLSTTTKPEKNFFFARDRQPEL SELVSPIQGYPLDGSVTIYLGRYLHIVTDLILTLPTPESLPVSTGTIQESDVRLGKQSYRLNIHRRMRSKEIHYLDHPKF GLIVYIE |
| 5134 |
PF10883 (DUF2681) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2681) (Length=94) MINLSMLGGFAIAVLILVGYVYWRVQKARRRERALADALEKMTQQNEQLKTEKAVAEKQVKNYKVKQRNEENANSLNRDS IIDGLRENNDLRPD |
| 5135 |
PF10827 (DUF2552) |
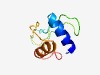 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2552) (Length=79) MEQKLKAMKNTAQNKTWVSFLNQNHPYTLLHWSIGGAESVKKDVWLLQDEMTFETQEFTTIDLAIEWIREHMDDITDVL |
| 5136 |
PF10655 (DUF2482) |
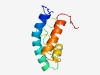 Estimated TM-score=0.49; hard target. |
>Hypothetical protein of unknown function (DUF2482) (Length=97) KNYKDMTQEELRDLLAEKNGELYELVNEIDEETEFDVLLFSTVGVNNGDCISSSHCALGNVVGLANLLKNRDGFQDVTNV IKMHELQKILGNDDDKE |
| 5137 |
PF10654 (DUF2481) |
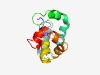 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF2481) (Length=126) TVMEITKSKARQREIISYIANNDVELDELLKLQKELNQLMNENTIEKQKTYWTKTFDRIVKKKKWAEITIREFADLRNAG LTCYAIAEHFKVSKAVVFNYTQRNKKEYYQIFDMNEYQKNKEIWND |
| 5138 |
PF10570 (Myelin-PO_C) |
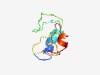 Estimated TM-score=0.49; hard target. |
>Myelin-PO cytoplasmic C-term p65 binding region (Length=65) LRRQAALQRRLSAMEKGKLHKPAKDSSKRGRQTPVLYAMLDHSRSTKAASEKKAKGLGESRKDKK |
| 5139 |
PF10567 (Nab6_mRNP_bdg) |
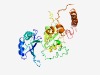 Estimated TM-score=0.49; hard target. |
>RNA-recognition motif (Length=327) VSYKMLPKGDDEYRTRSLLFENVNSSVDLYSFVSKYSKYGPVESIYMFRNRADKSKNSVLLSFLSRKICLDSYNSVLQRL AEFKTGLKSKSLTVSFVSLKYKRHVEPEEEDQIGEDNDPGDGDDAYNVSVVASLQYDIVNRGATRSLALNLDDECDKDEL LKNRLDFLSPSKNQRYVLESIYLIKAKEATRHFPKNYAILTFLNISMAVEVLDYLRNSEQNFESSQCFFVSVSPHSQPSF ESNTSFDHTGGAKNNSAGGLVGNAPNGNLGSYTSKNASLSSLNSFGSYISLDQEIDLLATSLQGSHTVIVAVDHYPQPFF AEYTDHL |
| 5140 |
PF10504 (DUF2452) |
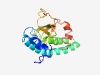 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF2452) (Length=155) VALVESNANPKGVQLVSTYQTNRMGDPSDLVELAQQVQKADDFIRANACNRLTVIAEQIRMLQEQARRVLEEAKRDADLH HAACNVVKKPGNIYYLYQRESGQRYFSILSPKDWGPSCPHEFLAAYKLQHDMSWTPYENIEKRDAEISIIDKLLN |
| 5141 |
PF10380 (CRF1) |
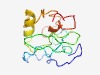 Estimated TM-score=0.49; very hard target. |
>Transcription factor CRF1 (Length=113) DESTDEDDNLPPPSSRSKNIGTKAKEVVSANVVGLKPPKLGTWETDSKPFTIIDGLSTKSLYPLMQEHQHLLEQQQRAQS QSPEYHSSHEATTVNGDELTLNELLNMSELEDD |
| 5142 |
PF10330 (Stb3) |
