

Go to page (83 pages in total): [First page] << < 57 58 59 60 61 62 63 64 65 66 67 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6101 |
PF15431 (TMEM190) |
 Estimated TM-score=0.44; very hard target. |
>Transmembrane protein 190 (Length=132) RSGIKKFFYPRSCQGKMWDREACGGQAAIEDPTLCLQLHCCYQEGICYHQRLDENMRKKHLWTLSLTCIGLLLLIFLACF FWWAKRRGLYKNMKMPARLSHFRKSKVSGNVSNSTTNSFSLQKKEQSPLLAR |
| 6102 |
PF15399 (DUF4620) |
 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4620) (Length=113) VPRWPHLSSQSGVEPPDRWTGTPGWPSRDQEAPGSMMPPAAAQPSAHGALVPPATAHEPVDHPALHWLACCCCLSLPGQL PLAIRLGWDLDLEAGPSSGKLCPRARRWQPLPS |
| 6103 |
PF15252 (DUF4589) |
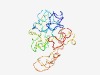 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4589) (Length=228) EVVSQIDKLTSDFDFELEPDDWTTATVSSTSSSDKAGVGGPFDLGHLDFMTADILSDSWEFCSFLDISTPSDSVDGPEST RPGAGPDYRLMNGGLPIPNGPRVETPDSSSEEAFSAGPVKGQPPQRTPGTRERVRFSDKVLYHALCCDDEEGDGEEEAAE EEGGLSPEPPHTEAPAGPLKPSSAPYKPRRSPLTGRRSGPTLVPEQTRRVTRNSSTQTVSDKSTQTVL |
| 6104 |
PF15226 (HPIP) |
 Estimated TM-score=0.44; very hard target. |
>HCF-1 beta-propeller-interacting protein family (Length=133) ILQQPLERGPQGRAQRDPRAASGASRGLDASSPLRGAVPMSTKRRLEEEQEPLRKQFLSEENMVTHFSRLSLHNDHPYCS PPMAFPPALPPLRSPCSELLLWRYPGNLIPEALRLLRLGDTPTPHYPATPAGD |
| 6105 |
PF15203 (TMEM95) |
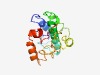 Estimated TM-score=0.44; hard target. |
>TMEM95 family (Length=151) CIFCRLPDHGLSGRLHQLCSQMEAQWKDCEASWNFPAFALDDTSMNKVTEKTHRVLRVVEIKGSLSPLPAYWRWLQKTKL PEYNREALCAPACRGSTTLYNCSTCEGFEVHCWPRKRCFPGSHDLWEARILLLSVSGTVLLLGFLSLSVEY |
| 6106 |
PF15176 (LRR19-TM) |
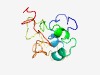 Estimated TM-score=0.44; hard target. |
>Leucine-rich repeat family 19 TM domain (Length=111) STTNPEHERIGKSWAFLVGVIVTVLATSLLIFIAIKCPIWYSMLLSYHHHRLEEHEAENYEDGFTGNMSYLPQTPEEETT VIFEQLHSFVVDDDGFIEDKYIDTHELQKEN |
| 6107 |
PF15152 (Kisspeptin) |
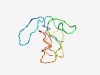 Estimated TM-score=0.44; hard target. |
>Kisspeptin (Length=76) PWEQGPRCAERKPAAAEANPRGTSSCQPPESSSGPQRPGLCTPRSRLIPAPRGAVLVQREKDLSAYNWNSFGLRYG |
| 6108 |
PF15151 (RGCC) |
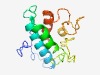 Estimated TM-score=0.44; hard target. |
>Response gene to complement 32 protein family (Length=142) MKSPTVQRSAGGAGAAGTGAAPSLDSVVAADLSDVLCEFDEVMADFSSPFHERHFEYEEHLKRMKRRSSASVSDSSSGFS DSESADSPYRHSFSCSDEKLNSPTVSTPSLSSPLVVPRKAKLGDTKELEDFIADLDRTLESM |
| 6109 |
PF15132 (DUF4568) |
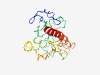 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4568) (Length=203) GGCGWKVWPWQGRAHFKPLGKKSVGKMCPVPSTVHQEPICCECQAKCRGYLPVPRFEATLPYWVPLSLRPRKQIQKMVQF NIPKTSEACPCPCHRFGGRLPMPRDQAVMPYWVPQVLRPHKKVVKRQQSFTGIQEPALGLRSWYNRWRICCDGHLLLKWQ QLQALHQHEPLAPGKGASIPAPLLPRSLLTLLQAVLRVVVAIR |
| 6110 |
PF15098 (TMEM89) |
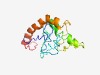 Estimated TM-score=0.44; very hard target. |
>TMEM89 protein family (Length=133) WSRPLWYQVGLDLQPWGCQPNSLEGCKGSLSCPGHWMGLGVNRIYPVAGVTVTTTMMLMLSRAVMQRRRSQATKSEHPQV TADPCAPWKRRAPISDRALLLGVLHMLDALLVHIEGHLQRLATQQRTQIKGTP |
| 6111 |
PF15079 (Tsc35) |
 Estimated TM-score=0.44; very hard target. |
>Testis-specific protein 35 (Length=205) MSAKKAELKKASLSKNYKAVCLELKPEPTKTYDYKGVKQEGPFTKPGGTHELRNELREVREELKEKMDEIKQIKGVMDKD FDKLQEFVEIMKEMQRDMDEKMDVLVNIQKNSKLPLRRGPKEQQELRLVGKTDTDPQLRLKKVDGADGTPLSLHKKMMAL QKTKKDPLDSLHQCGPCCEKCLLCALKNSYNQGKLPYQAWPPFSS |
| 6112 |
PF15063 (TC1) |
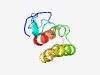 Estimated TM-score=0.44; hard target. |
>Thyroid cancer protein 1 (Length=76) YMSSESRRVSPSVHGNKFDTAHRKKAVANIFENVNQDSLMRLFQKTGDMKAEERVRSIFSYTQDPEETARALMALK |
| 6113 |
PF15021 (DUF4521) |
 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF4521) (Length=204) MATQEATPGSQSEESSALDLPTVYDIRDYVLQRPHQEADSEAFSSVEALSSPCSSDADPDTSNLNSEQNDSWTSENFWLD PSVKGQVKTKAEDDGLRKSLDRFYEVFGCPQPASGNALSAAVCQCLSQKINELKGQESRKYALRSFQMARVIFNRDGCSV LQRHSRDVHFYPSGEGSVSLDDEKPTPGLSKDIIHFLLQQNVMK |
