

Go to page (83 pages in total): [First page] << < 59 60 61 62 63 64 65 66 67 68 69 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6301 |
PF12820 (BRCT_assoc) |
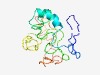 Estimated TM-score=0.43; very hard target. |
>Serine-rich domain associated with BRCT (Length=164) DPLYEKKEPSKQKPPCSENSRDTQDVPWITRNSSIQKVNEWFSRSDEMLTSDDSHDGGSESNAKVAGVLEIPNEVDGFSG SSEKIDLLATDPHNALISKRERICSKAVESNIEDKIFGKTYRRKASLPNLSHVTENLIIGAFAXEPQITQECPLTNKLKR KRRT |
| 6302 |
PF12768 (Rax2) |
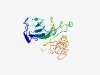 Estimated TM-score=0.43; hard target. |
>Cortical protein marker for cell polarity (Length=209) LATYDTKSQSWTAALGASAIPGPVTALTAANSDASQLWVAGTATNGSTFLMKFDGTTWNSVGDTLGSGTKIHGLQVLPLT EDHDSSDLISASQALMITGSLNLPGFGNASAVLFNGTSFQPFALTSSSSNTGGSLSQIFSQRQNFFNTPGGKLAKGFVVL IALAIALGLIFFLVIAGVVAERIRRKREGYVPAPTASFDRSQGLSRVPP |
| 6303 |
PF12761 (End3) |
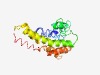 Estimated TM-score=0.43; hard target. |
>Actin cytoskeleton-regulatory complex protein END3 (Length=199) NKDACLAFLHILNNRHEGYRIPRTVPASLRASFERNQIDYQVGSARNAAQSRWATKADDETSTGRKAKFGDQYLTRLGRS GFKTAGTDFSNAQTDAEWEEVRLKKQLAELDEKIAKVEAAAERRKGGKRDSKPALVKRELEQLLDYKRKQLRDLEEGKAT NRPGGNLKSVHDDLQTVREQVEGLEAHLRSRQEVLEQLR |
| 6304 |
PF12457 (TIP_N) |
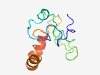 Estimated TM-score=0.43; hard target. |
>Tuftelin interacting protein N terminal (Length=98) DGEYEKFEVTDYDLDNEFNTNRPRRKMTKNQMIYGMWADNSDDDEDSGRPSFSKKKPKNYTAPVNFISGGVQQAGKKKEN EEKQDDDSEDEERQQIEQ |
| 6305 |
PF12300 (RhlB) |
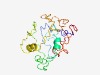 Estimated TM-score=0.43; hard target. |
>ATP-dependent RNA helicase RhlB (Length=175) EQKIPVEPVTTELLTPLPRTPRATVEGEEVDDDAGDSVGTIFREAREQRAADEARRGGGRSGPGGASRSGSGGGRRDGAG ADGKPRPPRRKPRVEGEADPAAAPSETPVVVAAAAETPAVTAAEGERAPRKRRRRRNGRPVEGAEPVVASTPVPAPAAPR KPTQVVAKPVRAAAK |
| 6306 |
PF11807 (DUF3328) |
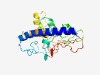 Estimated TM-score=0.43; very hard target. |
>Domain of unknown function (DUF3328) (Length=223) SSWRRHVSNPVILLLMTLCVLVMALIGISLTRKPTDNQCARQLSMWSPALEAVEYVTFDFDDAFNSTSKYRGPPTPEVEE AWFNLTYKHAVEIPEDKIAGLNRSEKDHLAHVPADVGTGYVALLEVFHQLHCLNMIRMFTWWQAGKYPGVPSGLYEAPED AFRNRMHIDHCLESLRIALMCWGDVTPLLVRLDGQGHDGRRADFDTHHKCRNWDKIESWMDEN |
| 6307 |
PF11768 (Frtz) |
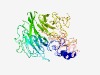 Estimated TM-score=0.43; hard target. |
>WD repeat-containing and planar cell polarity effector protein Fritz (Length=548) KRLYFESRGCKWSVDNKKPNRLRNALKLLEEELKQRRIAFCKWKNNVILQIMLSNGLLIHIFINPFNGDINRVYFDKYFI GKLISEQITDVIITQTHILISYNENQITFVHLQKPTARKNNLEKISLMDPKIYNVIIGGPTARKIPKHLVCSSSQEMVIV WTKSSQNEVYPWRPTVKDQDRANLHVYKLNGAKMELLCYYWTEYDPISVDFSLLNGFQVHSVEQKISKKGDVTIDSCIYQ IHKTKMRRVAVTSIPLQTQVCCNAFSPDHEKLMLGCIDGSVVLFDEGRGITHLVKAAFIPTFVSWHSDSSIVMIANEKGQ LQCFDIALSCVRTQLMSEDVVPSSILDLSNYFAHPYNLAKLCWSRKPEITEHFEKNAITDCFLLLAFENGPLGCVRFVGG TGLKGDVHTSGLTADVLIHKYISLNQVEKAINILLSLNWDTYGAMCLLSLHRIANYIFKQPLGTERELQLQKALGSFLVP VKPLCYETEVEFGDQVNDITLRFFHYLLRNKSYNKAFSLAIDINDADLFLKLHDKAKTDGDVELAKEA |
| 6308 |
PF11744 (ALMT) |
 Estimated TM-score=0.43; hard target. |
>Aluminium activated malate transporter (Length=455) WKVGKEDPRRVVHSLKVGMALTLVSLLYLMEPLFKGIGKNAMWAVMTVVVVMEFTVGATLSKGLNRGLGTLLAGSLAFLI EYLANAPGRIFRALLIGAAVFLIGAMTTYVRFIPYIKKNYDYGVVIFLLTFNLITVSSYRIDHVWDIAKDRMSTIAIGCG ICLVMSLLVFPNWSGEDLHNNTISKLEGLANSIQVCVVEYFYDSAKPEETRDDSSEDPIYQGYKAVLDSKASDETLALQA SWEPRYSRSCHRIPWNQYARVGAALRYFSYTVVALHGCLQSEIQTPKSIRALYKGSCIRLAEEVSKVLRELANSIRDNRQ FSPQKLSNNLKEALQDLDNALKSQPQLVLGSRNGRSTSTSTNTVEAVPPQADQKLEEDSRMSLSSVRNDSFSPRGCKSKE HSREQLKKVLRPQLSKIAIVSLQFSEALPFAAFTSLLVELVAKLDRVMDEVEELG |
| 6309 |
PF11718 (CPSF73-100_C) |
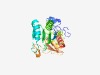 Estimated TM-score=0.43; hard target. |
>Pre-mRNA 3'-end-processing endonuclease polyadenylation factor C-term (Length=227) TLSGLLVSKDYSYTLLAPRDLRDFAGLSTCVVTQRQRLILGVGWELVRWHLEGMFGKVEEGADKDGIPTMRVMGTVDVKQ TAEHELTLEWDSSASNDMIADSTLALITGIDQSPASVKLTSHSHSHDHDHHHPHADDETESATVMRIQRLAMFLEAHFGE VELYMPDNAPDADREAGEDGENEPSLFVALDEADARINLATMTVTSSSEALRKRVEAVLEMAVSTVT |
| 6310 |
PF11682 (zinc_ribbon_11) |
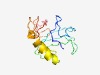 Estimated TM-score=0.43; very hard target. |
