

Go to page (83 pages in total): [First page] << < 50 51 52 53 54 55 56 57 58 59 60 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 5401 |
PF07713 (DUF1604) |
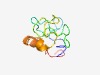 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1604) (Length=85) YVPIWKQEVTDERGRKRLHGAFTGGFSAGYFNTVGSKEGWTPSTFVSSRQNRAKDVKQQRPEDFMDEEDLREAEESRKLQ TAEDF |
| 5402 |
PF07596 (SBP_bac_10) |
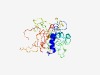 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1559) (Length=277) QSAREAARRMSCSNNMKQLGLAMHNYHDTMSSFPLGSYNLREAWPASGSNWRALILPFIEQGNIYDQLEFSYSSHFMAGG AAGADALLGNEVLKFLAVPTYACPSSAIELFENAPVSNNSEGAMNVHYVGIQGAARPVPGGDPNKGTADLGHGWSSNNGM LFANGKIKMRDVVDGTTNAIMIAEQSGWVDGVNRTSNYYGGWMGSRHPRPVDGPSSWDLWQTGTTCVRFAPNSQIVQTGA TDRMYRNNTVINSMHTGGINVVMGDASVQFITDSIDF |
| 5403 |
PF07590 (DUF1556) |
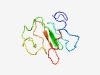 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1556) (Length=82) MNQGSETTERIRTSSEGRMGAICRDGNLKLKTHRAISVNPLPCNSSPAKLAESKRPIRKSPASEPTRLFVRERSREAIPK RD |
| 5404 |
PF07526 (POX) |
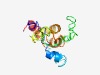 Estimated TM-score=0.48; hard target. |
>Associated with HOX (Length=139) ILNSQYLKAAQDLLDEIVSVRKALKQPGMEKQDNCRDIGLDGSKDSDGKSNSQSLQMSSGPNASSELSPSERQNLLDKKT KLLSMLDEVDKKYRQYCHQMQIVVSSFDMVAGCGAAEPYTTLALRTISRHFRCLRDAIS |
| 5405 |
PF07405 (DUF1506) |
 Estimated TM-score=0.48; easy target. |
>Protein of unknown function (DUF1506) (Length=127) MNGVRKRLSDMSFRMINVFKDPKPLKFYKGTVVKLENDSSYQRVFDKTKYIEFAGVIIDIKPQELAILYDSDMSDIQGYS KLYTYQDLNYEPKDRISIADLVYFEIFSIDSSIGYFTLVLKEFIWTN |
| 5406 |
PF07355 (GRDB) |
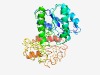 Estimated TM-score=0.48; hard target. |
>Glycine/sarcosine/betaine reductase selenoprotein B (GRDB) (Length=347) KIKVVHYINQFFAGIGGEEKADYKPEIREGVVGPGMALNKEFNGEAEIVATIICGDSYFNENVEEAKAEILKMVKEQDPD LFIAGPAFNAGRYGVACGTIADAVQTELGIPAITGMYEENPGADMYKKSVYIVSTKNSAAGMRDAVKKMAPLALKIAKGE EIGSPEEEGYIPRGIRKNYFTDKRGSERAVEMLLKKLKGEDFVTEFPMPDFDRVDPQPAIKDLSKATIALVTSGGIVPKG NPDHIESSSASKYGKYDIEGFDDLTSEDHETAHGGYDPVYANEDPDRVLPVDVLREMEKEGKIGKLHRYFYTTVGNGTSV ANAKKFAEEIGKELVADGVDAVILTST |
| 5407 |
PF07353 (Uroplakin_II) |
