

Go to page (83 pages in total): [First page] << < 56 57 58 59 60 61 62 63 64 65 66 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 6001 |
PF07887 (Calmodulin_bind) |
 Estimated TM-score=0.45; very hard target. |
>Calmodulin binding protein-like (Length=276) LRLIFSKNLSLPIFTGSKIVDIENNPLQVLLVDTSVAGTDRMSLVSFAHPIKIEIVVLDGDFPLGDCDAWTAEEYDSKIV KERTGKRPLLAGEVTITMREGVATIAGDIEFTDNSSWIRSRKFRIGAKVSPGSNHQGARIYEAMTEAFVVKDHRGELYKK HHPPMLNDEVWRLQKIGKEGAFHKKLQSEGINSVQDFLKLSVVDPQKLRTILGAGMSDKMWDATIKHARTCVLGNKLYLL KGNNLAIFLNPVCQLVKAEIDGHLYHTRDLNNLNRV |
| 6002 |
PF07797 (DUF1639) |
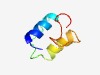 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1639) (Length=50) LSRKEIEEDFIAMTGKKPPRKPKKRPKIIQNELDAVFPGLWLSEVHPDRY |
| 6003 |
PF07760 (DUF1616) |
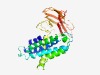 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1616) (Length=270) LLFTFLTTIFVTVPVLNGTVIKTVLELIFILFIPGYSLIAALFPRKQDLDGIERVALSFGFSIVLTLLIAFILNYTSFEI RLDPILIVLSLLSFILIVVAFLRRRRIPDQEKFKLNLGVHFHNLKSSFDQEKKLDKILSIILIISIILAISATIYIIVTP KEGEKFTEFYILGAGGKASDYPTNLTIGQSGTIMVGIVNHEYSNVTYNLLVKFNGTTVTSKNITLTNGQKWEQPLTFNAS TLGKNQKLELLLYKLPDQDKAYRSLHLWVD |
| 6004 |
PF07759 (DUF1615) |
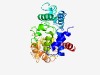 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1615) (Length=315) VRDRQGWATDIAAAFTAQDIPPTPENLCAVIAVTEQESTFQADPQVPGLGKIAWKEIDRRADQMHIPNFLVHTALLIKSP NGKSYSERLDKVRSEKELSAIFDDFINMVPMGQKLFGNLNPVRTGGPMQVSIAFAEAHAKGYPYKVDGTIRREVFSRRGG VYFGTMHLLGYPANYPQPIYRFADFNAGWYASRNAAFQRAVSRLTGIKLALDGDLINYGTDKPGSTELAVRSLGKRLDMS DSAIRRALEKGDSIDFEKTSLYKQVYALAEKSGGKKLPREILPGITLESPKITRQLTTAWFAKRVDGRWQKCMAK |
| 6005 |
PF07666 (MpPF26) |
 Estimated TM-score=0.45; hard target. |
>M penetrans paralogue family 26 (Length=135) NVNLFTKIKKHSKNMLIFYFTAIVLAIIGAILISVGAAKIALSLSNNYSGEYYDLSSFINGEWIVGVLFIVLTGIFYLVC FVIGIILVIKVNDAQGKHPEFQDITMHTVLLIIGIVILPICAIIDCFILYSKSKK |
| 6006 |
PF07587 (PSD1) |
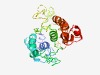 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1553) (Length=260) NRLGLAQWLVHPEHPLTARVTVNRMWQHHFGTGLVKTAEDFGVQGEYPSHPALLDWLAVDFIESGWDVKRLHKLLVMSAT YQQSSRVDAAKLAADPENRLLSRGARFRLDAEVIRDQALAVSGLLVPEIGGKSVRPYQPPGLWKPVGFGGSNTSVFKQDT GDKLYRRSMYTFWKRTSPPPSMTAFDAPDRETCQVRRARTNTPLQALVLMNDVQFVEAARKFAECVVRQGGSSVEERVTF AYRSLLSRRPTASELASVTK |
| 6007 |
PF07431 (DUF1512) |
 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1512) (Length=355) LYMLFFIVIIAISFIPGLQQQYQMMFMSKDVEKNLNTIENYLKDAKGIAEKLLKEKNFQDPKAFLDRVIDRFVIDPVSIE PTDIISRMKLLLRTNEDTVRQMITKYSPNIDPMSRSQIEVSIEVINALNMIYKVIRHYFLLAKKLKNLMIMYQLQAVAPI YAKLAEAYGKAQKVFLQGIPVGDGLGPLVASRFLMNINDKWSPSKDTIAGSMEFEGRKLIVVKAEGPMATVGEPGEAVQN IVEKEGNKVARIITVDAALKLEGEETGSIAEGMGVAMGDPGPEKIAIERVAAKYNIPIDAVIVKMSMEEAITEMRKEVYE AADKVVNYVKNIILERTKPGDTVVLVGVGNTVGIA |
| 6008 |
PF07326 (RCS1) |
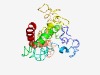 Estimated TM-score=0.45; very hard target. |
>Regulator of chromosome segregation 1 (Length=231) MASRWQNMGTSVRRRSLQHQEQLEDSKELQPVVSHQETSVGALGSLCRQFQRRLPLRAVNLNLRAGPSWKRLETPEPGQQ GLQAAARSAKSALGAVSQRIQESCQSGTKWLVETQVKARRRKRGAQKGSGSPTHSLSQKSTRLSGAAPAHSAADPWEKEH HRLSVRMGSHAHPLRRSRREAAFRSPYSSTEPLCSPSESDSDLEPVGAGIQHLQKLSQELDEAIMAEESGD |
| 6009 |
PF07325 (Curto_V2) |
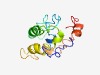 Estimated TM-score=0.45; hard target. |
>Curtovirus V2 protein (Length=126) MGPFRVDQFPDNYPAFLAVSTSCFLRYNKWCILGIHQEIEALTLEEGEVFLQFQKEVKKLLRLKVNFQRKCSLYEKIYKK YVYNVPEKKGESSKCVAEEEEDYYEFEEIPMEETCDKKQDSEVKDV |
| 6010 |
PF07312 (DUF1459) |
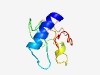 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1459) (Length=81) MFTKCIAAIFLCAMIIGSTHQQVVVPAYSAAYVPSAYAYPSVYSPYVAAAAYPSVWAWGSNKNKEDAPQAFARPSALVNN Q |
| 6011 |
PF07270 (DUF1438) |
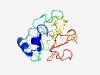 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1438) (Length=151) MMQSIPSFVKDTSDIEEHALPTAQVLPAQSTRCSKSETLCFSKEQSHCSEDGWIAEWDLYSFCVFESVDYLRSYRRLNSA MKKGTEVFQSESQRKPQVSPGDVENYKDKDTEEPDQPFPSLLREKGLDLATCDGGDCPDQDPVSDNSRHLG |
| 6012 |
PF07214 (DUF1418) |
 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1418) (Length=94) MRTIGVLPKSVLILEYLGMILLALALLSLNHYLTLPAPFNTPLAGVLMVFLGVVLILPAAVAMMWRIAQLLAPQLMKRPP DISSRSDREKHNDS |