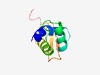 Estimated TM-score=0.49; hard target. |
>Putative Sin3 binding protein (Length=89) ITPAMLAKYHLPEILLQHGPLAIRHVMGYLTTSVPGFSGIPPAKARRLVVAALEGRGSDDKSDVVYEKVGWGRWDARRRG EPSRDSSRH |
| 5143 |
PF10295 (DUF2406) |
 Estimated TM-score=0.49; hard target. |
>Uncharacterised protein (DUF2406) (Length=63) AMNEAEPSEVAAHVKSSLASLRNVQHKDVFGNPIADPDRSNPTRSRWERPLDTIRSFEAAIDG |
| 5144 |
PF10218 (DUF2054) |
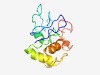 Estimated TM-score=0.49; very hard target. |
>Uncharacterized conserved protein (DUF2054) (Length=123) LTCRNSVQGKVLIVDDRGYVCSRGNVLPSGCCNIEAENTKRYSCETCKESGCCSIYEYCISCCLHPDKKQVLQNVLGKAS ETLNVLFASVTDHFELCLAKCRTSSQSVQHENTYRDPKAKHCY |
| 5145 |
PF10217 (DUF2039) |
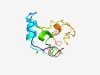 Estimated TM-score=0.49; hard target. |
>Uncharacterized conserved protein (DUF2039) (Length=88) QKYQNKTAFKNDLHDTSHRTKIINSLDITGVCKRCKDIIEWKIKYKKYKPLTAPRKCVNCEQKTIKHAYHMLCSKCATEK QVCAKCCK |
| 5146 |
PF10199 (Adaptin_binding) |
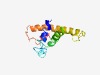 Estimated TM-score=0.49; hard target. |
>Alpha and gamma adaptin binding protein p34 (Length=119) ELEGMARIREVLHTNEWTSSNHGDLSANFLLESDEESGFDKEVSELEREMFGLRLAIEKGCDYNGDNEGEGGIRNDSDVD ELHVENMDGLMLRMQAIKDMAAELPEAERKAFAVKAIND |
| 5147 |
PF10062 (DUF2300) |
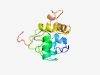 Estimated TM-score=0.49; hard target. |
>Predicted secreted protein (DUF2300) (Length=123) GRSREEVYCCENGQTIGRDRALVRSCGLYFEPQRLALQPEAWRGYWQARQAPAWLLDLERLQPQTRVPVKELLEQLQQLP AQGEARRVLLDVALQAGDGHAVASLGGRLRVKTWSWLDDHDPQ |
| 5148 |
PF09943 (DUF2175) |
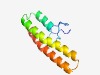 Estimated TM-score=0.49; hard target. |
>Uncharacterized protein conserved in archaea (DUF2175) (Length=101) TKWTCGFCGKPIYWDELFTFLSNKAVVHYTCLRERATKTSKINPEVLKVVLDSLEDELNKIVIYKQRLNQVGDNEDIKKV LDQTEKDAEKNAAQLTRLVEK |
| 5149 |
PF09929 (DUF2161) |
 Estimated TM-score=0.49; hard target. |
>Putative PD-(D/E)XK phosphodiesterase (DUF2161) (Length=116) ITVRLADQLVEVHADPAPFTPRKMKSRKDRLLREFARRVGDPNTGGSTRVTLVTAYRQDAMRCAQYLNENGPSRGAEVAK ATGVERATRIMADDHYGWFERVERGIYGLTPKGTEA |
| 5150 |
PF09880 (DUF2107) |
 Estimated TM-score=0.49; easy target. |
>Predicted membrane protein (DUF2107) (Length=73) FYAGIILAVVGTLGACWGPGVKDPVVRTMNTEVASVGVSLILLCYNHTLALLTMIATSVICTLILFRAISRLE |
| 5151 |
PF09872 (DUF2099) |
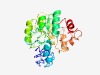 Estimated TM-score=0.49; hard target. |