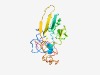 Estimated TM-score=0.48; hard target. |
>Uroplakin II (Length=179) PVQTLPLILILLAVLAPGTADFNISSLSGLLSPALTESLLIALPPCHLTGGNATLMVRRANDSKVVKSDFVVPPCRGRRE LVSVVDSGSGYTVTRLSAYQVTNLTPGTKYYISYRVQKGTSTESSPETPMSTLPRKNMESIGLGMARTGGMVVITVLLSV AMFLLVVGLIVALHWDARK |
| 5408 |
PF07290 (DUF1449) |
 Estimated TM-score=0.48; hard target. |
>Protein of unknown function (DUF1449) (Length=219) NWPFMVSLTIMVMIACLEGVSTLIGMGFSSMIESLIPDFNMDVDADMDVDANAGANSGIGPDHPNISTTGILTKTFGWLR IGQIPVLIILIAFLTIFGLVGFFIQSVVVKITGHLLPGIIASAISFLLTLPLVRAATGIIVKIMPKDETQAVAEKSFIGR IAVITLGKASKNNPAQAKVRDKFGTAHYVMVEPDIDDENFAQGDQVLIVKQTGFGYKAI |
| 5409 |
PF07253 (Gypsy) |
 Estimated TM-score=0.48; easy target. |
>Gypsy protein (Length=468) RITDFSHANYIPVFDGEVLVFDQRSYLRHSSNISEFVSMIDETERLSDSFPQSHMRKLLDVDTDHLRTLLSVLQVHHRFA RSLDFLGTALKVVAGTPDPSDFLRIRVTETQLVEANSKQILINSETQKQINRLTDTINKIISSRKGDLVDTPHLFETLLA RNRILNSEIQNLVLTITLAKASIVNPTILDHTDLKSLIEQDTPIVSLIEASKIKVLQSENIIHILIAYPKVEFRCQKVSV YFVSHQQTILRLDEDTLAECEKDTFAVTGCTVTTHNTFCERARRETCADSLHAGNIANCHTQPSHLKAIMPVDDGVVVIN EATAHVRTDDDAEVTVSGTFLITFERSATINGTEFVNLRKSLSKQPGIVRSPLLNIIGHDPALSIPLLHRMNINNLLSIR DFREEVVAAGSPKFWFAIGAVLNVGLIGSFFLFLVLRKRRASMQMQKAIDKYDMPEDGHHLRGGVVNN |
| 5410 |
PF07231 (Hs1pro-1_N) |
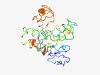 Estimated TM-score=0.48; very hard target. |
>Hs1pro-1 N-terminus (Length=200) MVDLDCTAKMVSPDMPKKSPKFSNKLQVSIPSAFRAGDLSSAPSPECSAYEYYLRLSELKKLWDSREFPNWKNESLIRPA LQALELTFRFVSTVLSDPRPYANRREWKRRLESLTTNQIQLVAILCEEEEEEGEMRGTAPISDLSLSNGVLARDGSCAEV WKVPGATPVVSRTSEASLLPRLATWPKSEDIAQKIMYSIE |
| 5411 |
PF07207 (Lir1) |
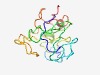 Estimated TM-score=0.48; very hard target. |
>Light regulated protein Lir1 (Length=133) MQAALSIAPPVFPVKPSKNLSQVILSPPQLGASSLAPRASPIKAAYATVAYDTSTVDYSSVISVFPAEACETLGGDACLA NIFPEVKLEARNDAAAARIASEPIEREYLEYNDAKTVFCAEACDDLGGEFCSR |
| 5412 |
PF07128 (DUF1380) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1380) (Length=137) MYGTCETLCRLLREQYPAETPLNLIIWSPADIEALADGMEYSVSEQDTRAVLARMDAIPEEQRLESGVSAGAVMDLIEQV KEAVPAVMVPADLLETLLTTAEQALWHREWTARDSNHPVPESVTRRLADAAKVRALL |
| 5413 |
PF07026 (DUF1317) |
 Estimated TM-score=0.48; hard target. |