| 6013 |
PF07175 (Osteoregulin) |
 Estimated TM-score=0.45; very hard target. |
>Osteoregulin (Length=160) NKEYSISNKENTHNGLRMSIYPKSTGNKGFEDGDDAISKLHDQEEYGAALIRNNMQHIMGPVTAIKLLGEENKENTPRNV LNIIPASMNYAKAHSKDKKKPQRDSQAQKSPVKSKSTHRIQHNIDYLKHLSKVKKIPSDFEGSGYTDLQERGDNDISPFS |
| 6014 |
PF06810 (Phage_GP20) |
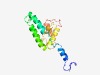 Estimated TM-score=0.45; hard target. |
>Phage minor structural protein GP20 (Length=143) EQAKKVMDSLDGNFVTKTRFNEVNEENKTLKQSVADRDKQLDDLKKSSGDNAELKKQIETLQQQNADQKKTHDAEMALLK LDNAVDAALTAAGAKNAKAVKALLDISKVKLGEDGKLSGWEEQLSSVQKSDSYLFEAKQNQPV |
| 6015 |
PF06708 (DUF1195) |
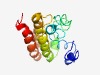 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1195) (Length=135) ANNNNNNHNNNSNKGNYNYKFWVLAAIIVLALWSMFTGSVTLKWSASNLTRFSHDLDHSITLQDLDVLEVEEREKVVRRM WEVYTHSTTAKVPRFWSDAFHAAYEHLVSDVPTIRDAAISEIAKMSIQFLSLQQQ |
| 6016 |
PF06668 (ITI_HC_C) |
 Estimated TM-score=0.45; very hard target. |
>Inter-alpha-trypsin inhibitor heavy chain C-terminus (Length=187) TGIVVNGQIIGDKKVIPGSKLHTYFGHFGIVHERFGIRLMVSTKEISLSEEGKQAKLSWTNNRTINGLNVDLQVIKDCSL TVTLRNSVKFLIILHKIWQKHPYHQDYLGFYTLDSHHLSHNVHGLLGQFYKGVNFEVSDLFPGKDPDKQDAIMFVKGNKL TVTRGWKRDFRKDVKNGDNVPCWFIHN |
| 6017 |
PF06663 (DUF1170) |
 Estimated TM-score=0.45; very hard target. |
>Protein of unknown function (DUF1170) (Length=214) TSLKKEKSAILDLYIPPPPAVPYSPRDEKGSFVYGGLSKCKQPLPGSKGSESPNSFLDQESRRRRFTIADSDQLPGYSVE ANVLPTKMREKTQSYGKPRPLSMPADGSWMGIVDPYARPRGNGRKGEDALCRYFSNERIPPIIEESSSSQHRFSRPVTDR HLVRGADYIRGSRCFINSDLHNSATIPFQEEGSKKNSVSSSKKSSSTEPSLLVS |
| 6018 |
PF06615 (DUF1147) |
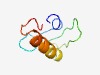 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1147) (Length=59) MYTSLWGHLGVAKANGLLILQTRKPHTGNHLETSGGMVTMVKKWLLLMTFMAGCRGMIY |
| 6019 |
PF06586 (TraK) |
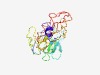 Estimated TM-score=0.45; hard target. |
>TraK protein (Length=226) ALPLLAALLASPAQADQFKQTADGGTIECSVSARELTRFALVGDQFASVSKLSTGYPYNDFAVTNEPVRGDIYVSVPETY AAAKISFFATTKKGYVYKLACPVERIEAQQVFITNPAIARNDATRWETETPLGETAVRLVQAMANNAAIDGYRIRQSAAL PARVGSLEVQLIADYRGAALTGKVIHLTNRGNRPMTITEHDLAPRDALAVAIANPALEPGASTTAY |
| 6020 |
PF06364 (DUF1068) |
 Estimated TM-score=0.45; hard target. |
>Protein of unknown function (DUF1068) (Length=171) AVLKAGLVIVGVCIAGYIVGPPLYWHFMEGLAAVSHSSSSSSLSSCPPCTCDCSSQPLLSIPIGLSNISFADCAKHDPEV NEDTEKNFVDLLTEELKLREAEAIENQQRADMALLEAKKIASQYQKEADKCNSGMETCEEAREKAEATLVAQRKLTATWE LRARQRGWKEG |
| 6021 |
PF06320 (GCN5L1) |
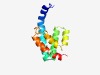 Estimated TM-score=0.45; hard target. |
>GCN5-like protein 1 (GCN5L1) (Length=113) LLKEHQAKQNERKELQEKRRKEAIAAATSLTEALVDHLNVGVAQAYVNQRKLDHEVKTLQIQAAQFAKQTTQWIGLVESF NQALKEIGDVENWARSIEKDMRIIATALEYVYK |
| 6022 |
PF06256 (Nucleo_LEF-12) |
 Estimated TM-score=0.45; very hard target. |
>Nucleopolyhedrovirus LEF-12 protein (Length=173) DAEFNRRLEYVGTIATMMKRTLNVLRQQGYCTQQDADSLCVSDDTAAWLCGRLPTCNFVSFRVHIDQFEHPNPALEYFKF EESLAQRQHVGPRYTYMNYTLFKNVVALKLVVYTRTLQANMYADGLPYFVQNFSETNYKHVRVYVRKRAMQVVATLPVYE QIIEDTINELVVN |
| 6023 |
PF06163 (DUF977) |
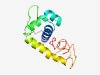 Estimated TM-score=0.45; easy target. |
>Bacterial protein of unknown function (DUF977) (Length=127) MAKPFTHEQREELKARIIGLVRKNERMTISQLERATGAGWHSVRRCLVDVLACGDLYMPGKYGVFTSEQVYRVWRKAAEK ATDQTLIRKLPDGEIRRYDRQQNIICGECRKSEVMLRVLAFYQGNFQ |
| 6024 |
PF06129 (Chordopox_G3) |
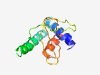 Estimated TM-score=0.45; very hard target. |
>Chordopoxvirus G3 protein (Length=107) LIYLLFFIIFLVLTYYFNKHPTNKLELSVDKLNRENKIIKQRDDAFPVVLNTTVFTRPETPVPTKVHTYYDSATGVVTML SNNKKRIFRLDFDDDVRTLLPILLLSK |
| 6025 |
PF06121 (DUF959) |
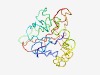 Estimated TM-score=0.45; very hard target. |
>Domain of Unknown Function (DUF959) (Length=188) LLTCCLVPSWADLLTLDWLWSTDLADSRDTLVSQPQGSLPVQPAAAPATHVAPQDDPAEQRTAPASPRPPLENPKAAPSR APGAPISAAASAAPAASPDTREENVAGVGAKILSIAQGIQSIVQLWNEATPTQSSAGSASAPTDPLTLAGPSGPPEESGT TLGPHRGAPGSLGTWTAESGTLAVPTQS |
| 6026 |
PF06116 (RinB) |
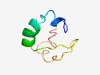 Estimated TM-score=0.45; hard target. |