>Probable zinc-ribbon (Length=125) RILNCYMANDSKGHFVTAKEAAKHNRQDVLCCVSCGCPLTLQRGNDGQPPWFEHDQMTVAEKILLRCTWLDPAEKEARRL HLQGMTVPDYTVKVRKWFCVMCDEDYEGEKCCPRCGTGVYSRAWG |
| 6311 |
PF11657 (Activator-TraM) |
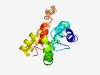 Estimated TM-score=0.43; hard target. |
>Transcriptional activator TraM (Length=142) TDKIEETIKEIAVKHGIAVGRDDPILILQTINERLMQDSAAAQQEILDHFKEELEAIAHRWSDDAKNKAERTLNAALTAS RETMTQTMQEGAKSAAELIRREIKAASAQFAAPIHEAKRVALMNLVAAGMTIIAAGLVLWAA |
| 6312 |
PF11352 (DUF3155) |
 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF3155) (Length=87) RRRKRKSRRRLEGRKILDHIPQFNLDSGEEKSVTAARKYIQEKSIQPPALILVRRNEHTVDRFFWAEKGLFGAQYVEENH FLFPSLR |
| 6313 |
PF11301 (DUF3103) |
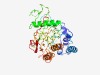 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF3103) (Length=354) KRQLAESFSQNYAAYAHTLKTQISEDNLSISVSELVESAPNTEMSQQLRSADKNVRTLKGIDQFTEQLLQLRLADASMLK EWQEGQSPLFAFEPSGNDDSWQYIEAYDVYGQIHQLDVYQLPDVPVFVVDNDSAVELKAGLQAMRAEMQRLGQSPQLSTQ ESSSIEASTRSLSRSASADTAPISTTVLKKIRLQDDKEPWISGRAEIYALVTGVDPSRDKPTIDLIDMPYLDYDKQDYFP NQVVIHWTRYRWGAADMILMEQDDGTDYKELAKKLVKVAEEVLKLIPDPEVQGYAIIAQITGKIIEAIPDGVLVNDDDFV DVFYTLMQDTQYTDHPGANGNATATFEPLTIYPT |
| 6314 |
PF11259 (DUF3060) |
 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF3060) (Length=55) VTGDGNSVSITGASDQVTLTGNSAVVAISGANDALVVSGSNETITTTSAAAITLT |
| 6315 |
PF11229 (Focadhesin) |
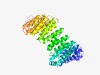 Estimated TM-score=0.43; hard target. |
>Focadhesin (Length=537) DTLRSLTEESQQTPGFSLALGLLVHGLAVSGHGKAEDIHPRLLAAWIKILLAEGCPTMQRLAAANGLVALVGSESCLIQL QTEMELSSQQQSRLNEVIRAITQIITFSGAIGLQSNSACLIGHLHLAHMSTSHTHTAVPQDFSYLPEKSVIRSIMDFISE AGKKGPEFSHPALVSTALSPLASVGASFQYPPVNWSAVLSPLMRLSFGEDIQHQCVVLAATQAQSSQSASLFLGSWLSPP LVHSLSHRTRAHLYETLGVWMKHMAEDKLQVYVQSLGLQQFQEDVRAQRVSLCQSLLQGLAQAMALPNPPNHCWTILCSS AEKIFGLLPNQIQDAEVDLYVGVAKCLSEMSDSEIDRIVQVSQAQMEKTCFVLAYLTSQGRVPLLSLNDVIAGVLRGWPS CRVGWILLQAFYQCRLAASANTGVSKRLEWLLELMGHIRNIGYGATPVSCGDARLATDFLFRLFAAAVISWGDHYMPLLF GIRPQWFPWQPASKPATLTHGLYGAVSLSELALSQCLLGVSHSLPLLLDKEPWSSQT |
| 6316 |
PF11124 (Pho86) |
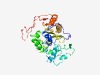 Estimated TM-score=0.43; hard target. |
>Inorganic phosphate transporter Pho86 (Length=307) QKEVDLNEPLDAHEAPTLSKTPLKPEFAKAALILHGDTYKQLQAKLNRFIFWHPIAIIAYFLLIPAALLYNLWDFVEVSD SAWEFFLLGKKNPKDFIYGIIAAFPMVAGIFATFGLITYIMGDDIGNITSAFVAQKYCDKVFGFDIKKFAQLTGHETDVD SIKLLKKGLNTQIIMYRESPIAICTVVVDEEQSTPESFIVEITGLHVRKVFENVDFHELLIEWAVLRTRELYEEYAKQKK VKTQEGTILITTDAYSFDKRLQTCLYNTNFGVMSKDVELNRFSDTLNKPKEILHKLLGVSRDTFGLA |
| 6317 |
PF11109 (RFamide_26RFa) |
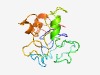 Estimated TM-score=0.43; very hard target. |
>Orexigenic neuropeptide Qrfp/P518 (Length=130) SLSCLVLLPLGACFPLLDREEPVAAMSGVGGEMSWANLPGGHRAPLPRGSSRWLRAPHPHGLLVTAKELQMSGRQRAGFR FRFGRQDDGSEATGFLPADGEKAHGPLGTLAEELSSYSRKKGGFSFRFGR |
| 6318 |
PF11044 (TMEMspv1-c74-12) |
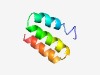 Estimated TM-score=0.43; hard target. |
>Plectrovirus spv1-c74 ORF 12 transmembrane protein (Length=49) MPTWLTTIFSVVIILGIFAWIGLSIYQKIKQIRGKKKDKKEIERKESNK |
| 6319 |
PF10954 (DUF2755) |
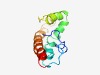 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF2755) (Length=101) MADFTLSKSLFSGKHRDTSSTPGNIAYAVFVLFCFWAGAQILNLLVHAPGVYEHLMQMQETGRPRVEIGLGVGTIFGLVP FLTGCLIFGVIAVILRWRHRH |
| 6320 |
PF10873 (CYYR1) |
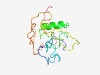 Estimated TM-score=0.43; hard target. |
>Cysteine and tyrosine-rich protein 1 (Length=148) RRSGVLLPKLVLLFIYAEDCLAQCGKDCRSYCCDGSTPYCCSYYAYIGNILSGTAIAGIVFGIVFIMGVIAGIAICICMC MKNNRGTRVGVIRAAHINAISSYPVAPPPYTYDHEMEYCTDLPPPYSPTPQASAQRSPPPPYPGNSRK |
| 6321 |
PF10867 (DUF2664) |
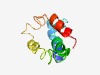 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF2664) (Length=89) ISKDLEVRHRDSLKIRLGERHPLSVHQHMIAARQIIKSDNAEQQHVISSLSGFLDKQKSFLRVQQKALKQLEKLDVDEII DTAAEVKAV |
| 6322 |
PF10839 (DUF2647) |
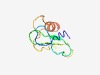 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF2647) (Length=70) AYSSCLNRSLKPNKLLLRRIDGAIQVRSHVDLTFYSLVGSGRSGGGTTAPLFSRIHTSLISVWRAISRAQ |