>Protein of unknown function (DUF1317) (Length=60) MKHPHDNIRVGAITFVYSVTKRGWVFPGLSVIRNPLKAQRLAEEINNKRGAVCTKHLLLS |
| 5414 |
PF06922 (CTV_P13) |
 Estimated TM-score=0.48; very hard target. |
>Citrus tristeza virus P13 protein (Length=119) MSIRRVWLKVIAVITVLWYCKEPSISEGYNALMNDDFKFIDTHFSNVSYAKKCYDLANFDLDFLRIVIIPLSGGTANESR SDRTNVSEIVESHVSDRDRMHILLRNKRIQIPSLLPCDN |
| 5415 |
PF06896 (Phage_TAC_3) |
 Estimated TM-score=0.48; hard target. |
>Phage tail assembly chaperone proteins, TAC (Length=114) MKINITKLGLRKKSAEVKTTVGVIEKAQNLQIQLLKSDAVDFTNDDPLAVLETQKKMVQGLLQFMIDIFKLSDKEVEKIK STLDFTDFQNFMAYVLFRFQGMSDEDWETVTKQQ |
| 5416 |
PF06882 (DUF1263) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1263) (Length=57) TPHVCPSSYNDFLAMVAMKPGMNLAGTDIPTPGVSTLAPARDECLEALIIPTGRGKA |
| 5417 |
PF06788 (UPF0257) |
 Estimated TM-score=0.48; hard target. |
>Uncharacterised protein family (UPF0257) (Length=236) MKKPILLTLLCMMLAGCDNPKSPESFTPEMASFSNEFDFDPLRGPVKDFSQTLMNENGEVAKQVTGTLSQEGCFDTLELH DLENNTGLALVLDANYYRDAQTLEKKVQLQGKCQLAALPSAGVTWETDDNGFVVSATGKEMKVEYRYDSEGYPLGKTTIN SQKTLSVTAKPSADPRKKLDYTAVSRVDDQQVGNVRQSCEYDAYANPVDCRLVIVDESVKPAVSHHYTIKNRIDYY |
| 5418 |
PF06770 (Arif-1) |
 Estimated TM-score=0.48; very hard target. |
>Actin-rearrangement-inducing factor (Arif-1) (Length=205) LAYLIVFVLSIMGVIDGRYAFLLEIEGKQSVINLSILIMLAFGMWILFYMFYFIWKIIVWTKNRSSSSNTNVNFNLKENF YIAITCITVNVITGLCWMLFAAFQIYVFKNGHLPALDVLYRHYDLESMCWNSIVYLEIDYANTETLSQNCVYVNIYKKCI MCRAIVSDHEPTVFNQNYPVIIMGVLTILAVQCWNLYVQLKEMRR |
| 5419 |
PF06752 (E_Pc_C) |
 Estimated TM-score=0.48; very hard target. |
>Enhancer of Polycomb C-terminus (Length=232) AFTAEQYQQHQQQLALMQKQQLAQIQQQQANSNSSTNTSQGFVSKTLDSASAQFAASALMTSEQLMGFKMKDDVVLGIGV NGVLPASGVYKGLHLSSTTPTALVHTSPSTAGSTLLQPSNITQTSSSHSALSHQVTAANSATTQVLFGNNIRLTVPSSVP TVNSITPINARHIPRTLSAVPSSALKLAAAANCQVSKVPSSSSVDSVPRENHESEKPALNNIADNTVAMEVT |
| 5420 |
PF06691 (DUF1189) |
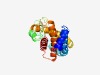 Estimated TM-score=0.48; hard target. |
>Protein of unknown function (DUF1189) (Length=244) FVQLWKSLYSPKDIARFRFQGIGKTIGYVFLLTFLSILPMAYYLTTSFVEGIRNTSALLQNELPSFTIENGQLHSSAKEP IHIDQHGFTIIFDSTGQVTKEDMERFNNAIGLLKHEVVLIANHQTQYYSYSTFPDVKITDEDVHSFIETMQSLLPVFIPL LFIIIYLFASASKFIGICILAFFGLILRSTLDKKLQYRHTWIMSAYAVTLSTVFFAVMEALQTVVPYAPFIHWFVSIAVL FLAM |