>Uncharacterized protein conserved in archaea (DUF2099) (Length=254) HVMEILGMARVVVKDGKVTEVGEPQLDWCPLFEKARGIKHINSEEIRKNMEFRIRDFGLFTPQRKLDMDVFVGFGASEVM MTGLNRGMLDATVTVCDGAGTVITANPRLVQGMGARISGLVETEPIPEVIAGIEQRNGIVLDPQHASIDPVQGLQKAAQI GYKKVAVSVIDALTASKLREIESAQGLDVCIIAAHTTGLTKEHASELLKYTDIVTACASRNLREIIKPIAQVGTAVPLFA LTQKGKELLLERAK |
| 5152 |
PF09869 (DUF2096) |
 Estimated TM-score=0.49; hard target. |
>Uncharacterized protein conserved in archaea (DUF2096) (Length=169) VEQTWFVLVELLTDLRKRDVDVPTSITEDVRLVRTSINFYKSDPENPEMMKELKRINDMLNSIQEELLELAETVSSDYPA QWIEKLKRAARGEEVHRPPQTKSKFIVGAPPGFAAARVHLREPLAEDRVQDIAETHSLIIEFEEDDLIAIYGDSEDIKKG LREIGSFFR |
| 5153 |
PF09853 (DUF2080) |
 Estimated TM-score=0.49; hard target. |
>Putative transposon-encoded protein (DUF2080) (Length=50) VPFNVATELKVNNICGFYKREVKPFGTSAKVDCPKEHLGKTVYLVILDND |
| 5154 |
PF09826 (Beta_propel) |
 Estimated TM-score=0.49; easy target. |
>Beta propeller domain (Length=547) FSTTNTQVQGVDEADIVKSDGTYLYQSTQNEVRIIKAYPATEMKIASRISYDNGKFVPQEMYVDDRRLIVIGQADETISY PPDPASKRGIPRYQGMQSVKTMIYDITDKKQPRLIRQLELEGQYMSSRKIGASLYVTANHYMDAYRILQEKAEVPGPTYK DSVHHNQYQTIPYSEIRYFPDHFRPEYLIVAGVNLDDTKQEMSLNTYLGAGENIYASTKNLYVAVTQNNVQPVAVKQKGA TTPLIAPPMESDTTIYRFAMNKGDIRYTGKGSVPGRILNQFSMDENDGYFRIATTSGNMWRDDEHTSKNNLYVLDNKLEV FGKLEGIAPGERIYSVRFMGDRAYMVTFKKVDPLFVLDLKKPSAPSVLGALKIPGYSDYLHPYDENHIIGFGKEAEADKD WAYYQGMKIALFDVSDVTKPKEKFKTVIGDRGTDSELLRNHKALLFSKEKNLLAFPVTVHERTSEQKAKNDMREYGHFTF QGAYVYQLDLQKGFQLTDQITHLAKEDIARAGEGWYDSSSNIKRILTIGDTLYTVSDDFVKTQQLST |
| 5155 |
PF09813 (Coiled-coil_56) |
 Estimated TM-score=0.49; hard target. |
>Coiled-coil domain-containing protein 56 (Length=99) DPLNAKNGNAPFAQRIDPSREKLTPAQLQFMRQVQLAQWQKTLPQRRTRNIMTGLGIGALVLAIYGYTFYSVAQERFLDE LEDEAKAARARALERERAS |
| 5156 |
PF09747 (DUF2052) |
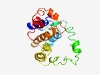 Estimated TM-score=0.49; very hard target. |
>Coiled-coil domain containing protein (DUF2052) (Length=197) NRRYQAMQQLLEKGEYFSEHEMMLRAPDLYQELVGQYLTEAEKKARDSYDVKNTSFSGILMNSLEKQQRDELLEGSKQEE IEEELPEATAPTPSAVDFEVPAACQKQWGGFDDEPIACSTSKNSTAVNASRIAKISTPEYYNPGERALLRNEFLSMMKER FLSGEDKDFDYTAVDDNAQLDDLKQIEQDEEDAYFED |
| 5157 |
PF09745 (DUF2040) |
 Estimated TM-score=0.49; hard target. |
>Coiled-coil domain-containing protein 55 (DUF2040) (Length=109) RLEMQKALEQDSTVYDYDAVYDDMQKQKLESTKKVLKGADKRPKYIHQLMKAVEERKKEQERRDERKIQKEREAEGEKFA DKEAYVTSAYKKKLEEQKEEEEREKRQAX |
| 5158 |
PF09600 (Cyd_oper_YbgE) |
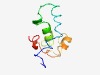 Estimated TM-score=0.49; easy target. |