>Transcriptional activator RinB (Length=51) KEILRLLFLLAMYELGKYVTEQVYIMMTANDDVEAPSDFAKLSDHSDLMRA |
| 6027 |
PF06103 (DUF948) |
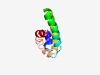 Estimated TM-score=0.45; easy target. |
>Bacterial protein of unknown function (DUF948) (Length=88) IAGLIAAIAFLILVIYLCRVLIRLVQTLNEVNDTVNTLTKDADSMLDQVNDLLDKANDLVEDVNGKVKTIDPLFTAVADL SESVSDLN |
| 6028 |
PF06101 (Vps62) |
 Estimated TM-score=0.45; hard target. |
>Vacuolar protein sorting-associated protein 62 (Length=521) ETTFQLPSPLPIWPPGDGFASGIINLGGLQVRQTSSFNKVWATHEGGPDNLGATFYEPSNVPQGFFMLGCYSQANNKPFF GWVLVAKDDSCGALKKPLDYTLVWSSESLKIKQDGNGYIWLPTPPDGYKAIGHVVTNSPDKPSLDKIRCVRSDLTDQCEA HTWTWGPGNTGDANGFNVYSLRPSNRGIQAMGVSVGTFVAQIGGVVSPLSNIACLKNAKSNLSCMPNLKQIEALVNAYSP WVYFHSEETYLPSSVSWFFVNGALLYKKGEESKPVAIEPTGSNLPQGGSKDGAYWLDLPIDEGAKERVMKGDLGNSQVCV HIKPMLGATFTDIAIWVFYPFNGPAKAKVKLVNVPLGKIGEHVGDWEHVTLRVSNFNGELQRVYFSEHSGGTWVDASELE FQNGNKVVAYASLHGHAFYSKPGLVLQGSGGIGIINDIEKSRLVMDTGANYSLVSAEYLGSAIVEPAWLNYSREWGPKIS YNIADEIKKAKKLLLGPLKSAFEKSVRSLPNDVLGEEGPTA |
| 6029 |
PF06047 (Nkap_C) |
 Estimated TM-score=0.45; hard target. |
>NF-kappa-B-activating protein C-terminal domain (Length=99) NYGKALRPGEGEAIAQYVQKNMRIPRRGEVGWSGQEIENLESLGYVMSGSRHKRMNAVRXRKENQVYTAEEKRALALINF EEKQQRENRIMTEFREMLT |
| 6030 |
PF05955 (Herpes_gp2) |
 Estimated TM-score=0.45; hard target. |
>Equine herpesvirus glycoprotein gp2 (Length=230) KNFMEASCTVETNSDLAIFWKIGKPSVDAFNRGTTHTRLMRNGVPVYALVSTLRVPWLNVIPLTKITCAACPTNLVAGDG VDLNSCTTKSTTIPCPGQQRTHIFFSAKGDRAVCITSELVSQPTITWSVGSDRLRNDGFSQTWYGIQPGVCGILRSEVRI HRTTWRFGSTSKDYLCEVSASDSKTSDYKVLPNAHSTSNFALVAATTLTVTILCLLCCLYCMLTRPRASV |
| 6031 |
PF05840 (Phage_GPA) |
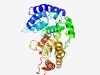 Estimated TM-score=0.45; hard target. |
>Bacteriophage replication gene A protein (GPA) (Length=324) HQIQTLATMTAAMFSGTFEKLCDGFGATDGELTMDVTLKAYQMLARMALHLHAMPPHYDALTTDKDRRNEPDTELLPGAI LRLTCAEWWKRKLWLLRCEWREEQLRAACLVSRKTSPYLSQDALSEFRAQREKTRDFLKSFMLENEDGFTIDLETVYYAG VSNPIHRKAEMMATMKGLELLAEARGDRAVFLTVTCPSKYHATTENGHPNPKWNGATMRDSSDYLVNTFFAAVRKKLNRD GLRWYGIRTVEPHHDGTVHWHMMVFAHPEEIDTIVSHTRDIAIQEDRHELGNDITPRFKVEYVDGSKGTPTSYIATYIGK NLDS |
| 6032 |
PF05756 (S-antigen) |
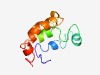 Estimated TM-score=0.45; hard target. |
>S-antigen protein (Length=89) MNRILSVSFYLFFLYLYIYKTYGKVKNTDHELSNIYGIKYYLRNGLSDKKNGKGQKYQDLEEDENDDEEDSNSEESNNDE ENKLIEGQG |
| 6033 |
PF05752 (Calici_MSP) |
 Estimated TM-score=0.45; very hard target. |
>Calicivirus minor structural protein (Length=165) MSWLVGALQTFGSLADVAGTVSNIVYQQRQAAQLEKQNELMETWMNKQEALQKSQMELTRDLSINGPAARVQSALDAGFD EVSARRIAGSGERVIWGNLDRPIMHAGTMDSIRQTRHLDSLSHSLATFKNGTPFGKPAPPTAKSGRPQATTAQITIGHNP GSTSV |
| 6034 |
PF05749 (Rubella_E2) |
 Estimated TM-score=0.45; hard target. |
>Rubella membrane glycoprotein E2 (Length=267) GLQPRADMAAPPTLPQPPCAHGQHYGHHHHQLPFLGHDGHHGGTLRVGQHYRNASDVLPGHWLQGGWGCYNLSDWHQGTH VCHTKHMDFWCVEHDRPPPATPTPLTTAANSTTAATPATAPAPCHAGLNDSCGGFLSGCGPMRLRHGADTRCGRLICGLS TTAQYPPTRFGCAMRWGLPPWELVVLTARPEDGWTCRGVPAHPGARCPELVSPMGRATCSPASALWLATANALSLDHALA AFVLLVPWVLIFMVCRRACRRRGAAAA |
| 6035 |
PF05745 (CRPA) |
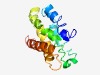 Estimated TM-score=0.45; very hard target. |
>Chlamydia 15 kDa cysteine-rich outer membrane protein (CRPA) (Length=146) MLTGVENSESGVIDLIKPGLDDVMKNETVQVTLVNSVLGWCKAHIVDPIKTSKIVQSRAFQITMVVLGVILLIAGLALTF VLQGQLGKNAFLFLIPAVIGLVKLLTTSVFMEKPCTPEKWRLCKRLLATTEDILDDGQINQSNTIF |
| 6036 |
PF05744 (Benyvirus_P25) |
 Estimated TM-score=0.45; hard target. |
>Benyvirus P25/P26 protein (Length=225) DIDHCMPVFDMAYSDDNHLPYYIQRSTHHVVRDVDYTGFICYPLQVDLNDNVEVGADIYHMKIKTMRFNVDIYNNDVATK FPGWVRFIVFCTPPVSSWVNDGCSSLFSPFVGVNSFIDPKLLKRDGHGITVLHDGIYCLCHQEHFTRSFEFNFRGPGNYT LTSDVCWSPATNVDSIYVACVASWCGDSAFMLQSDSVSWVHKRFWQRPVLEFGQCLDDLPDHDND |
| 6037 |
PF05546 (She9_MDM33) |
 Estimated TM-score=0.45; hard target. |
>She9 / Mdm33 family (Length=194) SKQFTNLMDNLQSNIFIVGQRLNDLTGYSAIEKLKKEIESHEARVREARSVVRKAKEAYSAAINRRSASQREVNELLQRK HAWSPADLERFTSLYRSDHANEVAEAEAQQALAEAEREVEEATAQLNRSILSRYHEEQIWSDKIRRMSTWGTWGLMGVNV LLFLIFQVLVEPWRRRRLVKGFEEKVKEAIENEA |
| 6038 |
PF05545 (FixQ) |
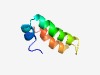 Estimated TM-score=0.45; hard target. |