| 5421 |
PF06622 (SepQ) |
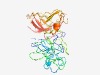 Estimated TM-score=0.48; easy target. |
>SepQ protein (Length=305) MNLLNSQLNMKINDFYLPLLSVIGTGRLYITPEGHACHAYFREVSGNGIRFTLTCSGYEGRFWISEEQFIQWCHELFPYS ESRLIPEDMIKLMILWVMQTALPEGDVSVDDVQFTMLNKDVYPVIENNNGENRLNVIILEITVQSLQYLINENWQFVPHS NTIFFDGYIAPGWTDYPVTELTVGDSLRLYHVDDSKERECWLVINNPLATVKLCDNNLFIADVQAADLLCTISNEAVMDR IYCVIGTIHVDIHMLRNAKKDDIIDSTGYHLFGGCRLIRNNTTIAYGSIVKINEDFYFTVSLVCD |
| 5422 |
PF06589 (CRA) |
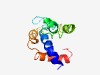 Estimated TM-score=0.48; very hard target. |
>Circumsporozoite-related antigen (CRA) (Length=121) NTKLYSLFVFICLIIVYCATVQGSKDNGKNGVYELKMIKKKHKKNNKDNKHVQRAGEDEDEDDNIDYTNLVKEEEKRNEE VKRLSKMHKAYTLGMSTIIFLSTLFLIRDCVLESISRPDSY |
| 5423 |
PF06584 (DIRP) |
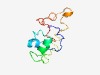 Estimated TM-score=0.48; hard target. |
>DIRP (Length=101) FYSAIDYPWFSNREFVEYLNHVGLGHIPRLTRVEWGVIRSSLGKPRRLSKRFLHDEREKLKQYRESVRKHYAELRTGVRE GLPRDLPRPLSVGQRVIAIHP |
| 5424 |
PF06388 (DUF1075) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1075) (Length=119) RLCERDASSSLRLTRNSDLKRINGFCTKPQESPKAPSQTYSHRVPLHKPTDWEKKILIWSGRFKKEDEIPETVSFEMLDA AKNKMRVKISYVMIALTVAGCILMVIEGKKAAKRHESLT |
| 5425 |
PF06362 (DUF1067) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF1067) (Length=97) MTTPTPQGHDMHTKTPLPRGANNYPHTHACIDIAFSTAQVPSPWHHQHVDQAASTTDMLTCAALIVSTAAKHTKPHRKQA VSHPPTKTPQHSNKRFS |
| 5426 |
PF06295 (DUF1043) |
 Estimated TM-score=0.48; hard target. |
>Protein of unknown function (DUF1043) (Length=122) ALIGLVVGFVIGALVMRFGNAKLRQQKALQTELENNKSELEEYRKELVGHFARSAELLDNMARDYRQLYQHMAKSSHELM PNMPVQDNPFNYRLTESEADNDQAPVKMPPKDYSDGASGLFR |
| 5427 |
PF06213 (CobT) |
 Estimated TM-score=0.48; very hard target. |
>Cobalamin biosynthesis protein CobT (Length=272) EAPAEPFKRSVAGCLRAIAKTPELEVTFASDRPALTGERARLAEPPRKMSAADAAIVRGQADSMALRLACHDAVVHRRLA PEGQQARAVFDAVEQSRVESIGSNRMAGVASNITAMLEDRYHRGAYHEVTDRADAPLEDAVAMIVRERLTGLKPPPAAQK IVDLWRPWVEEKAGKDLDKLASNLEDQRHFARSVREMLASLDMAEDSSLDPEDSEEDEESNEQSQDKEQQEGESEEQSAA ERAEMERSEEATEEMEEGQSETSEGPTGEMPD |
| 5428 |
PF06152 (Phage_min_cap2) |
 Estimated TM-score=0.48; very hard target. |