>Cyd operon protein YbgE (Cyd_oper_YbgE) (Length=78) LRALSLLMALLLAGCMFWDPKRFAANSSSLEIWQGLLMMWAVCTGVIHGVGFRPRRLRWQAFFSPLPAWIVLIFGLAF |
| 5159 |
PF09577 (Spore_YpjB) |
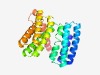 Estimated TM-score=0.49; hard target. |
>Sporulation protein YpjB (SpoYpjB) (Length=229) WKKLDQISDQALQLAKNGRFEEAKEVLTYFSDQFFQMNARDRLKTADELRAITITHENALKTVTASSLPEEERIDKVTQF RLVVDAIHSTHQPLWSEMETTVMGSFDQLEKAVEQGKEEAFTASLRQFLNHYELIEPSVKIDVQPEEAKKIDKEIEMLQT PAFQQLSQEDQQKHLKNMRADLQTLFDGVKKDEADPSLIWVIISTGGMIVLTLSYVGWRKYRGEKEKNR |
| 5160 |
PF09567 (RE_MamI) |
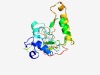 Estimated TM-score=0.49; hard target. |
>MamI restriction endonuclease (Length=182) HWSTITAQSSMIDTGYIAQHLVSLQTQIAGQGMRGKGDDLCDGSEVKSANFLDSLDKNGATAPRWNFMANTVDIMERFLN YTAIYLLSIDYSPEHNCRIRIWKVNIQKHTILQERYKEWMTKLGYPKFADTAKKPSVNFQLFPPNSGTNENYARHGNGRS NGFAKLHIPLENTPGSELIFKA |
| 5161 |
PF09535 (Gmx_para_CXXCG) |
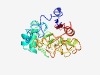 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (Gmx_para_CXXCG) (Length=238) RFFWLREDEAATAAFSGSFNAAHKWSLPGVKCSACGTTWSSWGNHYPLVDLSQLPERAAFEKARPEPFTEFSRLRELVRP LAPPDAELPPGAAFGPLVGTASGEFGPFTWQGTSLLLVRRDALERLQAEGVRGLLGGQTELRFRKKESPELLDLQLEPRG LLHPDCIPSDASPPCPTCGRNAFARPDAPILAAASLPADVDAFRLVNFATMIIGTDRFVDAVRRLGLEGIACRELPTR |
| 5162 |
PF09506 (Salt_tol_Pase) |
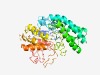 Estimated TM-score=0.49; very hard target. |
>Glucosylglycerol-phosphate phosphatase (Salt_tol_Pase) (Length=392) NLLIIQDLDGVCMGLVKDPLTRVMDSNYIEATRSFNGHFYVLTNGEHIGKRGVNLIIERAYGDPNLVKEKGFYLPGLAGG GVQWQDCHGEVSHPGVSDKELDFLARVPQYIEGSFKDFFANNNPNLDDTAIANYIQASILDNKVSPTANLNVFHEVFIEE EDYYFQLQQHIKQLMDDLLAKAGNEGLEDSFFIHYAPNLGRDEKGQEIMQEAKGKDSGTTDFQFMLKGGIKEVGVLGILN RYYYQRTGNYPLGEDFSVRNAPRNHHQLVALIKDNFDPEQMPTIVGVGDTVTSKAVKNNGKLSFGRGGSDRGFLQLIQEI GQEFNTGNLIVYIDSSAGEVKNRKPLKLEENNGQTYVIEGPGDPRDEDDPLTLNIAFPGGYQQYINCFQAAA |
| 5163 |
PF09472 (MtrF) |
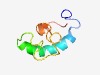 Estimated TM-score=0.49; hard target. |
>Tetrahydromethanopterin S-methyltransferase, F subunit (MtrF) (Length=63) GVPMVINPQMGPIEAVVEDIRYRAQLIARNQKLDSGVSATGITGFIAGFVFALVMVVIIPMFI |
| 5164 |
PF09429 (Wbp11) |
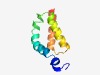 Estimated TM-score=0.49; hard target. |
>WW domain binding protein 11 (Length=79) ERNFNPVQAQRKADKAKAIKKGKSEVQNRRNEKLARRNPDRIQKQIDDLKAITTNGAKLSRHEEQVLEGLEKELRAVKK |
| 5165 |
PF09428 (DUF2011) |
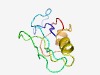 Estimated TM-score=0.49; hard target. |