| 6114 |
PF14982 (UPF0731) |
 Estimated TM-score=0.44; very hard target. |
>UPF0731 family (Length=78) FRFGTQPRRFPVEGGDSSIGLESGLSSSAACNGKEMSPNRQLRRCPGSHCLTITDVPITVYATMRKPPAQSSKEMHPK |
| 6115 |
PF14442 (Bd3614_N) |
 Estimated TM-score=0.44; hard target. |
>Bd3614-like deaminase N-terminal (Length=92) TSSVEDDDVAFLVAPSSGDADAPSVVWFATSRKTSPPWDSALVRLVLGAREAGGRANGNRWARARMLTTRALSDLDRAVV KVAAQRATAFDL |
| 6116 |
PF14389 (Lzipper-MIP1) |
 Estimated TM-score=0.44; easy target. |
>Leucine-zipper of ternary complex factor MIP1 (Length=81) RERKLALLEDVDKLKKKLRQEENVHRALERAFSRRLGTLPHLPPYLPPQTLELLAEVAVLEEEVVRLEEEIVSFKQGLYQ E |
| 6117 |
PF14287 (DUF4368) |
 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4368) (Length=67) ELQQEEKKSVDVKRFLAIVKKYTDLTELTPEILREFIDKIIVHAPDKSSGRRLQEIEIIYNHIGEFD |
| 6118 |
PF14238 (DUF4340) |
 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4340) (Length=191) WKIAGPFDASAPTPSVTAMLGNVSTLRAERYEVHAAKELDKYGLDKPYLKLVVREADKDKEAGKEHTLLIGKPTAEGAKT RYARVADGDAVIVVSEALIGAVDRGALDLLDRHLLKLDKEKIVRVESTGANPLTLNLVKDDKWEAETPQAKFAADNRVAA GVLTTWFNLTAQKFAAYGSKVDLAPYGLDKP |
| 6119 |
PF14073 (Cep57_CLD) |
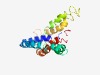 Estimated TM-score=0.44; hard target. |
>Centrosome localisation domain of Cep57 (Length=178) AILSALKNLQEKIRKLELERTQAEDNLRRLSQETSEYKKVLDKEIRQKDSSKTEVARQNRELSTQLAAAESRCTLLEKQL EYMRKMVKNAESERTTVLQKQASIGRERSLDHLELQTKLEKLDMLEQEYLKLTATQNIAQNKICLLEQKLREEEHQRKLV QDKAAQLQTGLETNRILL |
| 6120 |
PF14045 (YIEGIA) |
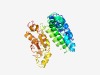 Estimated TM-score=0.44; very hard target. |
>YIEGIA protein (Length=280) ILFGVIIGTFTRLYMLRTDYRQYPTYLHGKVIHIALGFIAAALGTVAIPSIMEEEFTAITFLTLAASQFREVRNMERNTL TELDEFELVPRGKTYIEGIAVAFEGRNYLVIFTSFITTLAYIVVGWWAGIIAGVICLFISKKLMSGNKLNEIVDIEHHEL HFDGAGLYIDNIYIMNIGLPARQEEVLKYGMGFILKPKNFNVRSTIANLGQRQAILHDVSVALGVFRDSGTPALVPLAKR DLDDGRVGVFILPQEKDIEKAITVIGQVPVLENAIRMPTE |
| 6121 |
PF13968 (DUF4220) |
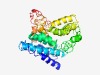 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4220) (Length=339) WLIYLSADWMATLVLSTLLRGSMELNNELIVFWTPFLLWHLGNPYNITAYSFEDNELWLRHFLGMVIQIAEAIYIYVRFR SNTILNAMALPLFIAGVIKCAERIWALRCASQKQLIKSFYSSPISKIPNENNVIGNLKEMIRTGLFDLSKKKDFIGDPSI PSEVKYLREADLSFHIFKPLFSDLPFQISQEFHDKMVYLSNYTIADEAFNFVGIELEFLYDLLYTKNPIQYGQHLASLIL RSIYFFFVISVLFAFSALLHKIKHSTVDTTVTYLLLSGAVLLEIYSFFMHLRCKWTMLRYAVPRDKLQKLYHRLVDHRLR SIRSRKGIQKMAQHDLIDY |
| 6122 |
PF13928 (Flocculin_t3) |
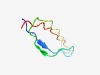 Estimated TM-score=0.44; hard target. |
>Flocculin type 3 repeat (Length=44) TAIITITSCHNDICEGTTVSATLGPVTTTVGGTTTIYTTYCPIS |
| 6123 |
PF13898 (DUF4205) |
 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4205) (Length=357) RMTVFGTSSAPPRGEWTRTPLAFGQAAQELPYGLRCPRNATRGLQSVVQAFIIKQFLFDNRPKAKSLPLDQMLKPTEAEQ QQSLWTAIAEILWTIGERQKVTIGLPGDIPHIQHSHSYFADGVTEKLYLFDFTSKEELQIFLKRYLYYFTEDPGPGAMLL LYAAIFTRGVPKVKTDLDAPKGAYLLGHLEEGSLNIVTLLLSGRATPYLHNGVVYVGDEDHYALPQFGILARATIGLLVF EGENEHLKGSSRQPGSRLKTPAYPIWVTLCCGHYGVLFNSNRELLRNYHAEKRFELHYYTCAGNCVSMTVDNRVQDFCPM TSIQRQSSSTNSKDKHEEVTNASPLERLIHCKWPEAK |
| 6124 |
PF13804 (HERV-K_env_2) |
 Estimated TM-score=0.44; very hard target. |
>Retro-transcribing viruses envelope glycoprotein (Length=169) PVTWMDNPIEIYVNDSVWVPGPIDDRCPAKPEEEGMMINISIGYRYPPICLGRAPGCLMPAVQNWLVEVPTVSPISRFTY HMVSGMSLRPRVNYLQDFSYQRSLKFRPKGKPCPKEIPKESKNTEVLVWEECVANSAVILQNNEFGTIIDWAPRGQFYHN CSGQTQSCP |
| 6125 |
PF13775 (DUF4171) |
 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4171) (Length=126) DTAMDLLKAITSPLATGAKPSKKTGEKSSSSSSHSESKKEHHRKKVSGSSGELSLEDGSSHKSKKMKPLYVNTETLTLRE PDGLKMKLILSPKEKGSGSVDEESFQYPSQQATVKKSSKKSARDEQ |
| 6126 |
PF13595 (DUF4138) |
 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4138) (Length=253) VQVTFAKTVHIIFPSAVKYVDLGSNYIIAGKADGAENVVRVKATTEGFPGETNFSVICEDGSFYSFNAKYAHEPEMLNIE MKDFLENEDTTDFSHTRMNIYFRELGNESPLLVKLIMQSIYKNNDREIKHLGCKRFGVQFLVKGIYSHNGLFYFHTQVRN SSNVPFDTDFIRFKIVDKKVAKRTAIQETVIDPVRSYNEILVIDGKSTVRTVYTVPQFTIPDDKVLVIELMEKNGGRHQT IRVENSDVVAAKV |