>Phage minor capsid protein 2 (Length=354) LSPEYIDHLPDRVVELYADLEIRILEDMARRISKTGALTETAQWQMWRLEQIGAEREFIRYHLQRLTGKTQGEINELLAE AGEKALYYDDQIYRAAGLSPKALRDSEALQKIIKAGSDKTMRLFENLTSTTADTASRQFENALDAAYMDITSGAFSYQDA VRSAVKSLSKAGIDAIIYPSGHTDKMDVAVRRAVLTGVNQTTARMQIARADEMECDLVETTAHMGARPEHMDWQGRIFSR SGKSRKYPDFVKSTGYGTGPGLCGWNCRHSFFPYFEGLSERAYSRAKLREYENKTVTYNGRTLPYYDATQQQRYLERQIR RWKREYLAMDAAGLDTSEASAKLAAWRAKQKDFL |
| 5429 |
PF05961 (Chordopox_A13L) |
 Estimated TM-score=0.48; hard target. |
>Chordopoxvirus A13L protein (Length=68) MISELLLLIICVVIVGFIVYGIYTKKKTTQHVPPSSEQYEKMENLKTGYVDKLKSAHLKSFYKLFSSN |
| 5430 |
PF05820 (Ac81) |
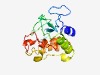 Estimated TM-score=0.48; very hard target. |
>Baculoviridae AC81 (Length=176) KIKYDSELLLHYLYDDHRNKNSDYANNNINVIKISKVKVKKTGASILAHYFAEIHVSTGYSFEFHPGSQPRTFQTIHTDG LIIKVLILCDECCKKELRDYIKGENSFNVAFRNCESILCRRVSFQTVLLTCAILLLLFNVEKFSMINLLIILLILISLFC HNNYIISNPYIEFCNH |
| 5431 |
PF05811 (DUF842) |
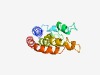 Estimated TM-score=0.48; hard target. |
>Eukaryotic protein of unknown function (DUF842) (Length=130) QRQRIERAVTEMVDDMDKTHLRKMQTDMHLCAAKCCQDMNSSIDNVQRCVDRCSAPLTRAQNYVQHELGEFQGRLQRCVM QCNDDVKVKMPPNPSESDIAKYTDQFERCAIQCVDKHVGLIPTMMKTIKS |
| 5432 |
PF05774 (Herpes_heli_pri) |
 Estimated TM-score=0.48; hard target. |
>Herpesvirus helicase-primase complex component (Length=127) WLESVQFGLNILLCTKCNQSICDVLETLLQLYFKNRHNAKFWLVPQHFLTSKPQKPNIPSDCVAPTLFFFTTDGPLCWHQ KLNVPLNINYEEYIHKAITVFEKVPSLTINKDMYIKIESWKNILCLL |
| 5433 |
PF05632 (DUF792) |
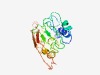 Estimated TM-score=0.48; very hard target. |
>Borrelia burgdorferi protein of unknown function (DUF792) (Length=184) PQEISQIIRDVITQIFALFGADNFLVLFPRMDLKGFGYVPQLFFIKPKTELITRTYNTSCSKRPVINYYDRKAEYVSYNP VMTGEHISLNGGILTSLYKDMLSLLKMTVFGNTMLRFDAHLVKEQLANRIQAQVPFSIYSPTFGLKELAVITSLSFKDTP FIDEVEVSLSIEIVKTFALEKYKG |
| 5434 |
PF05440 (MtrB) |
 Estimated TM-score=0.48; hard target. |
>Tetrahydromethanopterin S-methyltransferase subunit B (Length=93) VHVAPEVNLVMDPMSGLVATEREDIISYSMDPILEQLDELDKVADDMVNSLSPNMPLLNSNPGRERTSYYAGIASNAFYG IIVGLIFATIIII |
| 5435 |
PF05436 (MF_alpha_N) |