>Fungal protein of unknown function (DUF2011) (Length=90) FEFRLFSTPTNTTSEPIHAQKIILDDDDSGKGNGGFVIPRRNSDYYFTTSAQGEAKWGFEHAAVNGEDVLKAQKQRAWGL EVPWRVTVIK |
| 5166 |
PF09135 (Alb1) |
 Estimated TM-score=0.49; hard target. |
>Alb1 (Length=102) HSRAAKRAASPSLDADKSLASVPRPEPTATVKPHILSAQHNSGINKKSKPKHMTRAQKRRQEKGLERAELVMDRTEKKVA KSVGRGKTVQARRATWEELNQK |
| 5167 |
PF08822 (DUF1804) |
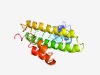 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF1804) (Length=167) MAHDEATRRAARSAYAYDQLPLERAAAKAGVPASTLARWKREAKAEGDDWDKVRAANALASGGVEDVARQMLTDYVVQHK TLIDHLRDETTGERMSPRDKADILASLADSFNKTVAACRRVMPETNELAVALEVLGLLGDYVREHFPGHAPALLEVLEPF GYELSKH |
| 5168 |
PF08812 (YtxC) |
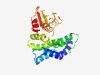 Estimated TM-score=0.49; very hard target. |
>YtxC-like family (Length=217) HYLADIISDIILTHWEDILLKDIIRENYYYFNEEEKKLIYDYALRHINREGQESGPTIYWANRKSRILHKLIDFLHYNNR IIIEGFIRFRLKEYVSELKEAADKAVDDFLIEREYQEFVQLLKYFVEIQEPKVEVVHVLIRGNGVFRLYDGNMQPIRSDY MEGFMIDLSDNEINYEDLLISALITIAPREITLHCKSSNGMSTTLDTIRNVFTGRVK |
| 5169 |
PF08735 (DUF1786) |
 Estimated TM-score=0.49; hard target. |
>Putative pyruvate format-lyase activating enzyme (DUF1786) (Length=260) MGGGASSKAIKEHLSYGYRVYATEEAALTLKDNLNKVKEIGVEIVESPPQNCDQIELKDIDISGLKEALALFEEELPADY ALAVQDHGFAPDSSNRLFRFGHWEKFLKSGGYLADLIYDENSVPDYFTRMHAVMEAVQTQSVRSGHKVWITDTGAAAILG ALEDNNVNREVTQNNGMIVNIGNQHTIAFLVVGQRVLGVFEHHTKKLTEEKLQHHLDRFVLGELSNEEVLEDKGHGCMRV PEADEHKFNFIAVTGPRRNL |
| 5170 |
PF08573 (SAE2) |
 Estimated TM-score=0.49; very hard target. |
>DNA repair protein endonuclease SAE2/CtIP C-terminus (Length=111) AFSEVVRNKDDRACLPGCTDMECCGKSFRAMALAERGNGRRTAEQLAEDKKLLEDYLGEEAHRLFSMTKEERDELWLEAK TKELANRIGKHRHRFSRMKTPPGFWRTDFPD |
| 5171 |
PF08552 (Kei1) |
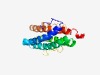 Estimated TM-score=0.49; hard target. |
>Inositolphosphorylceramide synthase subunit Kei1 (Length=190) TFLYVMSLRTGASLITLSLLLNKISGLYGLLALLTGYHLSPVQLSMYLYSLVALGLAALLFPHIRKQTPLQCLALAWLYV FDSVINAAYTAAFGVTWFLVISQHYDSDAASGPGSETIEQTAGFTNPKYDTSDYSSNDHEARSALDDGLSNAVSQPESFQ SIVCICLLWIVRAYFVFVMLAFARQALRRY |
| 5172 |
PF08283 (Gemini_AL1_M) |
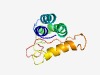 Estimated TM-score=0.49; hard target. |
>Geminivirus rep protein central domain (Length=106) GQQTANDECAEALNRSSKEEALQIIKEKLPKDFLFCYHNLVSNLDRIFTPAPTPFVPPFQLSSFTNVPEDMQEWADDYFG VSAAARPMRYKSIIIEGESRTGKTMW |
| 5173 |
PF08219 (TOM13) |