>Cbb3-type cytochrome oxidase component FixQ (Length=48) TYETLANFAQTWGLLYFVLIFAGVLVYTFWPSNKKRFDDAANMPLRED |
| 6039 |
PF05418 (Apo-VLDL-II) |
 Estimated TM-score=0.45; hard target. |
>Apovitellenin I (Apo-VLDL-II) (Length=79) KSIFERDRRDWLVIPDAVAAYIYEAVNKMSPRAGQFLVDISQTTVVSGTRNFLIRETARLTILAEQLMEKIKNLWYTKV |
| 6040 |
PF05315 (ICEA) |
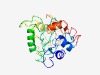 Estimated TM-score=0.45; hard target. |
>ICEA Protein (Length=218) TKTELFLELAKPDSNGVSRWVSVNEFIGKYKALQLGNGGSWCRASSSLAKTYTIEFDKTKTRGNSIDAIRLTGFNNSNTF NQSIRKEIKDFYKNKNCVMLGINGNSINTKIEIDHKDGRKDNDRVSNIKTQRLEDFQPLSKAANDAKRQICKRCKETNLR WNAKNLKGNPYSFYEGDEHYTQELGCVGCYQFDPVEYRKSSVIKISKEVTDNILKKLY |
| 6041 |
PF05084 (GRA6) |
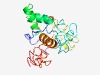 Estimated TM-score=0.45; very hard target. |
>Granule antigen protein (GRA6) (Length=215) MAHGGIHLRQKRNFCPLTVSTIAVVFVVFMGVLVNSLGGVAVAADSDGVKQTPSETGSSGGQQEAVGTTEDYVNSSAMGG GQGDSLAEDDTTSDAAEGDVDPFPVLANEGKSEARGPSLEERIEEQGTRRRYSSVQEPQAKVPSKRTQKRHRLIGAVVLA VSVAMLTAFFLRRTGRRSPPEPSGDGGGNDAGNNAGNRGNEGRGYGGRGEGGGED |
| 6042 |
PF04894 (Nre_N) |
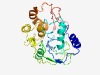 Estimated TM-score=0.45; hard target. |
>Archaeal Nre, N-terminal (Length=268) CGLSYCPLLARRMATYKLRRVVGSRELFGSSPPSVFVGRHGYPYVNIGPSIPPETGDTSIYDLPEKWLGLKLEKILDYRW SLITGSRKHRVNDLSDSFIEKIHEIILSIKPVDVEVYLEKPPKPLIIFSEYEPPQGPRAPVKNVRVVSNPKIPRIIDRVY QDKDLKATNAIIELYRHNIPVTIIQRIFSVGALGVQRKRRLVPTRWSITAVDDTISKYLIEKIKEYDEITEIEVYVRKYY DNLFIAILYPGKWSFEWMEAWWPGSTWN |
| 6043 |
PF04846 (Herpes_pp38) |
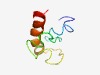 Estimated TM-score=0.45; hard target. |
>Herpesvirus pp38 phosphoprotein (Length=63) ENKNLRKKYNIILDVSTKILVFGTCIMFATGTLIGKGTKIQVPVCNSTNLALAFLAGFLTRHV |
| 6044 |
PF04741 (InvH) |
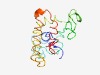 Estimated TM-score=0.45; very hard target. |
>InvH outer membrane lipoprotein (Length=147) MKKFYSCLPVFLLIGCAQVPLPSSVSKPVQQPGAQKEQLANANSIDECQSLPYVPSDLAKNKSLSNHNADNSASKNSAIS SSIFCEKYKQTKEQALTFFQEHPQYMRSKEDEEQLMTEFKKVLLEPGSKNLSIYQTLLAAHERLQAL |
| 6045 |
PF04725 (PsbR) |
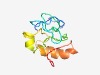 Estimated TM-score=0.45; very hard target. |
>Photosystem II 10 kDa polypeptide PsbR (Length=95) AKKIQTSQPYGPAGGVVFKEGVDASGRVAKGKGLYQFSNKYGANVDGYSPIYTPEEWSSTGDVYVGGKAGLLLWAITLAG ILVGGAILVYNTTSV |
| 6046 |
PF04530 (Viral_Beta_CD) |
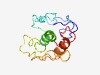 Estimated TM-score=0.45; very hard target. |
>Viral Beta C/D like family (Length=120) ILHSPNCSCQFCSSELPSTHTCGSQDRTVPLHVEATAAGHMEAKNFSLQYVLLVAFVSVLLGFSFCVYLKSMSNDEASDM TYYYQDLNSVEIKLGKNPLDPEVIKAIHSFQEFPYGNIPS |
| 6047 |
PF04370 (DUF508) |
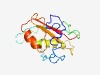 Estimated TM-score=0.45; hard target. |
>Domain of unknown function (DUF508) (Length=149) NPTITSSSASTSSSASSSVTSSATSDSTYMSIPDFSHPATLKSTGTGKLVTMVHVKFILLHHDILKRSVQSTFTDEFQSD CRLEDVILNFQQLYARQLRNARLQPRLSYCIGEISGINSKPVLSSDLGKTLAQLAASNTVFQFALIIDN |
| 6048 |
PF04293 (SpoVR) |
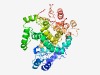 Estimated TM-score=0.45; hard target. |
>SpoVR like protein (Length=432) EWTFELIQQYDREISRIAERYALDTYPNQIEVITAEQMMDAYASVGMPIGYNHWSYGKHFLSTEKNYKRGQMGLAYEIVI NSDPCIAYLMEENTQCMQALVIAHACYGHNSFFKGNYLFRTWTDASSIIDYLVFAKQYIMQCEERYGIDAVEDLLDSCHA LMNYGVDRYKRPYPISAEEERQRQKEREELIQRQVNDLWRTIPRVGGKDKEQSMARYPSEPQENILYFIEKNAPLLEPWQ REVIRIVRKIAQYFYPQRQTQVMNEGWATFWHYTLLNDLYDEGLVNDGFMMEFLQYHTSVVYQPSFDSPYYSGINPYALG FAMYRDIRRICEEPTDEDRRWFPDIAGSDWLATLKFAMKSFKDESFILQFLSPKVIRDLKLFSILDDDQKDDLLVAAIHD EAGYRTIRETLAAQYNLGNREPNIQIWNVDRR |
| 6049 |
PF04283 (CheF-arch) |
 Estimated TM-score=0.45; hard target. |
>Chemotaxis signal transduction system protein F from archaea (Length=204) KVVDTRGKFTQVVKDGRELNDAEWTGGRILLSNKRLILAGNGGKRTIPLSKIESLEGRTDVNQLVAKVSGYVSLQLATED VVLVSAKEPESFERMLYRALLDRKVILARHPAVEGGVVTDAEWEKARLKIEEGAVDLAIADGSFVEIDLDDIGSMEANER SVKDEKRQVLEVEHSEEGTSVQTYISGSSRRCSILETLLRKGES |
| 6050 |
PF04207 (MtrD) |
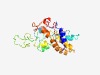 Estimated TM-score=0.45; very hard target. |
>Tetrahydromethanopterin S-methyltransferase, subunit D (Length=214) IAYIVMVTLGGMLISWSVHFVPVGGAPAAMAQATGIGTGTVQLAAGAGLTGLVTAGAVMNVSDSVALVVASGAVGAMIMI AVTMIVGTWVYVYGVGVPPASAKVDYDPITKDRQDLYVSQGTEGHGLPTVSYVSGVIGGALGGIGGAMIYYALMSTSTTL SQSDLVALASIFAVGIFFVNAVIPSYNIGGTIEGFHDPKFKKWPKAVVASFVAT |