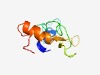 Estimated TM-score=0.48; very hard target. |
>Mating factor alpha precursor N-terminus (Length=88) MKFSTILSTAAFVSSSVMAAPVNVTETEQENLAIPSEALIGYLDLKGDDDISILPVSNSTNSGLLFLNSTILTQAMNENT EAALSKRE |
| 5436 |
PF05434 (Tmemb_9) |
 Estimated TM-score=0.48; very hard target. |
>TMEM9 (Length=139) RKLYIENVPPNKCDCTNVVLPRVGDDIKGKEKEFCPRCECKYESRNTTTIKVVVIIVIWIISLLVIYMLFLVCLDPLLNK KTTTQYQEHNNEEDESETHSQPLRASGAGVLNRVGQTTDKWKKTVQEQRRHIYDRHTIL |
| 5437 |
PF05393 (Hum_adeno_E3A) |
 Estimated TM-score=0.48; very hard target. |
>Human adenovirus early E3A glycoprotein (Length=100) MTGSTIAPTTDYRNTTATGLTSALNLPQVHAFVNDWASLDMWWFSIALMFVCLIIMWLICCLKRRRARPPIYRPIIVLNP HNEKIHRLDGLKPCSLLLQY |
| 5438 |
PF05324 (Sperm_Ag_HE2) |
 Estimated TM-score=0.48; hard target. |
>Sperm antigen HE2 (Length=69) LLPFVASLLLVALLFPGSSQARHVNHSATEALRELGEGAPGQGTNGSQLLHHPVKQDLLPPRTPPYQGT |
| 5439 |
PF05322 (NinE) |
 Estimated TM-score=0.48; hard target. |
>NINE Protein (Length=58) MARQRRSITDIICENCKYLPTKRSRNKPKPIPKESDVKTFNYTAHLWDIRWLRHRARK |
| 5440 |
PF05304 (DUF728) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF728) (Length=139) TCVLKGCVNEVTVLGHETCSIGHANKLRKQVADMVGVTRRCAENNCGWFVCVVINDFTFDVYNCCGRSHLEKCRKRVETR NREIWKQIRRNQAENMSATAKKSHNSKTSKKKFKEDREFGTPKRFLRDDVPFGIDRLFA |
| 5441 |
PF05267 (DUF725) |
 Estimated TM-score=0.48; hard target. |
>Protein of unknown function (DUF725) (Length=120) SIECMNYYIPKINDVANQFNENFLACYDEANLEIASIDDKTSENRTQINTSATNSCKLLESCSTIETSVDFFSCYASAGS ENARNMYTISADASELQAYVVEQIRLIEVNKYVCTNNSER |
| 5442 |
PF05122 (SpdB) |
 Estimated TM-score=0.48; hard target. |
>Mobile element transfer protein (Length=50) RIGPVQVGTYYDNRGQEKHTAACTAPRCGFSTDYDSRAAAELAARTHRCP |
| 5443 |
PF05100 (Phage_tail_L) |
 Estimated TM-score=0.48; very hard target. |
>Phage minor tail protein L (Length=197) TEVGGERYFFCNEQNEKGEPVTWQGWQYQPYPIQGTGFELNGKGSAARPTLTVSNLHGMVTGMAEDLQSLVGGTVVRRKV YARFLDAVNFVNGNSEADPEQEVISRWRIEQCSELSAVSVSFVLSTPTETDGSVFPGRIMLANTCTWTYRGDECGYSGPA VADEYDQPTSDITKDKCSKCLSGCKFRNNVGNFGGFL |
| 5444 |
PF05093 (CIAPIN1) |
 Estimated TM-score=0.48; very hard target. |
>Cytokine-induced anti-apoptosis inhibitor 1, Fe-S biogenesis (Length=97) IQPPECQPKAGKRRRACKDCTCGLKEKIEAEDAAKRANADKALNTMKLDTDDLAEVDFTVQGKVGSCGNCALGDAFRCDG CPYIGLPAFKPGEEVRL |