| 6323 |
PF10836 (DUF2574) |
 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF2574) (Length=93) MKKYLLMGIIVSAYGISVPVFASDTATLTISGKVTAPTCSTEVVNAQLQQRCGNTIHVSTLQTPAAMPMRGVTTQLYTVP GDSTRQIVVNHYD |
| 6324 |
PF10624 (TraS) |
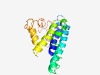 Estimated TM-score=0.43; hard target. |
>Plasmid conjugative transfer entry exclusion protein TraS (Length=162) MKNLAQIVLVTVVQFIACYLAEWGVAETGIILLFVLLWQGLFIWLFIQIRKKNNISDEFKFSKGIWYVIMPVCSLLSPLL SLMIFIGGTLYELRRVSGCISIKEWVKCQLDDQYDEDRGLDFESVEYRQTTYYNPSTGYPMHGGVDSAGNTFGSRWQDNN DR |
| 6325 |
PF10252 (PP28) |
 Estimated TM-score=0.43; hard target. |
>Casein kinase substrate phosphoprotein PP28 (Length=91) NPNRVTKKATQKLSAIKLDDGPGSEGGKGGAPKPELSRREREQIEKQKARQRYEKLHAAGKTTEAKADLARLALIRQQRE EAAAKREAEKK |
| 6326 |
PF10195 (Phospho_p8) |
 Estimated TM-score=0.43; hard target. |
>DNA-binding nuclear phosphoprotein p8 (Length=59) EAFFDEYDYYNFDHDKHIFSGHSGKQRTKKEVSEHTNHFDPSGHSRKIVTKLVNTENNK |
| 6327 |
PF10104 (Brr6_like_C_C) |
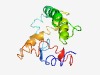 Estimated TM-score=0.43; hard target. |
>Di-sulfide bridge nucleocytoplasmic transport domain (Length=131) GYLQLFFNIFLVGVLLYFVIAFVRTIQRDVDQKVDEYSAEILQEMSLCSKEYTENRCSPDHRVPAMERACTAWERCMNRD PTVVGRAKVSAETFAEIINSFIEPISYKTMIFCILIIFGSVFVSNFAFGFF |
| 6328 |
PF10071 (DUF2310) |
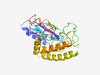 Estimated TM-score=0.43; very hard target. |
>Zn-ribbon-containing, possibly nucleic-acid-binding protein (DUF2310) (Length=255) MFLYEVRFECYSDTTISQAEKAINALLDCWRYNGQIIGREFPLVLKEAVFACRVVSPLQDSLLEKYQSNQVKRAKQGLHD VGLLSPKLKLLGRDINSDDSDECHTPKWQVLYTNYLQTCSPLRCGEHFTPIPLHRLPAVANGDHKQLIKWQEDWSACDQL QMNGSILEHGALNEISQINSRLFRRGYDLCKRLEYLTQVPTYYYLYRVGGDNLMSEQTRSCPSCGGDWRLEHSVHDIFDF KCDNCRLVSNLSWDF |
| 6329 |
PF09903 (DUF2130) |
 Estimated TM-score=0.43; hard target. |
>Uncharacterized protein conserved in bacteria (DUF2130) (Length=257) LKAKDDIIAYKDGEIERYKDMKARLSTKMVGESLEQHCETEFNKLRSTAFPRAYFEKDNDASEGSKGDFIFRECDEDGNE IVSIMFEMKNESDDSSHRHRNEDFFKKLDSDRTKKGCEYAVLVTLLEPESELYNQGIVDVSWRYDKMYVIRPQFFIPLIS ILRNAALNSLAYKAELAVVRNQNIDITHFEEQMEAFKNGFARNYDLASRKFQTAIDEIDKTITHLQKTKEALLSSENNLR LANNKAQDLTIKRLTRN |
| 6330 |
PF09815 (XK-related) |
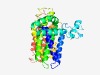 Estimated TM-score=0.43; hard target. |
>XK-related protein (Length=340) LWILAALAVYFADVGTDIWLAVDYYLRGQRWWFGLTLFFVLLGSLSVQVFSFRWFAHDFSTEDSAAAAAGHCPAAESKLL SLVHILQLGQVWRYFHTIYLGIRSRRSGENDRWRFYWKMVYEYADVSMLHLLATFLESAPQLVLQLCIVVQTRTLQALQG LTAAASLVSLAWALASYQKALRDSRDDKKPISYMAVIIQFCWHFFTIAARVITFALFASVFQLYFGIFIVLHWCIMTFWI VHCETEFCITKWEEIVFDMVVGIIYIFSWFNVKEGRTRCRLFIYYFVILLENTALSALWYLYKAPPISDAFAIPALCVVF SSFLTGIVFMLMYYAFFHPN |
| 6331 |
PF09575 (Spore_SspJ) |
 Estimated TM-score=0.43; hard target. |
>Small spore protein J (Spore_SspJ) (Length=46) MGFFNKDKGKRSEKEKNVIQGALEDAGSALKDDPLQEAVQKKKNNR |
| 6332 |
PF08730 (Rad33) |
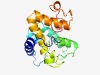 Estimated TM-score=0.43; hard target. |
>Rad33 (Length=171) SDQFEELLPEIEDEILEIYSDLVDDEEQDLYLNQLPQIFQTLRIPNCFSKDIIHCVDYYYGFIKDKNVQIDPMNKRQSNT INMIHAYTISSSNIKDIGSIIDIIDIDKLLRNLNRLIKFRNNYKHILASWKLFVTSANNGKPTMTTSSLESYQLTFPDLK QIKTNLNLDAD |
| 6333 |
PF08637 (NCA2) |
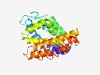 Estimated TM-score=0.43; hard target. |
>ATP synthase regulation protein NCA2 (Length=285) LLSSSTILRIVVNRRQEILQWVQDFGVTVRDFWFNWVVEPVRKVIGTIRHDSNSEIAIMSKDSLKADRESLERMVVDFAI DKPSAATGESSLSETQVAEIRAKVKEGDLTPVLRAYEKDLRSPFKGTVRGDLVRTLLIQVQKTKVDVEVAMSGIDALLKS QELVFGFVGLTPGILVSIGVMQYLRGAFGGRKGLNQSRRARRCVRVLRNIDRILSEALASPPQNNVLQYKDHGMLISEIH VLRRLARKIIPGDIEKEFLEDLDDLANLKGIGSQVRALERIRWAY |
| 6334 |
PF08524 (rRNA_processing) |
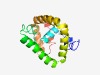 Estimated TM-score=0.43; hard target. |
>rRNA processing (Length=149) FRGKNPYTDRREFKSKEIKKSLVHRARLRKNYFKLLEKEGINHEPEQNGDESAESQNKSEEFKRSHIPDSRNQPSKRPMN FAERAKIAKERKEHSRQAKLKSIQDRRETIEKKSKERERRKDNLSKKTKSGQPLMGPRINNLLDKIKKD |
| 6335 |
PF08196 (UL2) |
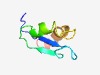 Estimated TM-score=0.43; hard target. |
>UL2 protein (Length=59) MAEDSVTILIVEDDDAYPSFGSLPASHAQYGFRLLRSIFLIMLVIWTAVWLKLLRDALL |
| 6336 |
PF08114 (PMP1_2) |