| 6127 |
PF13315 (DUF4085) |
 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF4085) (Length=206) YFTKEWFEEAQISRFLLFPETKKEWDENVAWHVSEGLHFEEIQLNRLQIAKKDLLRFLPAFFHPYIQDGTLNSQFPSEEL KKMADQWRMEYDERTKAKGISYRNYYNSIKCSLPENAIQLFEKSLHDARVTSIEMPMKDAFIIKLDCRGGYHYGTDVTLT FAGVKKLQPHSLSVGSIWLYDEVYLTDTGFELHVLFDGPLMEFTIS |
| 6128 |
PF13259 (DUF4050) |
 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF4050) (Length=165) SPFRFDSPDAIGAAVKASVADKRARRRRAVREEMAWNEGLACFEARRNAWTGARTRPSNEAATAVSDGSELSKDPSKELT AQKSKESQKSAASPASTTYPIETIVPLSAPLLPPGNPMRASITPAVYMSLYEKVIVHALTPSCPVNLSDMIRACVVGWKR DGEWP |
| 6129 |
PF13120 (DUF3974) |
 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF3974) (Length=126) MGFIKMVLLLIGTLLLIGFTIVVLLVYFGRKLYLSWAKPYKRAHESIEKLSNKSTPFLQEFTQHPLFYRWIRTEGKKEQK AFNTLFCVANQRTREQVFSMLPKDRQKKVHSMAKSTKKITNEDMDI |
| 6130 |
PF12526 (DUF3729) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF3729) (Length=107) HLWESANPFCGESTLYTRTWSTSGFSSDFSPPEAAHAVPVPDVGLPSGAPSSASDVWVFPPPSEGSAIVPPPETPVSKPA NPPCSITPRPPVRKPPTPPPARTRRLL |
| 6131 |
PF12444 (Sox_N) |
 Estimated TM-score=0.44; hard target. |
>Sox developmental protein N terminal (Length=83) SPGMSDDSRSLSPGHSSGAAGAGDSPLHGQQHQPQLVGLDEASAGCSSVKSDDEDDRFPAGIRDAVSQVLKCYDWTLVPM PVR |
| 6132 |
PF12403 (Pax2_C) |
 Estimated TM-score=0.44; very hard target. |
>Paired-box protein 2 C terminal (Length=113) ASTPSYCAYAHRGSSYGQYASQPLIAGREMASTTLPGYPPHVPPTGQGSYPTSTLAGMVPGSEFSGNPYTHPQYTTYNEA WRFSNPALLSSPYYYSAASRGSAPPTAATAYDR |
| 6133 |
PF12329 (TMF_DNA_bd) |
 Estimated TM-score=0.44; easy target. |
>TATA element modulatory factor 1 DNA binding (Length=67) LRETVKKLRETVDELRETVKKLRETVKKLRETVKKLRETVKKLRETVDELRETVKKLRETVDELRET |
| 6134 |
PF12294 (DUF3626) |
 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF3626) (Length=301) RIALHFHPDRLDSNMKSVAEALFEQGVYKSQFETLLSNGSVSAYPGGERDLWEKRIFGGAYQLEGVTNNQRPKYGALNLM LHPDGPAPRFGSCYFLLSPKVSSRSTYTYLDSHQDPKEKGTYEEFDMILAALLEETFVRDFAIGEGNLTPTILINHLLAN LKRPFIDVVSKEPARNLNHYIEAQIHGEISLKEDVEALVADPSFKGTDVGRVLEEICLKYAIDLYWHIGFVLAVDEVPMD FRGSKMPSLARRIARNHCIDANIIGSAVADLKRNPLSWSDRGTYEEVLQELKLLWHVLVRF |
| 6135 |
PF12144 (Med12-PQL) |
 Estimated TM-score=0.44; very hard target. |
>Eukaryotic Mediator 12 catenin-binding domain (Length=208) DLLHHQPGSTVSRLAYGQSPMGLYAQNQPLPAGGPRLDTSYRPVRMPLGKLVQSRPPYSGVLPSGMGSMMGLDPSYKPAV YRQQPPVSQGQILRQQLQAKLQGQGMMGQQTVRQMAPTPSYGALQPSQGYTPYVSHIGLQQHPSQSGTMVPPTYSGQPYQ NSHPSSNPALVDPVRQMQQRPSGYVHQQAPGYGHTLGNTQRFPHQSIQ |
| 6136 |
PF12115 (Salp15) |
 Estimated TM-score=0.44; hard target. |
>Salivary protein of 15kDa inhibits CD4+ T cell activation (Length=124) MKVVCIILLFVIVAANLSAANNTDSSKDGKDTNGENVKLKFPSFIHNPKKLAMELLDICKKNKTPPSSYNVINDKHVDFK NCTFLCKHGQHENVTLDLPKETPCGPSGQTCADKSKCVGHIPGC |
| 6137 |
PF12087 (DUF3564) |
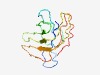 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF3564) (Length=119) MRLTVHLDTFDRVHPMAFAILWLDKDARKWSREGHLGLELPEWGTLHVDCGNTLVCGPHDTTPVCVLEGLDLNDRDGPFE GETGRAQWCAERGRTLMTGHWHVQCVDNEQVQPEHGLFA |
| 6138 |
PF12033 (DUF3519) |
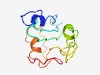 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF3519) (Length=112) ILRTIDKYNQEAITAFKQINGYAVVVEQAVNKKNELVLKTMYKNNGSYKNNNVYKEFSSTSLDANTPVPHGLSSHSGATD NPTPKPLNSQEDLLKRTELNNETTQEVKNLSP |
| 6139 |
PF12017 (Tnp_P_element) |
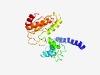 Estimated TM-score=0.44; hard target. |
>Transposase protein (Length=228) VLTKNLKETNLLLKQTQKTTTKQIELLSLNKSQTIENQAKKYLSSIFSANQLNLLMRKKKRVHWTRAEISKAFTLRYFSK RAYVYVKNELHYPLPGLSSLQRWAKSIEMRNGVLHDVLNLMKLNGEVLNNSEKLTVLMFDEVKVSSVMEYDVLHDEVVGP HNQMQVIMARGLASQWKQPIYVDFDKKMTKDILFDIIERLDQIGYKVVCCTSDCGGGNVGLWKSLNIS |
| 6140 |
PF11933 (Na_trans_cytopl) |
 Estimated TM-score=0.44; very hard target. |