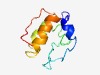 Estimated TM-score=0.49; hard target. |
>Outer membrane protein TOM13 (Length=83) PSDSENYDANHSNQDSSDEPTSDSSSPMILYQPPTIWSLMRGAAINLLLPFVNGLMLGFGELFAHEAAFRLGWSNTKVFP THR |
| 5174 |
PF08112 (ATP-synt_E_2) |
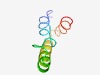 Estimated TM-score=0.49; hard target. |
>ATP synthase epsilon subunit (Length=56) MSEIDKSTIDKYINILKSKLDQKKNELLSKINMEYEKTLKQRLDELEKLKGNILKE |
| 5175 |
PF07893 (DUF1668) |
 Estimated TM-score=0.49; easy target. |
>Protein of unknown function (DUF1668) (Length=332) RQFVHMVTYSCNRGIYGLHRINPSRFFHPKDAAVGRPATAVEDVRLPRPAMSFLRPYETHIIPTEFMPLGHNNDKIVSVD ETGRSILYDVASHAIGTLPGRKAEPKFGCCVSLTVGDNLYVMQEIPRRAQCFEALVYCKSPKSDEEQENRWSWRPQPPPP YLHSVGYHCSGGDITAYAVVGDSHILVSTESYGTYSFDTVSSEWSKAGDWALPFNGRAEYVPEHGLWFGFSACDDGLLGA WDLSTVVTQQQPPVAHVLQDGFSVPEGFSSYVVHLGAGNLCVAKLFEKARRETSDQDCYLFETTRRFAMMTGVEVQRRGE ELNIIKHRSSRY |
| 5176 |
PF07816 (DUF1645) |
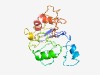 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF1645) (Length=190) AEELFDGGKIRPFRPPPRSPRKKKEIDTFATEVENTRERTKNDRGRERVSSLSSSNSGRRGARSLSPHRVSKYPWEEEQQ LQQKTEKKSPISSKPALSSTTSKGCKKWSLKDFFLFRSASEGRAADRDPLKKYIALFKRREEVKSSSVRSIEGSNSGSNS KRRGPISAHELHYTVNRAVSEDLKKKTFLP |
| 5177 |
PF07663 (EIIBC-GUT_C) |
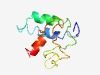 Estimated TM-score=0.49; very hard target. |
>Sorbitol phosphotransferase enzyme II C-terminus (Length=93) LSPLLGPGAVIAQVVGVLIGVEIGKGNIPPQYALPALFAIDPQVGCDFIPVGLSLGEAEPETVEVGVPAILFSRLITGPI SVLIAYLFSFGLY |
| 5178 |
PF07560 (DUF1539) |
 Estimated TM-score=0.49; hard target. |
>Domain of Unknown Function (DUF1539) (Length=124) ARRSNDFIRYLIENSPQRWTFLNRLKNNISASTSSLTLRREWFKMLDVISNKTSPEITDREVQDVARACLFKINSILKDP TVAPERKEELLKYVASYYDSSPDVWVEAIQQEVFLQNTLNPQII |
| 5179 |
PF07547 (RSD-2) |
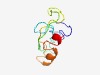 Estimated TM-score=0.49; hard target. |
>RSD-2 N-terminal domain (Length=82) FGDNPNYLTYNRDLGYADWYGIVDCSDSPPTGPGVYKAFIYVNDLNYQESGNPLFKVTGRLKFLANKDTSDTMMKLEIFE QN |
| 5180 |
PF07424 (TrbM) |
 Estimated TM-score=0.49; very hard target. |
>TrbM (Length=151) VLTGDTRLACEATLCLATSTRPSECSPSLSRYFSIHKRKWSDTVRARANFLSLCPVSDQTPEMRSLVNAMANGAGRCDAA SLNVTLLVWNSWDADGGRVLINNQMPGYCAAYTGHQYTDLGDLAPRYVGTPERGGYWVDARDYNAAVARYN |
| 5181 |
PF07406 (NICE-3) |
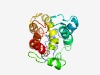 Estimated TM-score=0.49; very hard target. |