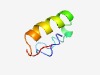 Estimated TM-score=0.43; easy target. |
>ATPase proteolipid family (Length=43) MLMSTLPGGVILVFILVGLACIAIISTIIYRKWQARQRGLQRF |
| 6337 |
PF08065 (KI67R) |
 Estimated TM-score=0.43; very hard target. |
>KI67R (NUC007) repeat (Length=112) KITKMPYKSLQPEPIDTPIRMKQRPKTSLGKVDVKGELLAVSNLTRTPGEITYTHTEPAGDGKSIRTFKESPKQILNPVA YVTGMKRWPRTPKEEAQSLEDLAGFKELFQTP |
| 6338 |
PF07792 (Afi1) |
 Estimated TM-score=0.43; hard target. |
>Docking domain of Afi1 for Arf3 in vesicle trafficking (Length=137) YILVASFDIDRGPVMEHQYPVAITGDEHMLAELMLPDQAHVRNQDWTIFFLHKDTSQEEEDEERRQKEQRRRRRRRKQNR AAGIISEEDEEELEEDEANDPDDWDDDESTDSEPEGGEGPPLIYVLNLVNTKQDKTV |
| 6339 |
PF07768 (PVL_ORF50) |
 Estimated TM-score=0.43; very hard target. |
>PVL ORF-50-like family (Length=116) IVKINGKPYKFTEHENELIKKNGLTPGMVAKRVRGGWALLEALHAPYGMRLAEYKEIVLSKIMERESKERKLERQRKKEA ELRRKKPHLFNVPQKHSRDPYWFDVTYNQMFKKWQE |
| 6340 |
PF07410 (Phage_Gp111) |
 Estimated TM-score=0.43; very hard target. |
>Streptococcus thermophilus bacteriophage Gp111 protein (Length=113) NVMARAWEIAKAAVLKFGGKAKEYFAQALVMAWAEIKTTKAIVEWDLPADTRKCRTWMARITGTHPVYKLDRQFVNPDGE DKYGDKLFDLTNGYYEAYNGKRRYFIQVIKGKA |
| 6341 |
PF07389 (DUF1500) |
 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF1500) (Length=97) MSSSVDVDIYDAVRAFLLRHYYNKRFIVYGRSNAILHNIYRLFTRCAVIPFDDIVRTMPNESRVKQWVMDTLNGIMMNER DVSVSVGTGILFMEMFF |
| 6342 |
PF07188 (KSHV_K8) |
 Estimated TM-score=0.43; very hard target. |
>Kaposi's sarcoma-associated herpesvirus (KSHV) K8 protein (Length=238) MPRMKDIPTKSSPGTDNSEKDEAVIEEDLSLNGQPFFTDNTDGGENEVSWTSSLLSTYVGCQPPAIPVCETVIDLTAPSQ SGAPGDEHLPCSLNAETKFHIPDPSWTLSHTPPRGPHISQQLPTRRSKRRLHRKFEEERLCTKAKQGAGRPVPASVVKVG NITPHYGEELTRGDAVPAAPITPPYPRVQRPAQPTHVLFSPVFVSLKAEVCDQSHSPTRKQGRYGRVSSKAYTRQLQQ |
| 6343 |
PF06797 (DUF1229) |
 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF1229) (Length=139) YSIPNRSVYYKFIIGFLILLISSSSIYFFLKETYIGQRIMNGIYSEKLNHHVDYWNTVKKDFFVYPKITIGSEYTFWYHN VFFDSHKTSGPITALILYIYSLFIFIIALKNSLKRNHRSFRYFHFYICFIPYLMTTIPW |
| 6344 |
PF06784 (UPF0240) |
 Estimated TM-score=0.43; very hard target. |
>Uncharacterised protein family (UPF0240) (Length=170) MGAAVARAIRNFNLENRAEREISRMKPSPAPRHPSTKNLLREQMSSHPEIKREIDRKDDKLLSLLKDVYVDSQDPVSSLQ GKDAATRQKPKEFRLAKDHQFDMMNVKNIPKGKISIVEALTLLNNHKLYPDTWTAKKIAEEYHLEQQDVNSLLKYFVTFE VKIFPPEGKK |
| 6345 |
PF06666 (DUF1173) |
 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF1173) (Length=383) YQIGGQSIHSDDPALSELLANIYSSRERPLCLCRTPGIEMYVAKVDDKYLIKRMPNSGSGHAPACDSYEPPPELSGLGQV IGSAIQENPDEGTTALKFDFALSKGASRSAPAPSGKETDSVKTDGNKLTLRGTLHYLWEEAGFNRWAPTMAGKRSWYVIR KYLLQAAEDKTAKGASLAGMLYMPESFNAEKKNEITQRRLAQMMRIATPEKGTRRLMLVVGEVKELAQSRYGHKIVLKHL PDCHFMMNDDIYKRLLKRFEVELGLWDALEGTHLMMIGTFSIGATGVASLEEVALMCVTENWIPFESTFEKMALDALTQQ NRRFMKGMRYNLPSTRPLACAVASDTEPEPTAMYVVPPGASEDYAAAVDELIAESKLPSWKWV |
| 6346 |
PF06469 (DUF1088) |
 Estimated TM-score=0.43; very hard target. |
>Domain of Unknown Function (DUF1088) (Length=168) EGRLLCHAMKDHIVRVANEAEFILNRQRAEDVHKHAEFESSCAQYAADRKEEEKMCDHLISAAKHRDHVTANQLKQKIVN ILTNKHGAWGIPCQSQLHDFWRLDYWEDDLRRRRRFVRNPFGSTHLDIACKSSQEYALKEEKVVKSKLVFRSPTLASQNP ETELVLDG |
| 6347 |
PF06336 (Corona_5a) |
 Estimated TM-score=0.43; hard target. |
>Coronavirus 5a protein (Length=64) MKWLTSFGRAVISCYKSLLLTQLRVLDRLILDHGLLRVLTCSRRVLLVQLDLVYRLAYTPTQSL |
| 6348 |
PF06223 (Phage_tail_T) |
 Estimated TM-score=0.43; hard target. |
>Minor tail protein T (Length=101) KLAREMGRPDWRAMLAGMSSTEYADWHRFYSTHYFHDVLLDMHFSGLTYTVLSLFFSDPDMHPLDFSLLNRREADEEPED DVLMQKAAGIAGGVRFGPDGN |
| 6349 |
PF06198 (DUF999) |
 Estimated TM-score=0.43; very hard target. |
>Protein of unknown function (DUF999) (Length=143) MVEDVCNVDLEQGLDLCKPEKVNKQSQRSRQSRQSLFTNTIKPQKDKMNIKTNKIKEFLNDLFTEFSKFHNSYYPDGRIS TRSNFRWPLLIIWSIIIVFAVDKKFEVQKFLSIWINENRFYSEIWVPIAIYVCLLVLMLLSLI |
| 6350 |
PF06147 (DUF968) |
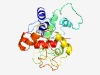 Estimated TM-score=0.43; hard target. |
>Protein of unknown function (DUF968) (Length=217) TERLSAIARSNVIAWIISVARGALAFDDTHELTLPELCWWAVRMDITDALPDSVARRALRLPPLPVQGVSRESDMVPGPS AAEIVQTKAQRAGAVKTLVNCDELQEQQPRVVALTIDPESPESYMLRPKRRRWENEKYTRWVKQQPCACCNQRADDPHHL IGHGQGGMGTKAHDLFVLPLCRRHHDELHRDTVAFEEKYGSQLELIFRFLDRALAIG |