>Cytoplasmic domain of voltage-gated Na+ ion channel (Length=214) KQKEQSGGEEKDDDEFHKSESEDSIRRKGFRFSIEGNRLTYEKRYSSPHQSLLSIRGSLFSPRRNSRTSLFSFRGRAKDV GSENDFADDEHSTFEDNESRRDSLFVPRRHGERRNSNLSQTSRSSRMLAVLPANGKMHSAVDCNGVVSLVGGPSVPTSPV GQLLPEVIIDKPATDDNGTTTETEMRKRRSSSFHVSMDFLEDPSQRQRAMSIAS |
| 6141 |
PF11802 (CENP-K) |
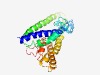 Estimated TM-score=0.44; hard target. |
>Centromere-associated protein K (Length=259) TADVDDVADAEKELIEECEEIWKDMEECQNKLSLVGTETLTNSNAQLSLLIMQAKCLAAELNQWQKETPKIIPLNEDVLL TLGKEEFQKLRHDLEMVLSTIQSKNEKLKEDLEREQQWLDEQEQILESLNTVYNELKNQVVPFSESRIFNELKTKMVNIK EYKETLLFTLGGFLEDHFPLPDGKVKKKKKNLQEPTAQLITLHEMLEILINKLFDVPHDPYVKISDSFWPPYIELLLRNG IALRHPEDPNRVRLEAFHQ |
| 6142 |
PF11693 (DUF2990) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2990) (Length=63) VCLAAISLAAPTHFLDDAYDWSDELAEFYGKVSQYINTARHNIRVPGACEASKIALPSYASGM |
| 6143 |
PF11488 (Lge1) |
 Estimated TM-score=0.44; hard target. |
>Transcriptional regulatory protein LGE1 (Length=71) NPFIYMMSNDDHSRTSKLKEVYKENDQIDNKLEELKLKIFKDELELGLMSTQCEKDALNVQLTQEKLDSLL |
| 6144 |
PF11296 (DUF3097) |
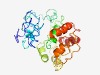 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF3097) (Length=258) WKRSAPVPEVPAEPDLVVEEVASGFCGAVIRCEKTAQGPTVTLEDRFGKHRVFPMEPRGFLLEGKVVTLVRPSAGGPARP TRTASGSVAVPGARARVARAGRIYVEGRHDAELVERVWGDDLRIEGVVVEYLEGVDDLPAIVAEFAPGPDARLGVLVDHL VPGSKESRIAAEVTGDHVLVVGHPYIDVWEAVKPSSVGIDAWPVVPRGQDWKTGVCRALGWPENTGAAWQRILSTVRSYK DLEPQLLGRVEELIDFVT |
| 6145 |
PF11263 (Attachment_P66) |
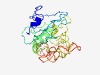 Estimated TM-score=0.44; very hard target. |
>Borrelia burgdorferi attachment protein P66 (Length=232) SDFSILGHISRKANTDDKDQFNPTENKLKFDNKRSTNFAFSVGTGIGLAWNTDDGEQESWSISGDNSYNTRIFGTQNKKS GIGLGITYGKNLYKPTSSNKIIQDIAAKSFQTFNAEISTYEDNTKGIIPGLGWIASLGIYDLLKEQPQSNDIITILTPKT NTQTQTQTVAFTEATKIGGALYIDYAIPVESISPTTHIIPYVGTHILGSFKGSDKSIYLKAGVELDNLIKLT |
| 6146 |
PF11110 (Phage_hub_GP28) |
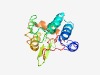 Estimated TM-score=0.44; hard target. |
>Baseplate hub distal subunit (Length=154) IPKMGLKHYNILKDVKGPDENLKLLIDSICPNLSPAEVDFVSIHLLEFNGKIKSRKEIDGYTYDINDVYVCQRLEFQYQG NTFYFRPPGKFEQFLTVSDMLSKCLLRVNDEVKEINFLEMPAFVLKWANDIFTTLAIPGPNGPITGIGNIIGLF |
| 6147 |
PF11027 (DUF2615) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2615) (Length=97) EGGFDPCECICSHEHAMRRLINLLRQSQSYCTDTECLQELPGPNSSSDNGISFAMIMMAWMVIALVLFLLRPNNLRGSNT AGKPTSQHNGQEPPAPP |
| 6148 |
PF10940 (DUF2618) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2618) (Length=40) KGRSLMAHIRRTRHIMMPSYRSCFSYSLFASQNKPSNRAL |
| 6149 |
PF10918 (DUF2718) |
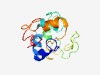 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF2718) (Length=131) MLCIFYIARLCNLIIYSLYSLLMFPMQKLISFMFGNLNPFDSVLDKDEKIQDNINTNHKPIESKEINNDLPLTVLDKKDT SDINIRNDNGVFDFIKIPNPFKKHYEYYCNQNTIKEPPRKGLVERMMNMVE |
| 6150 |
PF10814 (CwsA) |
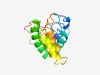 Estimated TM-score=0.44; hard target. |
>Cell wall synthesis protein CwsA (Length=133) LTPRERLTRGLTYSAVGPVDVTRGVVGLGVHSAQSTASELRRRYQEGRLARELAAAQETLAQELAAAQEVVANLPQALQD ARRKQRRSKRPYLIAGAAVVVLAGGAVTFSIVRRSMQPEPSPRPPSVDPQPRP |
| 6151 |
PF10812 (DUF2561) |
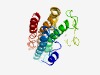 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2561) (Length=206) MVSRYSAYRRGPDVISPDVIDRILVGACAAVWLVFTGVSVAAAVALMDLGRGFHEMAGNPHTTWVLYAVIVVSALVIVGA IPVLLRARRMAEAEPATRPTGASVRGGRSIGSGHPAKRAVAESAPVQHADAFEVAAEWSSEAVDRIWLRGTVVLTSAIGI ALIAVAAATYLMAVGHDGASWISYGLAGVVTAGMPVIEWLYARQLR |
| 6152 |
PF10793 (Gloverin) |
 Estimated TM-score=0.44; hard target. |
>Gloverin-like protein (Length=161) QVSMPPGYAEKYPITSQFSRSVRHPRDIHDFVTWDKEMGGGKVFGTLGESDQGLFGKGGYNREFFNDDRGKLTGQAYGTR VLGPGGDSTSYGGRLDWANENAKAAIDLNRQIGGSAGIEASASGVWDLGKNTHLSAGGVVSKEFGHRRPDVGLQAQITHE W |
| 6153 |
PF10733 (DUF2525) |
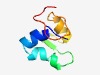 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2525) (Length=60) DVEALLEAIQDRSSGEVKEFMDNDRAHKVRVDGREYRSYTELADAFELDIRDFTVSEVNR |