| 6051 |
PF04129 (Vps52) |
 Estimated TM-score=0.45; easy target. |
>Vps52 / Sac2 family (Length=483) YIKESQNIASLHNQITACDTILERMEQMLNGFQSDLGSLSAEIQTLQEQSIAMNVKLKNRQAVRGELSQFVDEMAVNERM INIVLEAPVTDRQFLEQLHELNHKIRFVKEQAFKDALSCRDVEDILEKLKLKAIFKIREFILQKVNQMKKPMTNYQLQQN AMLKARFFYEFLLANERHVAKEVRDEYVDTMSKVYFSYFKGYINRLMKLQVLILNVLQKYQDDFVQTCFVKMLQCHTELG HAPGITYRLVAYQQDSLEAVIQGVIDGINVEHSKMEHTFESLFRSQHFALLDNCCREYLFVCDFFMVSGSNAQDLFTNLL GKTLSMFLKHMDTYTNECYDSIAVFLCIHIIQRYRVLMHKRSVPALDKYWETLFEMIWPRIKHILEPKSRITRRYAEYSG AIVNLNETFPDERVNRLLLKLQSEVENFILRMAAEFPSRKEQLIFLINNYDMMLNVITECTSEDSREAESFRQLLDARTQ EFI |
| 6052 |
PF04088 (Peroxin-13_N) |
 Estimated TM-score=0.45; very hard target. |
>Peroxin 13, N-terminal region (Length=139) SLTQTMNNSTQATFQMIESIVGAFGGFAQMLESTYMATHSSFFAMVSVAEQFGNLRQTLGSVLGIFTLMRWIRTAIAKLT GRPPPASSKELTPANFAAFSGASSSSAGPEGAPPRPSRKPFIFFLVAVFGLPYLMGKLI |
| 6053 |
PF03964 (Chorion_2) |
 Estimated TM-score=0.45; very hard target. |
>Chorion family 2 (Length=99) GYGNPGYDNGNPFYKPRKHHRRRHGRRGRHGRHGRHGRRHSDSSDSSSESRSRSSSGSSSSESRSYERDYYYVPRWPTAP SYGNTGNDVPAGTAYGSPA |
| 6054 |
PF03896 (TRAP_alpha) |
 Estimated TM-score=0.45; hard target. |
>Translocon-associated protein (TRAP), alpha subunit (Length=286) KLLIFALLVLPAVLLTVDSGSRLMAHATEDELDEELDVEVESDDASVTQTDEAETDDEPETTKSPDADTFLLFTRPLHSS GSQLELPAGYPVEFLVGFANKGSDDFIVETVEASFRYPMDFNYYIQNFSAIPYNREIKPGHEATVSYSFLPSESFAGRPF GLNIALNYRDASGNQFSEAVFNETVSITEVDEGLDGETFFLYVFLAAVVVLLLVLGQQFLGSYGKRKRSPAVRKTVETGT TNTKDVDYDWIPAETLKKIQNSPKGGKVSPKQSPRQRKAKRAAGSD |
| 6055 |
PF03859 (CG-1) |
 Estimated TM-score=0.45; hard target. |
>CG-1 domain (Length=113) AYTRWLRPNEIHALLCNHKFFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHV YYAHGNDNPTFVRRCYWLLDKSQEHIVLVHYRE |
| 6056 |
PF03470 (zf-XS) |
 Estimated TM-score=0.45; hard target. |
>XS zinc finger domain (Length=42) CPYCQEKSGRHLFYEDLLQHASGIGKSKIRTARDRANHLALA |
| 6057 |
PF03355 (Pox_TAP) |
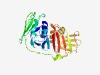 Estimated TM-score=0.45; easy target. |
>Viral Trans-Activator Protein (Length=260) MSVRIKIDKLRQIVTYFSEFSEEVSVNVDARSELMYIFSTLGGAVNIWAIVPLNANVFYDGGENCVFNIPVLKVKNCLCS FHNDAVVTIEPDLENDAVRLSSDHMVSVDCNHEPIPHRTGTVISLGIDQKKSYIFNFQRYEEKCCGRTVIHLELLLGFIK CISQYQYLSIGFADKNLVLKTPGTRDTFVRRYSMTEWSPALQAYSFKIAIFSLNKLRGFKKKVVMFETKIVMDTDDNILG LLFRDRIGTYRVNVFMAFQD |
| 6058 |
PF03324 (Herpes_HEPA) |
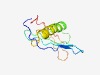 Estimated TM-score=0.45; hard target. |
>Herpesvirus DNA helicase/primase complex associated protein (Length=95) FADLVEVCEVGLRPRGHPQRVTARVLLPRGYDYFVSTGDGFSAPALVALFRQWHTTVHAAPGALAPVFAFLGPGFEVRGG PVQYFAVLGFPGWPT |
| 6059 |
PF03076 (GP3) |
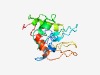 Estimated TM-score=0.45; very hard target. |
>Equine arteritis virus GP3 (Length=160) MGRAYSGPVALLCFFLYFCFICGSVGSNNTTICMHTTSDTSVHLFYAANVTFPSHFQRHFAAAQDFVVHTGYEYAGVTML VHLFANLVLTFPSLVNCSRPVNVFANASCVQVVCSHTNSTTGLGQLSFSFVDEDLRLHIRPTLICWFALLLVHFLPMPRC |
| 6060 |
PF03041 (Baculo_LEF-2) |
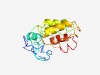 Estimated TM-score=0.45; very hard target. |
>lef-2 (Length=165) NVWSPLISASCLDKKATYLIDPDDFIDKLTLTPYTVFYNGGGLIKISGLRLYMLLTAPPTINEIKNSNFKKRSKRNICMK ECAEGKKNVVDMLNSKINMPPCIKKILGDLKENNVPRGGMYRKRFILNCYIANVVSCAKCENRCLINALTHFYNHDSKCV GEVMH |
| 6061 |
PF02948 (Amelogenin) |
 Estimated TM-score=0.45; very hard target. |
>Amelogenin (Length=175) PLPQHPGYINFSYEVMTPLKWYQSLIGHQYPRYGFEPMGGWMHHAAGPTMHQTTFQSHPSVHSTLHQMQPPHPALNPHMQ PSGHNPFGSMPGQNTLMPQFQPAHGGPIHHPFQPHAGEHPMHPQQTGNPVHPMHPQQPANPNSPIYPVQQLPPLISDTPL ESWPPADKTKQEEVD |
| 6062 |
PF02389 (Cornifin) |
 Estimated TM-score=0.45; very hard target. |
>Cornifin (SPRR) family (Length=123) QQQQVKQPCQPPPQNPCVSPIKEPCHTKLPEPCHPQVPEPCHPQAPEPCHPTVPEPCQPQMPEPCHPKAPEPSQSQVPEP CHPKVPEPCQPQVPEPCHPKVPEPCQPQVLEPCQSKMPEPCQP |
| 6063 |
PF01169 (UPF0016) |
 Estimated TM-score=0.45; hard target. |
>Uncharacterized protein family UPF0016 (Length=75) WSAFSMIIVSEIGDKTFLIAAILSMRHARIEVFSGAFSALALMSVLSALLGAAFPTLLPKSLTTLLAAGLFLFFG |
| 6064 |
PF00803 (3A) |
 Estimated TM-score=0.45; very hard target. |