| 6351 |
PF05874 (PBAN) |
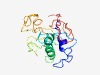 Estimated TM-score=0.43; very hard target. |
>Pheromone biosynthesis activating neuropeptide (PBAN) (Length=184) FNVLALALFSIFFASCTDMKDESDRGAHSERGALWFGPRLGKRSMKPSTEDNRQTFLRLLEAADALKFYYDQLPYERQAD EPETKVTKKIIFTPKLGRSVAKPQTHESLEFIPRLGRRLSEDMPATPADQEMYQPDPEEMESRTRYFSPRLGRTMSFSPR LGRELSYDYPTKYRVARSVNKTMD |
| 6352 |
PF05816 (TelA) |
 Estimated TM-score=0.43; hard target. |
>Toxic anion resistance protein (TelA) (Length=326) VADFAAKIDLRSSNAILQYGAGAQKKIADFSESALENVKTKDLGEVGDMLAGVVTELKSFDEEEEEKGIFGFFKKGGNKL ANMKAKYDKAEVNVNKICDALEGHQIQLMKDIAMLDKMYELNTTYFKELSMYIAAGKKKLQDVATTELPELEAKAARSGL PEDAQAVNDLNALCNRFEKKIHDLELTRTISLQMAPQIRLVQSNDTVMSEKIQSTLVNTIPLWKSQMVLAIGVENSSRAA KAQREVTDMTNELLRKNAEKLKLATVETAKESERGIVDIETLKATNESLISTLDEVMKIQQEGKEKRRTAEAELNRIENE LKQKLL |
| 6353 |
PF05540 (Serpulina_VSP) |
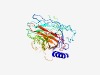 Estimated TM-score=0.43; hard target. |
>Serpulina hyodysenteriae variable surface protein (Length=371) KKVLLTAMALLTIASASAFGMYGDRDSWIDFLVHGNQLRARMDQLGFVLGNGTIKGTFGFRSQSAVTKIGNILSGNTGNV DLQTTISAGIGYTSEPFGIGVGYNYTYVNPRLGVHTPVLMINALNNNLRIAVPVQIAVSHDPFNDSAKFPYSSSTKDYMG ISTDIQLRYYTGIDAFNAIRVYFKYGQAGYKTADGANEAFAQSLGFEARFYFLNTPVGNVTINPFIKVAYNTALQGYNRT VRAGEAVTYDASKEYAGLGAGSFDAKDQKWDSNPYDVKAQAVLGITANSDVVSLYVEPSLGYQATYLGKHTSENIDSKVQ HSLAWGAYAELYVRPVQDLEWYFEMDVNNGGTRQPSNIPVYFATTTGITWY |
| 6354 |
PF05520 (Citrus_P18) |
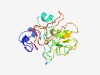 Estimated TM-score=0.43; very hard target. |
>Citrus tristeza virus P18 protein (Length=167) MSGSLGNSTHVDLLRSDSRFLSGWWSFIVNVGDIIVRFALHVPSEDMLNSFSAISNCTIIADGSALLKDNTVVDRLESMN PLAYLLKLAKTTTTICFTMSNKVLFGTTKSEPLSCLAITSDRVLFKVIMGTNVDDSRCGCSIWFYDNGTFQNGLTRCNNL VALFSAT |
| 6355 |
PF05414 (DUF1717) |
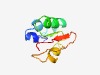 Estimated TM-score=0.43; hard target. |
>Viral domain of unknown function (DUF1717) (Length=78) DLLNFLVNEDISDELLDCIEEDKGLSHEMIEEVLITKGLSMVYTSDFKEMAVLNRRYGVNGKMYCTIKGNHCELSSKE |
| 6356 |
PF05407 (Peptidase_C27) |
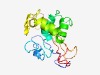 Estimated TM-score=0.43; very hard target. |
>Rubella virus endopeptidase (Length=164) CRGWHGMPQVRCTPSNAHAALCRTGVPPRASTRGGELDPNTCWLRAAANVAQAARACGAYTSAGCPKCAYGRALSEARTH EDFAALSQRWSASHADASPDGTGDPLDPLMETVGCACSRVWVGSEHEAPPDHLLVSLHRAPNGPWGVVLEVRARPEGGNP TGHF |
| 6357 |
PF05397 (Med15_fungi) |
 Estimated TM-score=0.43; hard target. |
>Mediator complex subunit 15 (Length=115) NNLPEKIRQLYNDIAKNAPAAEPVALSPEQKAAMTQQLRECTDMLGRMDTLAQWFSKIPGQEKNVRSLLAMRIQLMRQFK PGPDWTINDQFTIAPEYLTGTINYIRKLFHAMITR |
| 6358 |
PF05358 (DicB) |
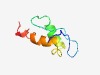 Estimated TM-score=0.43; hard target. |
>DicB protein (Length=62) MKTLLPNVNTSEGCFEIGVTISNPVFTEDAINKRKQERELLNKICIVSMLARLRLMPKGCAQ |
| 6359 |
PF05298 (Bombinin) |
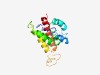 Estimated TM-score=0.43; hard target. |
>Bombinin (Length=141) MNFKYIVAVSFLIASAYARSEENDEQSLSQRDVLEEESLREIRGIGTKIIGGLKTAVKGALKELASTYVNGKRTAEDHEV MKRLEAVMRDLDSLDYPEEASERETRGFNQEEIANLFTKKEKRILGPVLGLVGSALGGLIK |
| 6360 |
PF04999 (FtsL) |
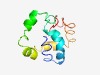 Estimated TM-score=0.43; easy target. |
>Cell division protein FtsL (Length=96) LAKLIVVDLFTVGRVPLVLLILIFASAMGVVFTTHHTRQAISEKDQAFMERDRLDNEWRNLILEETALAEHSRVQELAQK DLEMKRPDADKEVVVN |
| 6361 |
PF04917 (Shufflon_N) |
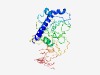 Estimated TM-score=0.43; very hard target. |
>Bacterial shufflon protein, N-terminal constant region (Length=329) DYIQTKGWQTEARLVSNWTSAARSYIGKNYTTLQGSSTTTTPAVITTTMLKNTGFLPSGFTETNSEGQRLQAYVVRNAQN PELLQAMVVSSGGTPYPVKALIQMAKDITTGLGGYIQDGKTATGALRSWSVALSNYGAKSGNGHIAVLLSTDELSGAAED TDRLYRFQVNGRPDLNKMHTAIDMGSNNLNNVGAVNAQTGNFSGNVNGVNGTFSGQVKGNSGNFDVNVTAGGDIRSNNGW LITRNSKGWLNETHGGGFYMSDGSWVRSVNNKGIYTGGQVKGGTVRADGRLYTGEYLQLERTAVAGASCSPNGLVGRDNT GAILSCQSG |
| 6362 |
PF04786 (Baculo_DNA_bind) |
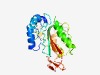 Estimated TM-score=0.43; very hard target. |