| 6154 |
PF10713 (DUF2509) |
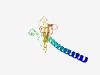 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF2509) (Length=131) MLMALSLMMLKALHYQQESAMLMMMDEKKYLNAFQRAESSLEWGKVQSWQFNMQEKENWFCQQQPEFHLQSCLKHYADNL FLLKGVAQFASGETLELYQWMKQMKKANRDTLIPIESGWLDFCPVPQAEFC |
| 6155 |
PF10653 (Phage-A118_gp45) |
 Estimated TM-score=0.44; hard target. |
>Protein gp45 of Bacteriophage A118 (Length=62) MAERIFRKQTIFGNSEIFIDDRTKMIANPAFRQKIALIETGCEKMTDYIEELKLKGYEEVTR |
| 6156 |
PF10288 (CTU2) |
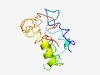 Estimated TM-score=0.44; hard target. |
>Cytoplasmic tRNA 2-thiolation protein 2 (Length=120) SIHRLMEAFILRLQTQFPSTVSTVYRTSEKLVKAPRDGPATGDHSPRCLLCMCALDIDAADSATAFGVQTSSHPSQIQLT PQGPCCFPGGPRAQGCCQGASRREDPQACIVEQLCYGCRV |
| 6157 |
PF10131 (PTPS_related) |
 Estimated TM-score=0.44; easy target. |
>6-pyruvoyl-tetrahydropterin synthase related domain; membrane protein (Length=621) FRYWAPFPYYIFALLEFLTGGNVSQAYIMFIGLMFFVGAFGWLLWGNREKRVFICFIFGVLWFLLPENIRIFFFEGNIPR VVIAALIPYLFYFLWGFVEKGKKASIIAMAGIMLLIIMCHLMVAAMLGITTFIFLLIYAVLSKKFLRSIQSIAAMLLCFA IAGIWLYPALKGGLVSMDTDSTSEVMRALSTPFTTSLDPLIRLTAAGRSGFFYYGVSIFVISCLGIFLSNKKSVPGFLTV IIVFLGTTTAFVPVLLKLPLNQLLWMMRFTPIAYGLFIMSLINWGKCKKVFMAIFLSLIILDSSLSFNFVLFPSEKSNTV ESIIESAKKVTNQRIALLDSSMFGSYPSFYIPSIGKETAYSYGWAWQGASTSKNIVLLNTALENGYYNYMFDRSIEMGCD TVVIRKASLEKKKSSFKLIDEAAKKLNYFLYNETVEGYVYHRETPKTFGVVTKYYGISIGRSAEEIPLEYTGFKSGASYS IDDYRLEELQKYKIIYLSGFNYKNKAKAENMISKLSKMGIKVVIDMNRIPSDETTNRMSFLGVTAQPITFDKKLPDLFYE NEKYVSGAFKKEYTKWNTVYLQNVSDIKGFSWIKDKKLTFMGKGTGDNKNITFVGFNLMFH |
| 6158 |
PF10107 (Endonuc_Holl) |
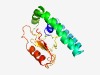 Estimated TM-score=0.44; hard target. |
>Endonuclease related to archaeal Holliday junction resolvase (Length=164) LLFLAVLVLFYKYSRIQGQVERKAREIFESWRQREREDIENWKEQELKRLSDEKAKILFETWRQDEEGNIRTDAIKRSQS VTRGRVTECLIPYFPDFPYNPKDARFLGTPVDLIIFDGLSDADEVQNVVFVEIKTGKAANLSKRERAVRECIKAGRVQYF TIHQ |
| 6159 |
PF10083 (DUF2321) |
 Estimated TM-score=0.44; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2321) (Length=157) TYRTAQICLNGHEITTSANTSPELMENFCSKCGAATITNCPICNTPIRGSYHVEGVLSLGHEYKIPSYCHNCGKPYPWTK TALESANALIEEDENLNSDEKKQFCETLPDLIVESPTPKTQLATARFKRLISKAATYTTDGVKGIIVNIASEAIKKS |
| 6160 |
PF10047 (DUF2281) |
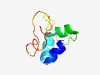 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2281) (Length=65) ELLETLTQMPESLQQEVLHYARYLNEKYSRAHTTEQPKKRRSGILKGTFKLPLPKDFNAPMELLD |
| 6161 |
PF09972 (DUF2207) |
 Estimated TM-score=0.44; hard target. |
>Predicted membrane protein (DUF2207) (Length=519) TIDQYDVDIQLQKDGSANYEERILYQFDGAFNGVFYDLNLTGRRNDSDISAVEVRTQPFKGKETAPYKENSSGENQTYQL TKNNGIAKFKVFNQVTNAKQTVIYRYHLRNVVTNYRDIAELNQMVIGKEWQDTLNNVQVTIHLPEGTTSGDLRAWAHGSL NGNVKLENNHTVRLTSNKNPANSPIEARVIFPTSITSDNQNKVDSNQLDKILSSEKTAAENANKKRKTVMGVSYSLAGLA VIILIVGSFHVRKIAKKEKFSLLVPEHLYDIPVDITPAVMYHVTTNQLPRTTDLTATIMDLVRKGQLTIEEIEIDIPSIR RNKKREKTFLIKKVTTPKPVQLLEHERYLQTWLIEEVGNGQEVTLKQIEDYGKKDVKKAEIFTKKYSNWQQKVKQASINL GYIKSNSQKALAVAMTYGVVLVLLGIGTILTMLMLNGFYPIVAVAVSVAAILSFVQWIGYFPVRTLEGETAIKQWAAFKQ MLKDVSNLKMADVGSVILWDHFLVYAISLGVSKEVINAL |
| 6162 |
PF09926 (DUF2158) |
 Estimated TM-score=0.44; easy target. |
>Uncharacterized small protein (DUF2158) (Length=52) RIGDVVTLKTGGPRMTVTYAGPVVFDTGEWVICQWFDEHGEFRQEMFPNETV |
| 6163 |
PF09725 (Fra10Ac1) |
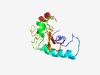 Estimated TM-score=0.44; very hard target. |
>Folate-sensitive fragile site protein Fra10Ac1 (Length=117) TDLDVIRENHRFLWSDDDDADITWEKQLAKKYYDKLFKEYCLADLSRYKENKIGMRWRTEKEVVDGKGQFICGNKKCAEN EGLRSWEVNFAYMEHGEKKNALVKLRLCPDCSYKLNY |
| 6164 |
PF09595 (Metaviral_G) |
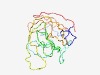 Estimated TM-score=0.44; very hard target. |
>Metaviral_G glycoprotein (Length=183) LIGITTLSIALNIYLIINYTIQKTSSESEHHTSSPPTESNKEASTISTDNPDINPNSQHPTQQSTENPTLNPAASVSPSE TEPASTPDTTNRLSSVDRSTAQPSESRTKTKPTVHTRNNPSTASSTQSPPRATTKAIRRATTFRMSSTGKRPTTTSVQSD SSTTTQNHEETGSANPQASVSTM |