>3A/RNA2 movement protein family (Length=230) QQSSAATSDELQKILFSPEAIKKMATECDLGRHHWMRADNAISVRPLVPEVTHGRIASFFKSGYDAGELSSKGYMSVPQV LCAVTRTVSTDAEGSLRIYLADLGDKELSPIDGQCVTLHNHDLPALVSFQPTYDCPMETVGNRKRCFAVVIERHGYIGYT GTTASVCSNWQARFSSKNNNYTHIAAGKTLVLPFNRLAEQTKPSAVARLLKSQLNNIESSQYVLTNAKIN |
| 6065 |
PF18862 (ApeA_NTD1) |
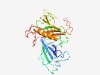 Estimated TM-score=0.44; very hard target. |
>ApeA N-terminal domain 1 (Length=288) NLDEAIEWAGLWWLPDDPDKQVPGVLRYDGQGSPSLSLIGAFEERIFSNPAPGMTVYHEGARTWDVIHGVAEQREVTLLG CIPTSTKRALGARVKSPDTQTVRAKIAIIGAHISGQEDAAFAAAEVSVEDLGFWAASSVFEGSFGISDGKIDGTGTISVK PVEARTVTVDGTEYCLVHTHTLPFFDQRMGGTVGRMRDTASIRLCPAESFTLGAALEAASLIRDLIALATHRAAGVIWLR LEVAGIESSRPNGQPLARRCADVLYSPVALGKHDTKVVDPHRAFFTCD |
| 6066 |
PF18754 (Nmad3) |
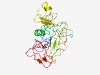 Estimated TM-score=0.44; very hard target. |
>Nucleotide modification associated domain 3 (Length=261) RIILSRKGFDSSSGGCPSPILPDGRMVSLPIPDSRSKIRYRDLKLVDVNAGRLVSQLTRKKIIGSSGAHLDPDLSIGHLP RLLGWRPLLGQTGSAQGHLHKQGIEPDDIFLFFGLFRDAEIHNRRWRFVPGSKPRHIIWGWLQIDEILPVDSLDNDAYSW LNYHPHLHGEADKGNTLYVARDYLSLPGIDKVPGSGIFDRFDPRLQLTQTGSEKPSQWQLPKWFFPGDRQPLSYHAKAGR WQQNKSHCLLSAVARGQEFVL |
| 6067 |
PF17724 (DUF5568) |
 Estimated TM-score=0.44; very hard target. |
>Family of unknown function (DUF5568) (Length=204) MMRQVASNTNDYCMSGRASCLAEDGRHAASHFDLCTSQPNKFYPPVPPPSLQLPLPPLPPLPQSAMKSMPCQRPDVRNEP HPQSVGLKNSDESEDPKKKKGMGRSGRRGRPSGTTKSAGYRTSTGRPLGTTKAAGFKTSPGRPLGTTKAAGYKVSPGRPP GSIKALSRLNKLGYGNCSGAAFPYTMMHKRGLCEPSGKEKEPNE |
| 6068 |
PF17703 (DUF5549) |
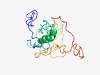 Estimated TM-score=0.44; very hard target. |
>Family of unknown function (DUF5549) (Length=181) MEQQKEGTMPERNNLFLPSPGTESWPPAPFPALHPSLLGTPDPAHLGLPESLASVTVPIRLDALSYFLHSALMGAYSYQQ SLPSCPCTSPPPQGTARRLYRGRGGWEVRCRPGRGQGQRRWGLGRAEQAERGWMEGSRAGPKNPPATPPSSTPPAQDGKK ETQGLEPPLETPPAEDWEAEY |
| 6069 |
PF17671 (DUF5531) |
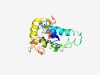 Estimated TM-score=0.44; hard target. |
>Family of unknown function (DUF5531) (Length=152) TLHEILDSSGTIGALVAWMISYKPALFGFLFLLLLLSNWLVKYEVKPTPTEPQQEKTEKRKIPEAEPNCNAMSTREVERL HACFALQDKILERLMFSEMKLKVLENQMFIVWNKMNHHKRSSGQQMFAERKHRPRRQESPFSIDSEGTTNSP |
| 6070 |
PF17626 (IncF) |
 Estimated TM-score=0.44; very hard target. |
>Inclusion membrane protein F (Length=104) MGDVMIQSVKTESGLVEGHRGICDSLGRVVGALAKVAKLVVALAALVLNGALCVLSLVALCVGATPVGPLAVLVATTLAS FLCAACVLFIAAKDRGWIASTNKC |
| 6071 |
PF17603 (DUF5499) |
 Estimated TM-score=0.44; very hard target. |
>Family of unknown function (DUF5499) (Length=97) MGKTYRRKDLKVRDYDYFGKRKAPDGVSHKDMVENIFRSDKWRRMKGIDSEVKDELNRQLRGEVRKLKKSVYIDDDFDYN TSQRVAKRKSNECYRYS |
| 6072 |
PF17562 (Styelin) |
 Estimated TM-score=0.44; hard target. |
>Styelin (Length=59) GWLRKAAKSVGKFYYKHKYYIKAAWQIGKHALGDMTDEEFQDFMKEVEQAREEELQSRQ |
| 6073 |
PF17343 (DUF5373) |
 Estimated TM-score=0.44; hard target. |
>Family of unknown function (DUF5373) (Length=186) MVKFYDPSQPEQYLSSPKLFGVLPIKLIVISMQILYMLLQIVWLTASGVTSKVSILLVIASVILTAFTVAVFVVENKSLM QAHIWVASLFLFFPLAVFVLMIAQFFFDIKFLATFFGEGIYTHPITSLTVAAVFLCAYIAYLFLCHKLVEAKDQQPDDFP ELDTTQGLFHYNRDAYDGSERVNLIY |
| 6074 |
PF17337 (Gal_GalNac_35kD) |
 Estimated TM-score=0.44; hard target. |
>Galactose-inhibitable lectin 35 kDa subunit (Length=221) YNVKYYPESNVYTASNENYNRLKCTTCCRVLFASEYNYAKNAKFTDADDIKDYTRFVMDNEFDDRSLVRAYEGGWEQNIL LRPLKPGNELQFWEFAPYKMYISFPIPRRVHDIRGTADPGNTLIIWNKKPPLAEGTKNQRFVYVHPYNGYPSNEKYSIYK QHFYIPFQTAYSLCYQTKTGGETRSTWDGNRNLQTTTTSYQIKAERCSPTDPKQMFVPVFA |
| 6075 |
PF17261 (DUF5327) |
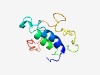 Estimated TM-score=0.44; hard target. |
>Family of unknown function (DUF5327) (Length=102) MNISVQKLLEKMEDELQSAKRSAKGESLRERVYSIKILCELILEEKADMPVQAAVSNPPVVQQPMVVQPAFQQPMFQQPA AINQPKKLDMGDDANGDSLFDF |
| 6076 |
PF17017 (zf-C2H2_aberr) |
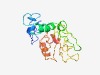 Estimated TM-score=0.44; easy target. |
>Aberrant zinc-finger (Length=172) YKCLHSRCDLFYDSVLGLRRHISHYDHNLYYNAGYSLKCPVRKCDMSICELDNTDFLYSHFEKEHAPRLVAQLKDTLVRI KKTYETTNINYLELNASDNCVNLSIVMRENDLLKDVMCFVYPNGVMKINDIDKPDLIFCGIPGCGKHFKSVMAYKYHCQT YLHSFITLFDSY |
| 6077 |
PF17002 (DUF5089) |