>ssDNA binding protein (Length=247) WVDELRYNLQAKNLTVLRCDVAFNKLFECLSFLRESLPLDNYLDSFLPEVAADIGILKPKAPAVVYLVGMLVKGGVEPFY VFDMAKVRRCMSNFGEFLSIRWSKQYIHNDAYANVIIKYKGFDCDKMKLQNSACVNLPSDDAVGRKTTFVRKFFDIKHHN NEKNYMTGRLIKSVKCEPFTVKRFNELFQFEQDSKSSDEVEMLVGIQIDGFKQGKNDIEFDTLVNNKKVSEKSYSLAIKP MVFFHIE |
| 6363 |
PF04771 (CAV_VP3) |
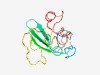 Estimated TM-score=0.43; very hard target. |
>Chicken anaemia virus VP-3 protein (Length=121) MNALQEDTPPGPSTVFRPPTSSRPLETPHCREIRIGIAGITITLSLCGCANARAPTLRSATADNSESTGFKNVPDLRTDQ PKPPSKKRSCDPSEYRVSELKESLITTTPSRPRTAKRRIRL |
| 6364 |
PF04753 (Corona_NS2) |
 Estimated TM-score=0.43; hard target. |
>Coronavirus non-structural protein NS2 (Length=105) IWHVSDAWLRRTRDFGVIRLEDFCFQFNYSQPRVGYCRVPLKAWCSNQGKFAAQFTLKSCEKPGHEKFITSFTAYGRTVQ QAVSKLVEEAVDFILFRATQLERNV |
| 6365 |
PF04584 (Pox_A28) |
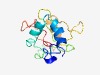 Estimated TM-score=0.43; very hard target. |
>Poxvirus A28 family (Length=140) MNVVTIFFIVVATVAICLIFFQGYSIYKNYDNIKEFNNAHSVLEYSKTVNVISLDRRVQDPNDQIYDVKQKWRCNKFEND YVSISIFGFKSDGNNIRKFRTLEDCINYTFSESTHSNIFNPCVSPNDPKSKECIFLKSIL |
| 6366 |
PF04220 (YihI) |
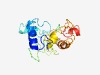 Estimated TM-score=0.43; very hard target. |
>Der GTPase activator (YihI) (Length=155) KAKARRKTREELNQEARDRKRDKKHRGHAPGSRATGASASGKSSGQSTQKDPRIGSKKPIPLGVSDAQETRPQKHQPQPK SEKPMLSPQEELDMLENDERLDALLERLEEGETLSADDQAWVDSRLDRIDELMQQLGLSYDDEEEEQQEDMMRLL |
| 6367 |
PF03669 (UPF0139) |
 Estimated TM-score=0.43; hard target. |
>Uncharacterised protein family (UPF0139) (Length=100) NSSDPRRPDKEIRYKPSVVNSQVQPGDDLTPDYMNILGMIFSMCGLMMRLKWCAWVALYCSCISFANSRVSDDTKQILSS FMLSISAVVMSYLQNPQPMT |
| 6368 |
PF03286 (Pox_Ag35) |
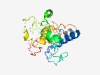 Estimated TM-score=0.43; very hard target. |
>Pox virus Ag35 surface protein (Length=195) MSWSINLGSGSGDNFKTLDEIRAHVKSTTETAIADDIFPEDIKIPSPKQSKTKRITTPRKKPVPKTSKKKEEELLPPPYK EEVKESEEEEEEEAVVEESPKSHQESTSNTQNTTPMTDTRDTMDTSDLKLVTEQIIKEMKALNAKVSAVSTILETIQAAS VGRQYTMLTKNIEELRMLVVCGKEQVVRKKTRHTT |
| 6369 |
PF03042 (Birna_VP5) |
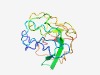 Estimated TM-score=0.43; very hard target. |
>Birnavirus VP5 protein (Length=135) SDDKPARSNPTDCSVHTEPSDANNRTGVHSGRHPREAHSQVRDLDLQFDCGGHRVRANCLFPWLPWLNCGCSLHTAEQWE LQVRSDAPDCPEPTGQLQLLQASESESHSEVKHTPWWRLCTKWHHKRRDLPRKPE |
| 6370 |
PF03040 (CemA) |
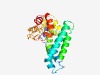 Estimated TM-score=0.43; very hard target. |
>CemA family (Length=221) ASTNSAKFLALLIVVPWVIDFIVHDYVMMPFLERYVQKVPLAAELLDVRRTQKLQMVKALKIEKARYRLEVEIGKSPPLS DEEVWIELRDDWRLENRNAFANIWSDTVYGIVLFLLMCFNQSKVAMLKFTGYKLLNNISDSGKAFLIILVSDILLGYHSE SGWHTLVEVILEHYGFEADEAAVTFFVCLVPVALDVYIKFWVYKYLPRLSPSVGNVLDEIK |
| 6371 |
PF03034 (PSS) |
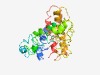 Estimated TM-score=0.43; hard target. |
>Phosphatidyl serine synthase (Length=276) PHPAIWRIVFGLSVLYFLFLVFIIFLNWEQVKNVMYWLDPNLRYATREADIMEYAVNCHVITWERILSHFDIFAFSHFWG WGMKALLIRSYGLCWTISITWELTELFFMHLLPNFAECWWDQLILDILLCNGGGIWMGXXXXLTENDTYHWASIKDIHTT TGKIKRAALQFTPASWTYVRWFDPKSSLQRVTGVYLFMIIWQLTELNTFFLKHIFVFPACHALSWCRILFIGIITAPTVR QYYAYLTDTQCKRVGTQCWVFGAIAFLEALACVKFG |
| 6372 |
PF02053 (Gene66) |
 Estimated TM-score=0.43; very hard target. |
>Gene 66 (IR5) protein (Length=201) VDGEESGAGTGTGAGADGLYPTSTDTAAHAVSLPRSVGDFAAVVRAVSAEAADALRSGAGPPAEAWPRVYRMFCDMFGRY AASPMPVFHSADPLRRAVGRYLVDLGAAPVETHAELSGRMLFCAYWCCLGHAFACSRPQMYERACARFFETRLGIGETPP ADAERYWAALLNMAGAEPELFPRHAAAAAYLRARGRKLPLQ |
| 6373 |
PF01350 (Flavi_NS4A) |
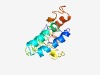 Estimated TM-score=0.43; hard target. |
>Flavivirus non-structural protein NS4A (Length=144) EVLVVLSELPDFLAKKGGEAMDTISVFLHSEEGSRAYRNALSMMPEAMTIVMLFILAGLLTSGMVIFFMSPKGISRMSMA MGTMAGCGYLMFLGGVKPTHISYIMLIFFVLMVVVIPEPGQQRSIQDNQVAYLIIGILTLVSVV |
| 6374 |
PF01309 (EAV_GS) |
 Estimated TM-score=0.43; very hard target. |
>Equine arteritis virus small envelope glycoprotein (Length=196) WWRGVHEVRVTDLFKDLQCDNLRAKDAFPSLGYALSIGQSRLSYMLQDWLLAAHRKEVMPSNIMPMPGLTPDCFDHLESS SYAPFINAYRQAILSQYPQELQLEAINCKLLAVVAPALYHNYHLANLTGPATWVVPTVGQLHYYASSSIFASSVEVLAAI ILLFACIPLVTRVYISFTRLMSPSRRTSSGTLPRRK |
| 6375 |
PF01054 (MMTV_SAg) |
 Estimated TM-score=0.43; very hard target. |