| 6165 |
PF09566 (RE_SacI) |
 Estimated TM-score=0.44; hard target. |
>SacI restriction endonuclease (Length=278) REEAEKILKKAYAVASYNPESTCTHEELIDYVIDNTHLTYKYVLFTALLSKATDEKINPLCLQKKSTLPGAYDARTICHK VIVPFEMEVLKKAMGGSNEPFLNKPARFPELSKTNAVRRGNDQNILNALCDNLPTIKTSQDAFDCLVYLLCKLIKLRDAQ KKSLNFYVRETSNTPALLWTYIIKALKESFEGEILTLMVAGTYHLIYKDRPGARVEVHPVNQSGASGREVSDLDIYVDNE LISSNELKDKNFSEPDVRHAADKVITAGGNHMLFIFGP |
| 6166 |
PF09495 (DUF2462) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF2462) (Length=74) MAQGAIKSSSRPAPTGKPKQTTKKQSKVNKPQKAKANSAADKLQKKYCAGLITKTEKMLGERAGHLELIGKGKK |
| 6167 |
PF09432 (THP2) |
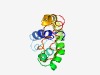 Estimated TM-score=0.44; hard target. |
>Tho complex subunit THP2 (Length=129) LDYVNLLQRLSVDLAKQIEISDPEVSEFVVDNWSPPDGMQSILEQLANPDKDSTHLQSQLDQYLDQIKMERAKYTIENKY SLQETLNEVNKEVNYWRRNWNAIENLMFGDSAHSIKKMLQSIDLLRAKL |
| 6168 |
PF09372 (PRANC) |
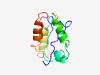 Estimated TM-score=0.44; hard target. |
>PRANC domain (Length=95) GSYSVYDLVFRRSRGIVPVSVTKHPSFTKFQTSILYGSTIRNIILNSLEFDVCVNEAVDVLNVYCKKRNYWGSLPVEIQK NILQYLTTYHIKYIL |
| 6169 |
PF09338 (Gly_reductase) |
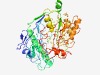 Estimated TM-score=0.44; very hard target. |
>Glycine/sarcosine/betaine reductase component B subunits (Length=426) MRLELGNIYIKDVQFGNETKVENGVLYVNKEELISIISEDEHLKSVDVELARPGESVRIMPVKDVIEPRVKVEGKGGIFP GVISKVETVGSGRTNVLKGAAVVTTGKIVGFQEGIIDMTGPGAEYTPFSKLNNVVLVCEPVDGLKQHEHEKAVRMAGLKA AEYLGKAAKNVEPDDVETYEFLPLLKSIEKYPNLPKVAYVLMLQSQGLLHDTYVYGVDAKQILPTLISPTELMDGAIISG NCVSACDKNTTYHHLNNPIIHDLYKKHGKELNFVGVIITNENVYLADKERSSNWTAKLAEYLGVDGVIISQEGFGNPDTD LIMNCKKIEQKGIKTVIVTDEYAGRDGASQSLADADPLANAVVTGGNANEVIELPPMDKVIGHIEPANKIAGGFDGSLKE DGSIVVELQAITGATNELGFNKLTAK |
| 6170 |
PF08872 (KGK) |
 Estimated TM-score=0.44; very hard target. |
>KGK domain (Length=124) DNFKLIECNDDDVIECNDWAYKVSKLKQALEKLGGSDGFLQTLWSGLREQKIYYLEGNYTYDNKSFKEGMDVKILNLGSK SWKKGKLKFKLSIEFYVEEELEESSEKDVIKEPESPLDDLRRMI |
| 6171 |
PF08658 (Rad54_N) |
 Estimated TM-score=0.44; hard target. |
>Rad54 N terminal (Length=173) PQSVDKLVKPFRCPGSATITRTSDKPTRKRRKVDYGGADGDKDDSDKPWTNEDRLALANRDANKFPIFQVKDKDTMFRKR FSVPLINKNAGGYNPSRPPPTLGLRQGAVFIAKALHDPSGEFAIVLYDPTVDEKPRTIQEARKDDEKKEEEPEKPKLDAP LAHKSLAEILGIK |
| 6172 |
PF07958 (DUF1688) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF1688) (Length=433) YLRSIHAVRERTRLVLEEARVNALNHFDVDMSKFKDTADYVVAIIKRDFQGDYASIPPHGRWQHFEVGGRPRVTQLLQSW PTSIDNQERARRLIDLFLVSVLLDAGAGTKWSYKSKESGKVYSRSEGLAVASLEMFKAGAFSSDPNQPHQVDAAGLNKMN MESLAKGMQVSDTNPMSGLKGRTGLLIRLASALQQNAEIFGVEGRPGNMIDYLLSHPTTQAASVPVILLPTLWSVLMDGL SSIWPATRTTIQGVSLGDAWPCRAMPKSPPAQPWETIVPFHKLTQWLCYSLMVPMTKLLNVHFAGAELMTGLPEYRNGGL LVDTGLLTLKDEDQQRGLKTYHETVGDGNAVEVVPTFEPSDDVIVEWRAVTVGFLDELLVAVNAGLGLQGAQQLTLAQML EAGTWKGGREIAQVSRPITGGPPIGIISDGTVF |
| 6173 |
PF07818 (HCNGP) |
 Estimated TM-score=0.44; very hard target. |
>HCNGP-like protein (Length=94) IPPEPAGRCSSQLQEKIFKLYERRVHGDFDTNSHIQKKKEFRNPSIYEKLIQFCGIDELGTNYPKDMFDPHGWPEDSYYE ALAKAQKVEMDKLE |
| 6174 |
PF07798 (DUF1640) |
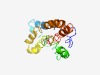 Estimated TM-score=0.44; easy target. |
>Protein of unknown function (DUF1640) (Length=168) VRKLESQGVPTKQAEAITSAITEVLNDSLESISESFVSKAEMQKSEMLQEANISKFKSQVQSSQENHFSLLQRETEKLRG DIDKMRSELKYEIDKVTAGQRLDLNLERGRIRDELAKQNEETTDLTTKLDKEIHSLKAQLEAAKYDVIKYCIGTIVSISA VGLAVLRI |
| 6175 |
PF07628 (DUF1589) |
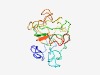 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF1589) (Length=164) MLWNKASRPAGHARLSDVRPFTPPGTTWPTFLSQPDASAKDTSEYWRSHQRRPGHYLAPSRCFGTKRAVQPVTQDSAMSV HSPRQVQPGLHICPNPTRQRETRLNSGEATNVGQVITWHPADALEQSGPSSRSRKTQRCPSIHPARYNLAYIFVPTRRVS ERHV |
| 6176 |
PF07376 (Prosystemin) |
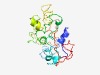 Estimated TM-score=0.44; very hard target. |