>NICE-3 protein (Length=182) NWLSGVNVVLVMAYGSLVFVLLFIFVKRQIMRFAMKSRRGPHVPVGHNAPKDLKEEIDTRLSRVQDIKYEPQLLADDDAR LLQLETQGNQSCYNYLYRMKALDAIRTSEIPFHSEGRHPRSLMGKNFRSYLLDLRNASTPFKGVRKALIDTLLDGYETAR YGTGVFGQNEYLRYQEALSELA |
| 5182 |
PF07403 (DUF1505) |
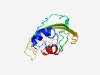 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF1505) (Length=122) MKFFILVASFLVILVAGAPTSTSDTTENLVTQNVKNCEEKKSTENEKAVIFFKTCTRAYTWQTRHNDECNISTYYKKTVT TTPETSTEPLNGVAQCTKTPCDASEKITVDCATAFGERLSEI |
| 5183 |
PF07380 (Pneumo_M2) |
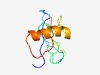 Estimated TM-score=0.49; hard target. |
>Pneumovirus M2 protein (Length=89) MTKPKIMILPDKYPCSISSILISSESMIATFNHKNILQFNHNHLDNHQRLLNNIFDEIHWTPKNLLDATQQFLQHLNIPE DIYTIYILV |
| 5184 |
PF07334 (IFP_35_N) |
 Estimated TM-score=0.49; hard target. |
>Interferon-induced 35 kDa protein (IFP 35) N-terminus (Length=76) LIDEITKKNIQLKKEIQKLETELQEATKEFQIKEDIPETKMKFLSVETPENDSQLSNISCSFQVSSKVPYEIQKGQ |
| 5185 |
PF07327 (Neuroparsin) |
 Estimated TM-score=0.49; easy target. |
>Neuroparsin (Length=102) MKPAAALAAATLLIAVILFHRAEANPISRSCEGANCVVDLTRCEYGEVTDFFGRKVCAKGPGDDCNVYASCGIGMDCRNK KCSGCSVKTLECYFESFPMSEE |
| 5186 |
PF07324 (DGCR6) |
 Estimated TM-score=0.49; hard target. |
>DiGeorge syndrome critical region 6 (DGCR6) protein (Length=193) RFGGAGYEVAELSRQQERHYRLLSELQELVKALPSSCQQRLSYTTLSDLALALLDGTVFEIVQGLLEIQHLTEKNLYSQR LKLHSEHRGLKQELFHRHKEAQQCCRPHNLPLLRAAQQREMEAVEQRIREEQRMMDEKIVLELDQKVIDQQSTLEKAGVS GFYITTNPQELTLQMNLLELIRKLQQKESESEK |
| 5187 |
PF07255 (Benyvirus_14KDa) |
 Estimated TM-score=0.49; hard target. |
>Benyvirus 14KDa protein (Length=123) MGMVDSLCVFVGRVITEGSESVEGVERFSIKFSDWKLFTTAVYVEYRQLGEKECSLKDVGRLHFNMSCVKCCQKLKCKKQ NKNHSKHVQNGYLRKVRNFSILGVCGDCCESFTLADEKHHVIV |
| 5188 |
PF07242 (DUF1430) |
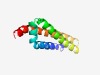 Estimated TM-score=0.49; easy target. |
>Protein of unknown function (DUF1430) (Length=100) IIGIASSLLLFYSVNLLYFEQFRREILIKRISGLRFFETHAQYMISQFASFVFGASLFIWRSRDVVIGLVTLSIFLVSAI LTLYRQAQKESRVSMTIMKG |
| 5189 |
PF07216 (LcrG) |
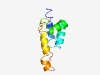 Estimated TM-score=0.49; hard target. |
>LcrG protein (Length=91) HFDEYDKTLKQAELAIADSDHRAKLLQEMCADIGLTPEAVMKIFAGRSAEEIKPAERELLDEIKRQRERQPQHPYDGKRP RKPTMMRGQII |
| 5190 |
PF07052 (Hep_59) |
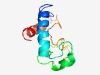 Estimated TM-score=0.49; hard target. |
>Hepatocellular carcinoma-associated antigen 59 (Length=100) QFSAETNIGDTDEEMMKYIDEQMKKRNGITIDKDGETISKYLTPEEAALLALPAHLRECSTKKSEEMLSNQMLNGIPEVD LGIETKIKNIEATEEEKQKL |
| 5191 |
PF07047 (OPA3) |