>Mouse mammary tumour virus superantigen (Length=173) LCLALPTDSKPISTTEAPILNITQSPSLNISSPSTLEPSEPLKNCTTFLDLLWQRLGENASIKDLMLTLQREEVHGRMTT LPSPRPSSKVEEQQLQRPRNLLPTAVGPPHVKYRLYNRLWEAPKGADVNGKPIQFDDPPLPYTGAYNDDGVLMVNINGKH VRFDSLSYWERIK |
| 6376 |
PF18852 (LPD34) |
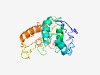 Estimated TM-score=0.42; very hard target. |
>Large polyvalent protein associated domain 34 (Length=213) MPYSSYDHDKLETAETMRIERRIYFEAKDGDIAPYVSLPLEQLHAMREESAAAEQAIFNDLSTRAAAWEQQAGKTLLLDK AIEYTRTTVVQHTSNEWQKGEYDRYTRSNRVYQMNYHIYENTRYDREKQQSVPYSYSLTWGVYTNSPNRNGQAKIAGQDR KVFSDKAAMEKYLNGRIKAYDRLFTEISPPIPQEYAEHFKVNGMLLPGYTIEG |
| 6377 |
PF18555 (MobL) |
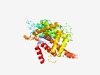 Estimated TM-score=0.42; hard target. |
>MobL relaxases (Length=402) SPGIVSVTHFKQASNKKFGDYVKYIDRDDATRVKYFNKWNFVERDGYNEYMGNPEKSTGLFASFSDHLGKDERHKVALEF GKAQKAGSNMWQTVVSFDNPFLEKYGLYNSKTDVLDEEKIRAATRIAMEEMLKREGMSSSALWTAAIHRNTDNIHVHIAT VEPVPSRPLQPFNDPDTHEKYWARRGSFKPSTFSAFKSKFANQILERDQELAKISTLIRDRMPPDTKLWRAAPTVQMNLL YQEIYKKLPRNKQLWRYNDNAMKEFRPEIDRLTSIYMEKYKPVELAEFRKAVAEEVEVRRDVYGTGQEGKELGRYEDYAD NKFHELQARMGNSVLGDMRNFDAELKAKYHDKTPGKFDERLAEGGSPLHQTLGRMGGLKRLLATDMKNMRLAKQQHKRME RE |
| 6378 |
PF18187 (RIF5_SNase_1) |
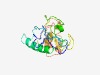 Estimated TM-score=0.42; very hard target. |
>TbRIF5 SNase domain 1 (Length=159) KRDDVLVSALSVEWIAKNVFLVAVPSYRRKVSCELTAVDGPRGTAPALTERVCDVLQRVNSAMRLYLRQDFYSNFPLEGY GDVIFSAETKQDTPYELHWCSLQETLVRAGVACVVRPSVIRQEALLQAEEEPTQEVKSSGASSWLHELQQREHSPLFCL |
| 6379 |
PF17734 (Spt46) |
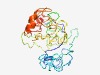 Estimated TM-score=0.42; very hard target. |
>Spermatogenesis-associated protein 46 (Length=228) MENFSLLSISGPRISSSALSTFPDIMSSRATSLPDIAKTALPTEASSPAQALPPQYQSSALRHGVHNTVLSPDCTLGDTQ SGEQLRRNCTIYRPWFSPYSYFVCADRESHLEACSFPEVQRDEGRGDNCLPEDMAESICSSSSSPENTCPREATKKSRHG LDSTDYITSQDIIAASKWHPAQQNGYKCAACCRMYPTLHSLKSHIKGGFKEGFSCKVYYRKLKTLWGR |
| 6380 |
PF17711 (DUF5556) |
 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5556) (Length=178) MELQGAQEDLGISLSSPRRNHETRPGSKAKGRSSICLQASVWMAGGKLRLRASEHLTQGHQQELRDWNLGEDASLLFSKS TFGAGKLIQAPAHVFRQCWVQGNAWISCITKFDSKRSPEVASSPSYLTVPRRSPLPVFLRPSDRCVCGGCYLGKSTRRTA CQSLLSDPLGTFPAQTRP |
| 6381 |
PF17707 (DUF5553) |
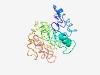 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5553) (Length=223) MRPLGKGLLPAEELIRSNLGVGRSLRDCLSQSRKLAEELGSKRLKPAKFGTEGKERVEQRTERQRTGSSKEPRMQIICRR RRREPPPRLLWGCLTPWAQPLRHVTAYENTGHWERLASVVSSKTQQPTVISHSSISITFSHYPPATLDSFLVLEPIKLFP VSSLRSPLCLNCGSCRESIRISGELIGNAHSPAPPRTPELETLGWDKQAVLSGAQVILVCAEV |
| 6382 |
PF17688 (DUF5536) |
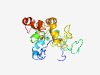 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5536) (Length=189) MKGTPSSLDTLMWVYHFHSSTEVALQPPLLSSLELSVAAAHEYLEQRFRALKSLEPREPKVQEMSPAPKPTLGLVLREAA ASLVSFGATLLEISALWLQQEARRLEGGDSSAGPALDIGDPSAALAQVAQAAGQGARKAGAAVGTGTRLLVQGAWLCLCG RGLEGSASFLRQWQQQLGLGIPREPVSSG |
| 6383 |
PF17685 (DUF5533) |
 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5533) (Length=139) MPVIPIPRRVRSFHGPHTTCLHSACGPVRTTHLVRTKYNNFDIYLKSRWMYGFIRFLLYFSCSLFTSILWVALSVLFCLQ YLGIRIFLRFQYKLSIILLLLGRRRVDFSLMNELLIYGIHVTMLLVGGLGWCFMVFVDM |
| 6384 |
PF17669 (DUF5529) |
 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5529) (Length=186) SQSPTMSQRPAPPLYFPSLYDRGISSSPLSDFNIWKKLFVPLKAGGAPAGGAPAAGGRSLPQGPSAPAPPPPPGLGPPSE RPCPPPWPSGLASIPYEPLRFFYSPPSGPEAAASPLAPGPMTSRLASASHPEELCELEIRIKELELLTITGDGFDSQRYK FLKALKDEKLQGLKTRQPGKKSASLS |
| 6385 |
PF17664 (DUF5526) |
 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5526) (Length=168) METRPSGGPSSRKESPETCPQGLLVFTGSSEQDANLAKQFWIAASMYPPNESQLVLSRGSSQRLPVAGSSKSSGLGKNYL QPFFLEKDKNTDVIAETLKIQESEEKEKYLQKAKRRDEILQLLRKQREARISKELISLPYKPKAKVHKAKKVISESDKED QEEVKALD |
| 6386 |
PF17620 (ORF45) |
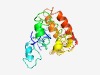 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (Length=191) MLPYEMVIAVLAYLSPAQILNLNLPFAYQKSVMFASNSAKVNELIRRRARDDDDNPYFYYKQFIKFNFLTNKIINVYSKT EKCIRMTFDGRYMVTRDFLMCFVNKSYIKQLLREVDTRITLQQLVKMYSPEFGFYVNSKIMFVLTESVLAFICLKHSFGK CEWLDKNIKTVCLQLRKICINNKQHLTCLSY |
| 6387 |
PF17594 (GP57) |