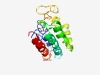 Estimated TM-score=0.44; easy target. |
>Domain of unknown function (DUF5089) (Length=190) EDILKEAQQVEEENYQTAKDIRMQIGSAAIEQKGATEEMRRQGETLDAAKRAAVGVHREARRGDVLADDIEREGHFFNFR CACLEKLFGWFRKDPNDEKVEEIANKEVKSMGDSSYEREDFVPAEKEMVPGQNKTDKELNEILNTVKGIRSEAELQGEEA QRQKTVVKDISAVNEQAEKVIKRTDTKLKK |
| 6078 |
PF16982 (Flp1_like) |
 Estimated TM-score=0.44; hard target. |
>Putative Flagellin, Flp1-like, domain (Length=49) DFWKDESGMGTVEIILIIVVLIGLVIVFKKQINDLVNKIFKKINSDSNK |
| 6079 |
PF16929 (Asp2) |
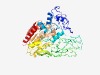 Estimated TM-score=0.44; hard target. |
>Accessory Sec system GspB-transporter (Length=509) FRVLQIGGDDLEPIFQHKKGVSWDYFDIGLFEFDSGYVEAIEAIVEAEGRFDFIYIQAPYSETLTNLLQMISEPYNTYVD ESFWSVEYEQDENFQKYVVQPLHYRNIEERNNKLEAVSFSGQYGDKVSPKLALVHPNFKGDVVYQGNSELTLSGEFGKEF KPIASWQNNLVYDKDKVIQIWPEFDIDGAVELQYTFRLIQTGADGALIEQIVLTDDMLDSPLEIPAKPFDAYISVTVKAR GNGTVHLGPIHKRWSRLDMGQFLLGGSRFVDSQRQEFIYYFHPGDMKPPLNVYFSGYRTAEGFEGYYMMKRMNAPFLLIG DPRVEGGSFYIGSSEYEQGIINVIDETLEKLNFKSHELILSGLSMGSFGALYYGAQLNPQAIIVGKPLVNIGTIAEHMRL LRPEEFGTALDVLVSNEGDTSQASIQALNQKFWQTFQKKSLSQTVFAIAYMQHDDYDPHAFQELLPVLTAHQARVMNRSI PGRHNDDSPTIASWFVNFYNIILEDKFGR |
| 6080 |
PF16638 (Tristanin_u2) |
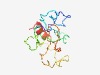 Estimated TM-score=0.44; very hard target. |
>Unstructured region on methyltransferase between zinc-fingers (Length=131) KLDVFSRTRGRGRGRGKRRFGPGRRPGRPPKFIRLEITSENGDKCDDGTQDLLHFSTKEQFDEAEPATLNGLDQPEQTTL PIPQLPQETQPSLEHEPETHSLHLQPQHEESVLPTQSTLTADDMRRAKRIR |
| 6081 |
PF16620 (23ISL) |
 Estimated TM-score=0.44; very hard target. |
>Unstructured linker between I-set domains 2 and 3 on MYLCK (Length=161) QGLDNANRSFVRGTKAANSDIRKEVTNGIAQEPKMDSLETIAESKNCSSTQRGGSPTWATSTQPQPLRESKLDLFEDSSR KALRTPVLQKTSSTITLQATKVQPEPRAPVSGPFSPGEKREKPAAPPPATLPTRQSGLRSQEVVSKVATRKIPMESQRDS T |
| 6082 |
PF16492 (Cadherin_C_2) |
 Estimated TM-score=0.44; hard target. |
>Cadherin cytoplasmic C-terminal (Length=83) LTLYLVIALASVSSLFLLSVLLFVGVRLCRRARETSMGGCSVPEGHFPGHLVDVSGVGTLSHSYQYEVCLTGDSGTTDFK FLK |
| 6083 |
PF16441 (DUF5038) |
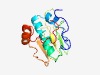 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF5038) (Length=143) ASQEETTQPEELTEMDFLSFEELSQFFSSAQTASLKEQFPVYLTRIGRTDVTSAEYLPDETGYPDTTTVELHFLLSDDST LPVFYDTMTGAFSFGEDRTVLGTSGKTYEKPVVDTLPPVSSSEVEQRQEGGYADVTAPESPAP |
| 6084 |
PF16328 (DUF4961) |
 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4961) (Length=312) IKIFFLLVLSLIVFGCSFLIKEVNVKQENEAGEMVAYIKAGEIATFTFSGEINIDGDASNETFIVGFLAPRSWNVRQNAT VTYREDRYETEVDHKMTVIPDTEQPANYKGMSWSAALKKKYGVRGNVLNDMEWIAFKSDNYPSVNGTIHYTVTIKCNSGK SNLKFRPSFFINHSSDGIGGDEAHYSVKDADDCFEVVEGSGTVIDFCSTHYYQIEPLSALQDDYVTFTFQGDINTNELIK AENVYIEATAYTIEGKIYTVNEKSAKTLMKRETKLPRYNVTLWPGGFFNIPDGETISRIEYIFTNEDGTVSI |
| 6085 |
PF16115 (DUF4831) |
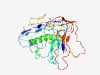 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4831) (Length=311) YGVTYALPKTAINIEVKVNKVTYTPGEFSKYADRYLRLTDVSGEPQEYWELVSVKAKSVGIPDSEHTYFVKLKDKTVAPL IELTEDGIVKSINVPLSPKKSAPMQPATTQKKKINPRDFLTEEILMAGSTAKMAELVAKEIYNIRESKNALVRGQADNMP KDGEQLKIMLANLEEQEAAMTEMFSGTLNKDEKIFNIRLTPDKEMDNEVAFRFSKKLGIVANNDLAGEPVYITLKNLKTV NVPEDDGKKKVDGIAYNVPGKAQVTLTEGKKQWFNEELPVTQFGTIEYLAPALFNKKSTVQVTFNPDTGGL |
| 6086 |
PF16062 (DUF4804) |
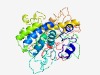 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4804) (Length=432) EFKRKSSNFPLEFGTNTCRVLSQPKERYPFIQKQIASAYPVIHERVLQLYLDFLEHKCKYGTALEKELYEKLSLICFIQR LLTQRCASFFGRNDKFLLLSRERGCSGFMDVGTAQEKPPLILKNVLSYDEIKLSALLSVSSHTELINNGNRQNCGVIEKN KSLIERSGVVVGIIGARLTRRDVMEFQDIIIAKTQNTKENGYGFGIAEDTTNSKHKDYRRIWKQFYEEPDFLYNQVIKDG KRFGASKNKDDIFDNLIMKKRYTISFDTLLLEAEARAKSEGKQAYVHVVGIGLGVWKAAEQQEQIFLQTFTQRVKYLLPQ LQHIGVLHFSWFSLSECGELKNGYVFKSETHPQGGIRCFLSKRNPADKLKTPENDNMLLVISYAWDGNALPGNEFWMKML KSTGDSSTACSTLITELHNPHINNEYVNGHPL |
| 6087 |
PF16054 (TMEM72) |
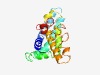 Estimated TM-score=0.44; hard target. |
>Transmembrane protein family 72 (Length=184) KWQAFWTGLEYFCRFLGISTAAVLIGVGVETFLQGQFKSLAFYLLFSGTAVSICEGSYFVAKLLSLCFRCQPESLAYVLW EKACRLGCFQKFLGYVLLSVACFLHPVLVWHVTIPGSMLIITGLAYFLLSKRKKNKASCTPEQYTDPSASAVSTTGYGDT EQTYTFHSSFREGPTSLFGHMKSI |