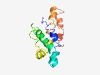 Estimated TM-score=0.49; hard target. |
>Optic atrophy 3 protein (OPA3) (Length=125) FPAAKLLILVVRQISKPIANRVKQRAKNSQFFRNYIIMPPAQLYHWMEVRWKMRMLNFGNPEQVPKLNEQMAIELGAELL SEGILMLVGVACLLAEYYRSSRKEAAKEAEKQRILNELENRLYEL |
| 5192 |
PF07038 (DUF1324) |
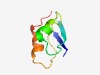 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF1324) (Length=59) MKCTLVFQSRFCIFPLTFKSSASPRKFLTNVTGCCFATVTRIPLSNKVLTAVDRSLRCP |
| 5193 |
PF06994 (Involucrin2) |
 Estimated TM-score=0.49; hard target. |
>Involucrin (Length=41) EPQEQELHLGQKQKQKLHEPELQLGKQQHQKPSEPELPLGK |
| 5194 |
PF06886 (TPX2) |
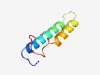 Estimated TM-score=0.49; easy target. |
>Targeting protein for Xklp2 (TPX2) (Length=57) FRCNERAEKRKEFYSKLEEKIHAKEEEQNTMQAKSKEHQEAEIKMLRKSLMFKATPM |
| 5195 |
PF06643 (DUF1158) |
 Estimated TM-score=0.49; hard target. |
>Protein of unknown function (DUF1158) (Length=79) MKHPLETLITAGGILLLAFLSCLLLPAPRLTLTLAQHLMTMFHLVDLNQLYTIIFCIWFLLLGAVEYYVIRFVWRRWFS |
| 5196 |
PF06502 (Equine_IAV_S2) |
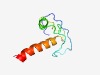 Estimated TM-score=0.49; hard target. |
>Equine infectious anaemia virus S2 protein (Length=67) MGLFGKGVTWSALHSVGVSQGEYQPLSPNKQNQQTHRKEIIWYINPIVIMIAIKKKWQRQETQDTKK |
| 5197 |
PF06363 (Picorna_P3A) |
 Estimated TM-score=0.49; hard target. |
>Picornaviridae P3A protein (Length=97) PYIDEYLNIEMSTLIEQMEAFIEPKPSVFKCFASRVGDKIKEASREVVKWFSDKLKSMLNFVERNKAWLTVVSAVTSAIG ILLLVTKIFKKEESKDE |
| 5198 |
PF06361 (RTBV_P12) |
 Estimated TM-score=0.49; very hard target. |
>Rice tungro bacilliform virus P12 protein (Length=110) MSADYPTFKEALEKFKNLESDTAGKDKFNWVFTLENIKSAADVNLASKGLVQLYALQEIDKKINNLTTQVSKLPTTSGSS SAGAIVPAGSNTQGQYKAPPKKGIKRKYPA |
| 5199 |
PF06323 (Phage_antiter_Q) |
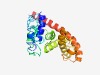 Estimated TM-score=0.49; very hard target. |
>Phage antitermination protein Q (Length=217) QYLQYVREQLMVATADLSGETKGQLLAWLENAQFDTKNYPRKKQRIWDEETESWITLNNPPIPGKQSLAKGSAIPLVKPV EYSTASWRRAVLSLDEHYKAWLLWNYSENTCWEHQVEITQWGWSAFAAQLDGKKMAGKTQERLRALIWLAAQDVKSELAG REVYQYKELAGLVGVSEKNWSETFTRHWLTMRAIFLRLDQASLLSVSESRSEQVAFN |
| 5200 |
PF06270 (DUF1030) |
 Estimated TM-score=0.49; very hard target. |
>Protein of unknown function (DUF1030) (Length=53) MVFIIHLGFKWGVFKIKFSELYIHGYTDIVVLVVYTVFERSAEAYMVYISSSL |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
umich.edu
| +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417