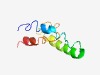 Estimated TM-score=0.42; hard target. |
>Phage Tail fiber assembly helper gene product 57 (Length=75) MSEQTVEQKLSAEIVTLKSRILDTQDQAARLMEESKILQGTLAEIARAVGITGDTIKVEEIVEAVKNLTAESADE |
| 6388 |
PF17539 (DUF5456) |
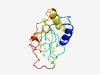 Estimated TM-score=0.42; hard target. |
>Family of unknown function (DUF5456) (Length=152) MAIYSLHEMAGLLKTGLWQGGEVEKLGYIGTAVPHASPAPACGLTGVYISYDTMLERCEVSMQGGGCELFISQYHPYTGT YMTYVLESNLSDRRLLLAKLEDILADRRNPYLWSHNLYKRNSFGIERRYDTGHSSMWGGRIAAPRQSKFFGR |
| 6389 |
PF17503 (DUF5435) |
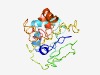 Estimated TM-score=0.42; very hard target. |
>Family of unknown function (DUF5435) (Length=208) MDQKRRRRIHALRPTTINAEVRPRRESLRRQLLSQLFFKRMAQLFKKKRQKKPSTVWYDKVAFPPSAWQGPARVSPYGYE SDSENEEYTCISSASLNNVLTESLTPTQNATANNCSNTYSESPAVLHLERIPRDLRETDALDSEEVYLDGFVTTRSRSNS FENMGPVKRRLYSKKFCRQFAAINIILLMLNILVLTGIVYRGFGKGCL |
| 6390 |
PF17439 (DUF5418) |
 Estimated TM-score=0.42; hard target. |
>Family of unknown function (DUF5418) (Length=151) MVNEYKAHSSFILKVVITLIGYWIASILAIIIYSMFFKIETNTFLLCLLLPTPIIWFNILIGMGLTYRCMENLTIYDKHK LWCVFVRDLTLTILATILATLTTMELYQIEHPLKPIEFVFIVGLVLIVGFTIITTLIIKYLKIIKNLKKIS |
| 6391 |
PF17382 (Ycf70) |
 Estimated TM-score=0.42; hard target. |
>Uncharacterized Ycf70-like (Length=89) MVYGYGKSNMPHPNRKRKGTDTQYDYWEELLVMVSGLYALFCVFLVLFIFFDSFKQESNKLELSGKEEKKKLNGENRLSR DIQNLLYIK |
| 6392 |
PF17377 (DUF5399) |
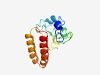 Estimated TM-score=0.42; hard target. |
>Family of unknown function (DUF5399) (Length=134) TTIDQLDIGIYVQYARRTQLIEQINQQYHLDEASSIPPQILLVDIYPKLSEMDLLLGIVPIQTPWAYFFPPPRFRFQRRS PFGFFRVAPSLGSFEKEDEDERKLEEVECETEEDLKEKKAIKSCFDQIDKINNW |
| 6393 |
PF17335 (IES5) |
 Estimated TM-score=0.42; hard target. |
>Ino80 complex subunit 5 (Length=115) LEKEYQKLSLRNDELVKQDSTLRKEYTTLLRKASSLASVLKVMDSKLAEQCEIEQPKLIGDRALKLVPGLQWYNDQINLV TQSFDNDNEEIEIPKELLDSYTLCKDTPLLYKDSQ |
| 6394 |
PF17245 (CDC24_OB2) |
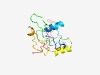 Estimated TM-score=0.42; hard target. |
>Cell division control protein 24, OB domain 2 (Length=133) LGDFWSSNTINLYLHRRYYDLVDPDSPDNGILKKGREIFLTGCCLRSATEGSGCRRLLPTEYLVILLDENQDEDAMLLGA QFCSDSFSSISLDSVKSGASYSFYARIESIGLLEIQGKFGSLQRKQITLVDND |
| 6395 |
PF17229 (SMG1_N) |
 Estimated TM-score=0.42; very hard target. |
>Serine/threonine-protein kinase SMG1 N-terminal (Length=106) SDSLSASQDGVKCSVSRDRGSEVLFEILPSDRPGTPPLHNDAFTATDALEEGGGYDADGGLSENAYGWKSLGQELRINDV TTDITTFQHGNRALATKDMRKSQDRP |
| 6396 |
PF17213 (Hydin_ADK) |
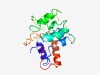 Estimated TM-score=0.42; hard target. |
>Hydin Adenylate kinase-like domain (Length=194) AGKTSTSVTLAKHYGTPCLNIDSMVLEAVSNGNSPVGLRARELCARAAVEQAQRKAEDAAAEAAAAAAGAQAPGFLSVEA VTKHTAEGSQTSEGKQGPHSSISTRNKASTVGGKGKTESSHTHKQQTSEHIAVSQSLSVPVPAGLPQRRLSVSASVAGEL GLLSCTLPEELLVDILTERLQLNDCHRGVVFDSL |
| 6397 |
PF17156 (GAPES2) |
 Estimated TM-score=0.42; hard target. |
>Gammaproteobacterial periplasmic sensor domain (Length=204) IYTQMVKERVYSLKQSVIDTAFAVANIAEYRRSVAIDLINTLNPTEEQLLVGLRTAYADSVSPSYLYDVGPYLISSDECI QVKEFEKNYCADIMQVVKYRHVKNTGFISFDGKTFVYYLYPVTHNRSLIFLLGLERFSLLSKSLAMDSENLMFSLFKNGK PVTGDEYNAKNAIFTVSEAMEHFAYLPTGLYVFAYKKDVYLRVC |
| 6398 |
PF17055 (VMR2) |
 Estimated TM-score=0.42; very hard target. |
>Viral matrix protein M2 (Length=235) MDAPINPSSPTRSSYETALERQWLIKDEPSHHVDLGCHFTLEIKAFSPKMIELLETGSLTLGSVYQGLYNLIKDKDCKGL ELDRDTTHGNKLHVLAAAFLCVLRKYENQVRFYNVKTKDSLTGEIQYMSRVHIGDDNLGLFDPDTYRMIGGSFPEGIYRL NLRTQLTPRGEDDGLEIIMGTILTVPVRGPADRQPLGVLMDRQLRQFPVGTNPWSSLGEAKPKGRSLSLLRGILM |
| 6399 |
PF17043 (MAT1-1-2) |
 Estimated TM-score=0.42; very hard target. |
>Mating type protein 1-1-2 of unknown function (Length=152) TLAILYATYTIGPHLEGFSFNHLSHMPISKSRELFLRVFVSISGNVYNDDEVFTTPPAFEFGAAQGNVRICQQGKKLLVR SLSEQFYREAPDWHPYRRVPGSPWNNFIRNTELPVFSTNENPTPHAKTKYILPYNAMVLAGQFKAKYNLVRA |
| 6400 |
PF17024 (DMAP1_like) |
 Estimated TM-score=0.42; hard target. |
>Putative DMAP1-like (Length=113) KNFLNLLEKVEELSSKRKQILERHFYELTKYNIDDLEDRDSIITRIVGKAPSHPNAFLASSLGFTKSSWNKKYENMFLKF GLDRKLKHSTYRNMLHFEKLKLLVIKYHEVNKK |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218