>Prosystemin (Length=202) METPSYDIKNKGDDMQEELKVKLHHEKGGDEKEKIIEKETPSQDIKNKDTITSYVLRDDAPEIPKVEHEEGGDGKEKIVE KETISKCIIKIEGDDAQEKLKVEYEEEEYEKEKIVEKGSPSQDINNKGDDPQEKPMVEHEEGDEKETPSQDIIKIEGEGA QEITKVVCEEREKIVVQVDLAVHSTPPSKRDPPKMQTDNNKL |
| 6177 |
PF07193 (DUF1408) |
 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF1408) (Length=71) MHTQIMNGREVLTVPTVIGYKHYDLEKREVVGEVIESTYRRKDGTMYIIRRSRTEREKAAMLNSCLSDWGY |
| 6178 |
PF07064 (RIC1) |
 Estimated TM-score=0.44; hard target. |
>RIC1 (Length=261) LENSLWGWDGSKIRVWLDALTIEKVKVDAKRDAYEAVAESVAIALDFYPLAVLMDKGIIIGVDQETSLRRSLDFAIFRII TTTHLFLHHVLRFHLTKSQGKEAVVFASHYQHLVYFAHALEILLHAVLEDEADARPSRPSTPAPAGEIAADENAVLPRVV EFLDHFDEALQVVVGCARKTEVARWSFLFDVVGKPRDLFEKCISAGFLKVAASYLLVLHNLEPLEQSSKDTVRLLKTAMD AGEWTLCRELLRFLYSLDRSG |
| 6179 |
PF07041 (DUF1327) |
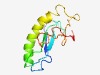 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF1327) (Length=112) MKKKYELVVKGINNYPDKITVTVALEIGGYPSLLLPDVAISLDRTEGATLEFYEAEAKKQAKQFFMDVAAGLCEGDGPLP ENRPVILEAQDVLITYKGKLPGRITCSLKMPP |
| 6180 |
PF06959 (RecQ5) |
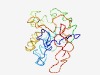 Estimated TM-score=0.44; hard target. |
>RecQ helicase protein-like 5 (RecQ5) (Length=203) KSCSAQVEHPEPTEYDIPPASQVYSLKPKRVGAGFPKGSYPFQTATELMEKMRPKEQAPQPVWGGKQEPPSRPCGLQDED STEPLPGPRGEAPGSSASCGESSPEKKAKGPTGGSPIAKARVNKKQQLLAAAALKDSQNITRFFCQRAKSPPPLMSALPA EGASPSSEGVRGPPTAPEKCAGEEDGALGHLAVPAQTEECTRE |
| 6181 |
PF06729 (CENP-R) |
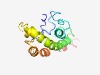 Estimated TM-score=0.44; hard target. |
>Kinetochore component, CENP-R (Length=138) RKRSITNYSPTTGTCRMSPFASPTSSKVQEYKNGPSNGKRKKLNHLSLTKRKESTTKDDEFMVLLSKVEKSSEEIMEIMQ SLSIIQALEGSRELENLIGISCTSRFLKREMQKTKELMTKVMKQKLFEKKSSGLPNKD |
| 6182 |
PF06706 (CTV_P6) |
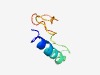 Estimated TM-score=0.44; hard target. |
>Citrus tristeza virus 6-kDa protein (Length=51) MDCVIQGFLTFLVGIAVFCAFAELIIIVITIYRCTTKPVRNASPYGTHATI |
| 6183 |
PF06693 (DUF1190) |
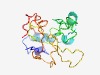 Estimated TM-score=0.44; very hard target. |
>Protein of unknown function (DUF1190) (Length=165) DETVSLYQNADDCSRANPSMSEQCTTAYNNALKEAEKTAPKYANREDCVAEFGESQCTQAPAQAGMAAQSQQSGGSFWMP LMAGYMMGRMMGGGYAQQPLFTSKNAASPANGKFVDASGKNYGPATAGGRTMTVPKTAMAPKPAVTNTITRGGFGETVAK QTSMQ |
| 6184 |
PF06625 (DUF1151) |
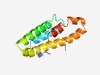 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF1151) (Length=113) GNPDLIKPKKLLNPVKASKSHQELHRELLMSHKRGLGFDNKPELQRVLEHRRRDQLIRQRKEEEEAKRMQSPFEQELLKR QQRLEELEKEEEKVKGEQENAPEFIKVKENLRR |
| 6185 |
PF06549 (DUF1118) |
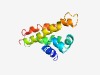 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF1118) (Length=112) KLEKRKVLSNVEKAGLLSKAEELGFTLSSIEKLGLLSKAEELGLLSLLETAAGFSPAALASASLPILVAAILAVVLIPDD STVLVVAQTVVASTLAATATAKTPDAFIKSTV |
| 6186 |
PF06300 (Tsp45I) |
 Estimated TM-score=0.44; very hard target. |
>Tsp45I type II restriction enzyme (Length=263) NYWTKLSIEYANQKSYLDDLFKVYPTIPNGIRDIDNKIWENIKTSFNNQNNKKLIENLLKLDLFPIKDSYIAYLRKDKTA IDRNPETIDRLAGRIYDMGIDEVYKQCTKPKETNRQIGPMFKEWVNKGVLGKKPIEIDEFINTENNAILSGSDQEMEEFA EDYCDYEYAGNTKKGLDFIARFNKKYVIGEAKFLTDIGGHQNAQFNDALNVLDSNVSEKVTKIAILDGVLYIKNERTKMY RTTVQKNDNIMSALILNEFLYSI |
| 6187 |
PF06054 (CoiA) |
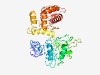 Estimated TM-score=0.44; hard target. |
>Competence protein CoiA-like family (Length=357) FTARNNIGETFTITKMNVERIKQEERLFCKSCASPVMIKAGQIKIPHFAHEKTMKCAFASEGESEEHLAAKKQIMAWFCY QRIPVEIESYFPEINRQADIFVNGKTVIEFQRSSISISEMIQRTMDYLSIGLEVHWILGQVVKKEKGRICLSAFQQAFIQ SDKKLGYHFWHYSAKREICTLYYHLTFEKGNRFFASEMKFSMKSPLREWNKKLARIISRHAYIKRDREIERQKICFYYAK YKKHGQFMQKLFNAGYYLHYLPKEIGVDVEEQFLVLTPAVEWQFDLWEKFFKGLEKGDTFSVESFLESFKSVVSQILTIW LSPNESLKLGKSYLSYLIGEGVLSTIPDNRYRVKRKM |
| 6188 |
PF05959 (DUF884) |
 Estimated TM-score=0.44; very hard target. |
>Nucleopolyhedrovirus protein of unknown function (DUF884) (Length=188) MNVNLYCPNGEHDNNISFIMPSKTNAVIVYLFKLDIEHPPTYNVNAKTRLVSGYENSRPININVRSITPSLNGTRGAYVI SCIRAPRLYRDLFAYNKYTAPLGFVVTRTHAELQVWHILSVRKTFEAKSTRSVTGMLAHTDSGPDKFYAKDLIIMSGNVS VHFINNLQKCRAHHKDIDIFKHLCPELQ |