| 6088 |
PF15936 (DUF4749) |
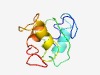 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4749) (Length=92) KVLHVQFNSPLQLYSQSNIMDTLQGQISSVSPDFPSLDQHNQTPGSIAIDRQSEVYKMLQENQESNEPPRQSASFLVLQE ILESEEKGDPTK |
| 6089 |
PF15865 (Fanconi_A_N) |
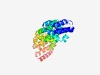 Estimated TM-score=0.44; hard target. |
>Fanconi anaemia group A protein N terminus (Length=353) LWKVQNSLLLEALWHLHVQNIVSLQELLESHADLQAVVAWLFRNLRLLCEQIEASCQHTDIARAMLSDFVQMFVSNGFQK NSDLRRNVQPGQMPQVAVAVLERMLILALEALDAGLQEESPTHKAVRCWFSVFSIHTFCRTISTDSLKKFFSHTLTQILT YKPVLRVSDAVQMQREWSFARTHPLLTSVYRRLFVIVSPEELISHLQEVLETHEVNWQHVLSCVSTLVICLPEAQQLVND WVARLLARAFESCDLDSMVTAFLVVRQAALEGPSVFLSYADWFQASFGSAKGYHGCSKKALVFLFKFLSDLVPFEAPRYM QVHILHPPLVPSKYRSLLTDYITLAKTRLADLK |
| 6090 |
PF15804 (CCDC168_N) |
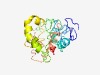 Estimated TM-score=0.44; very hard target. |
>Coiled-coil domain-containing protein 168 (Length=207) RQREDEVNIAVEKGIMRPKYLALKAKKSPLSQILNTKELRLTIKEQEKRGQEGKGKTVVILSKTCTFATSSTHLKLDTIK EEGELGIIRDYVPHLELWESLASGQRASTESVESNILSPTKKRKRHLQQKEDLVKTATNGLMHPKDTVFKAKTSPLPFMF SITEDGSLTKRKEPQWSIKRKVELELERKGEPDVILTKTHTSMPSAS |
| 6091 |
PF15789 (Hyr1) |
 Estimated TM-score=0.44; hard target. |
>Hyphally regulated cell wall GPI-anchored protein 1 (Length=44) IVSQSGSSFTTLTTFPAAEEPVETEFTSTWEVTNTDGSVSTESG |
| 6092 |
PF15765 (DUF4694) |
 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4694) (Length=157) LDKPDQLPPTFLQGRNGESAAWNAQGQAPSLLPPSVESQPPTWPHHDTGATQKPLRVSGGSLRNPWSSESGLLSLQQSSL GKCSDSDISLLSSGYAGDEENSEVSLLGSTPILRLQRRLLTQAGSALTSQLCAVYSPSQIGNRLASQAHSVKQTGGE |
| 6093 |
PF15722 (FAM153) |
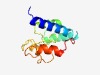 Estimated TM-score=0.44; hard target. |
>FAM153 family (Length=114) MVDKDTERDIEMKRQLRRLRELHLYSTWKKYQEAMKTSLGVPQRGDLEDLEEHLPGQTVSEEATGVHMMQVDPATPAKKL EDSTITGSHQQMSASPSSAPAEEATEKTKVEEEV |
| 6094 |
PF15684 (AROS) |
 Estimated TM-score=0.44; very hard target. |
>Active regulator of SIRT1, or 40S ribosomal protein S19-binding 1 (Length=127) QKKKQQTPSSAKVMELVSTKRQGVTKQVKRLQGRLGPGKSKATVKDKRVKSAVEEFRKKQGKSHMSDNLKYFMETGYKAT DSDTSKILSHNSGRQSRNRPEQPAKKAKESQSLFTEEEFQQFQKEYF |
| 6095 |
PF15621 (PROL5-SMR) |
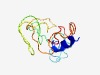 Estimated TM-score=0.44; very hard target. |
>Proline-rich submaxillary gland androgen-regulated family (Length=109) MKSLTWILGLWALAACFTPGESQRGPRGPYPPGPLAPPPPPRFPFGTGFVPPPHPPPYGPGRFPPPLSPPYGPGRIPPSP PPPYGPGRIQSHSLPPPYGPGYPQPPSQP |
| 6096 |
PF15559 (DUF4660) |
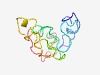 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4660) (Length=110) SDDEGPKSSKTKDGASASRSDVASQQGTKRAAGGMPLPKPDELFRSVSKPTFLYNPLNKQIDWESRIVKAPEEPPKEFKV WKTNAVPPPESYATEEKKKGPPPGMDMAIK |
| 6097 |
PF15555 (DUF4658) |
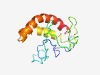 Estimated TM-score=0.44; very hard target. |
>Domain of unknown function (DUF4658) (Length=123) PGAGREGYKRCPPSILRQSRPERRSRGAEPQKTARHVRFREPPEVAVHYIASRESTTATKALGRPRSRGGSLLLRLSVCL LLVLVLGLYCGRAERVTLALEDLRARLLVLALRLRHTAVTCWH |
| 6098 |
PF15549 (PGC7_Stella) |
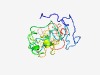 Estimated TM-score=0.44; very hard target. |
>PGC7/Stella/Dppa3 domain (Length=170) MDPLQKCNPTLISESSPVISEETSREVTAASPPSFSELLMMGLGDLKSSPGPKYPAPLPEGLLQQQYRDDKTLQERRWER SASPQRKRTLLGHMRRRHLDHVAPYRVERNARISSSGDRDQNGFRCECRYCQSHRPNVSGMPAERKGAPHPSSWETLVQG LSGLTLSLGT |
| 6099 |
PF15443 (DUF4630) |
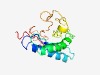 Estimated TM-score=0.44; hard target. |
>Domain of unknown function (DUF4630) (Length=155) GAALVGVLVAEAGPEDAVAPALRLLEALLRAVFGRQGGGPVQAAAYCPGLPASCLAVQAAACRALQAAGAGQPVEGAWER PGVPRLLACFSWGPWSRRKNQDIAACRSPAQEDFQEPEEELALTAVFPNGDCEDFGRGSKACDGVIHTPAEPTGD |
| 6100 |
PF15438 (Phyto-Amp) |
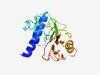 Estimated TM-score=0.44; very hard target. |
>Antigenic membrane protein of phytoplasma (Length=195) MQNQKNQKSLVAKVLVLFAAVALMFVGVQVFADDKLDLNTLECKDALELTAADAADAEKVVKQWKVQNTSLNAKVTKDSV KVAVADNKVTVTPADGDAGKALSGSKILNLVGVCELNKLTLGTEKKLTLTVKDGKVDAEAGLKALKEAGAKVPATVNKDD VTFTVGKDDNANKVTVKAVDGKTTVSGQVVFEFTV |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218