| 6189 |
PF05842 (Euplotes_phero) |
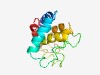 Estimated TM-score=0.44; very hard target. |
>Euplotes octocarinatus mating pheromone protein (Length=133) FKMTSKVNTKLQSQIQSKFQSKNKLASTFQTSSKLKGECDTIIPDFTGCNANDACPLSFTCSATGNDEQLCNAAGQNVID MIFAHWSTCWNTYGNCIEFARQTYAIYNAPELCGCDYVDEETWINTLESVCPY |
| 6190 |
PF05810 (NinF) |
 Estimated TM-score=0.44; hard target. |
>NinF protein (Length=58) MLNPIQTQAYEHQSIARALCAGCSKQLEPDETHCCEECVAQAIYYRDPNHFMAEDEDE |
| 6191 |
PF05687 (BES1_N) |
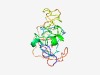 Estimated TM-score=0.44; very hard target. |
>BES1/BZR1 plant transcription factor, N-terminal (Length=145) GGTRVPTWRERENNRRRERRRRAIAAKIYTGLRAYGNYNLPKHCDNNEVLKALCNEAGWVVEPDGTTYRRGCKPPPQARP DPMRSASASPCSSYQPSPRASYNPSPASSSFPSSGSSSHITLGGGNNFIGGVEGSSLIPWLKNLS |
| 6192 |
PF05680 (ATP-synt_E) |
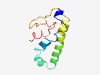 Estimated TM-score=0.44; hard target. |
>ATP synthase E chain (Length=97) MSSNASKAGSNVLRYSALAAGIFYGFTHQRSISAAEKAAAAQREYEHKQELINKAKEAYAKSKQPVSAVSTPSGQTVNQD PMSPNFDIEAYFSALLA |
| 6193 |
PF05615 (THOC7) |
 Estimated TM-score=0.44; hard target. |
>Tho complex subunit 7 (Length=142) ELHKARLLNVEEKPFKRITKRLSTVSSIVSKITDDEEQSATTAPNTEQLREDLTLDFAAFDSSIARLQFLHDANERERTR YDDDKQRILDECQAVRAKNAELREQLDGARAKLAQRKKFDELADKITSNRLLRPREDQLANL |
| 6194 |
PF05582 (Peptidase_U57) |
 Estimated TM-score=0.44; hard target. |
>YabG peptidase U57 (Length=283) IGDIVVRKSYGKDITFKVIDIREEKGKKIYILKGINIRIIADSPEEDLERTDSEEAGKKDEVFNRKVKESIKNILTSRSE SVEVMSGLTLKNKELKNKELYFGRPGKVLHLDGDSQYLEICMKTYKRIGIDVVGKVISEKEQPSKVVELVKEFKPDIVVI TGHDGMLKDTKDYMDLNNYKNSKYFVESVSELRNYERGYDELVIFAGACQSCYEAILGAGANFASSPNRVLIHCLDPVFV CEKVSYTNIEKIVSIEEVIENTITGTKGIGGLQTRGKYREGFP |
| 6195 |
PF05476 (PET122) |
 Estimated TM-score=0.44; hard target. |
>PET122 (Length=260) RRFLSEATRNTLYLDCMNRKFDDVLRVVRSTSANEMDYAFLQLYLSRSCQWGHIASVTHIWDKYVLRLKMMTVHPSLLCE IGNLALNEGKHFLPPQIYHHYVKFFSGTMPIDEEQRIEYELLRIKVESFAKGTMLKTRFREKWKVLLEDMDHHLSSAMEF RVRDFPYLTKSVQLEDNEMLMKLLFDQNKLDIKNRSTAPLLLNIILSQPQQSIESKISLFEKFYDTHRSLKYDDTLTILF RLCKGDGYRLSKLGQMVEEK |
| 6196 |
PF05335 (DUF745) |
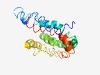 Estimated TM-score=0.44; hard target. |
>Protein of unknown function (DUF745) (Length=181) GNPKQKATNIAQKAAQEATAASEAQNAAGAAAAHQVKMQLAEKAMQAAKAAEAALAGKQQIVEQLEQEVREAEAVVQEEA SSLDSAHSNVQAAIRASQQAQKQLKTLTEAVQTAQANVGNSEQAATGAQAELAEKAQLMEAAKTRVDQLLRQLSGARQDY SNTKQSAYRAACAAHEAKVNA |
| 6197 |
PF05323 (Pox_A21) |
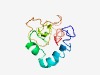 Estimated TM-score=0.44; very hard target. |
>Poxvirus A21 Protein (Length=111) MITLFLVLCYFILIFNIIVPAISEKMRKEYDAYLKYAHLKKDAVCVDDRLFTYDFKTSGVVAKMFIDSNGKPLPCSRTRD IRSNNAIYCDNDENVLDFRKSCSKAYLDLFF |
| 6198 |
PF05320 (Pox_RNA_Pol_19) |
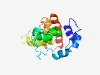 Estimated TM-score=0.44; hard target. |
>Poxvirus DNA-directed RNA polymerase 19 kDa subunit (Length=170) MADTDDIIDYESEDDMSEYEEEEDDEDNETKSLESSDVISLKQSNYKAELSTNLEEEVPQPGQSSKNVNSKILAIKKRYT RRISLLEITGIIAESYNLLQRGRLPLISNLSDETMKQKILHIVIQEIEEGTCPIIIEKNGELLSTNDFDKTGLKNHLDYI ISIWKNQNRY |
| 6199 |
PF05281 (Secretogranin_V) |
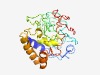 Estimated TM-score=0.44; very hard target. |
>Neuroendocrine protein 7B2 precursor (Secretogranin V) (Length=234) DLLNRMDNDMQVGYYDVDNEGDLAAVAAAAAKDNVDLVSRSEYARLCNGGSDCLLQSNNAHPSLRDQEFLQHSSLWGHQY VSGGMGEVPNRYNTIVKTDASLPAYCNPPNPCPEGYDTEQQAGNCISDFDNTAIYSREFQAAQECTCDSEHMFDCADQEN AAGVGPGNSDLNSAVEKFLLHQFGNENVLDNANVVAKKAGHMPALRMDSIPNPFLQVAPGDRLPIAAKKGNMLF |
| 6200 |
PF05006 (PIF3) |
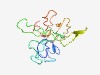 Estimated TM-score=0.44; very hard target. |
>Per os infectivity factor 3 (Length=148) RGIDCSLNRLPCVTDQQCRDNCVISSAANELTCQDGFCNASDALVNAQVPDLIECDPRLGLVHVFSAGGDFVVSQTCVST YRDLVDDTGTPRPYLCNDGRLIMNLNTVQFSPDACECSSGYEKMLFRQTALARTIPVCIPNRMANLYR |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218