| 5445 |
PF04956 (TrbC) |
 Estimated TM-score=0.48; hard target. |
>TrbC/VIRB2 family (Length=96) SSVTTARSIASVALAAAIVSVAMVQPAFAQATGGIESVLQNIVDMLTGNVAKLLATIAVIIVGIAWMFGYLDLRKTAYVV LGIGIIFGASEIVSTI |
| 5446 |
PF04930 (FUN14) |
 Estimated TM-score=0.48; hard target. |
>FUN14 family (Length=97) IIIGTTSGWLTGFVTMKVGKIAACAVGGGIIILQIAAHQGYIRINWDKIQKKAEKITDKVEEKVTGEGPKLLDKVGLWCR HNTLVATSFVGGFIIGL |
| 5447 |
PF04872 (Pox_L5) |
 Estimated TM-score=0.48; hard target. |
>Poxvirus L5 protein family (Length=80) EIIMLISYKDDISINPIDNFSKSWLSCDGNKLVINNLPNTNGTKTALAINRQDITYKDCNFLLQSINGSSKISLNDVLRR |
| 5448 |
PF04697 (Pinin_SDK_N) |
 Estimated TM-score=0.48; very hard target. |
>pinin/SDK conserved region (Length=133) MAVAVRTLQDQLEKAKESLKNVDENIRKLTGRDPNELRPGQARRLTIPGPGGGRGRGIGLLRRGLSDSGGGPPAKQRDIE GALLRLAGDQRARRDSRHDSDAEDDDDVKKPALQSSVVATSKERTRRDLIQDQ |
| 5449 |
PF04640 (PLATZ) |
 Estimated TM-score=0.48; very hard target. |
>PLATZ transcription factor (Length=79) RRSSYHDVVKVSELEDILDISDVQTYVINSSRVVYLTERPQLRSCGVSNTKLSSSQTYKCEICSRTLLDDFRFCSLGCN |
| 5450 |
PF04589 (RFX1_trans_act) |
 Estimated TM-score=0.48; very hard target. |
>RFX1 transcription activation region (Length=154) SSSKTAGAPTGTVPQQLQVHGVQQSVPVTQERSVVQATPQAPKAGPVQPLTVQGLQPVHVAQEVQQLQQVPVPHVYSSQV QYVEGGDASYTASAIRSSTYPYPETPLYTQTASTSYYEAAGTAAQVSTPATSQAVASSGSMPMYVSGSQVVASS |
| 5451 |
PF04578 (DUF594) |
 Estimated TM-score=0.48; hard target. |
>Protein of unknown function, DUF594 (Length=53) DKQMDKWKVMCKVWVELLSYAASHCRADTHAQVLSKGGEFVTVVWLLMAHLGI |
| 5452 |
PF04537 (Herpes_UL55) |
 Estimated TM-score=0.48; very hard target. |
>Herpesvirus UL55 protein (Length=172) TATPLTNLFLRAPDITHVAPPYCLDATWQAETAMHTSKTDSACVAVRSYLVRASCETSGTIHCFFFAVYKDTHHTPPLIT ELRNFADLVNHPPVLRELEDKRGVRLRCARPFSVGTIKDVSGSGASSAGEYTINGIVYHCHCRYPFSKTCWMGASAALQH LRSISSSGMAAR |
| 5453 |
PF04527 (Retinin_C) |
 Estimated TM-score=0.48; hard target. |
>Drosophila Retinin like protein (Length=65) VQPAVLTKTAFVDNSASSAITHQSFTNLEKKVPVVHSYAAAPVVKTVSYESAPVVKSVSYSAPVV |
| 5454 |
PF04522 (DUF585) |
 Estimated TM-score=0.48; very hard target. |
>Protein of unknown function (DUF585) (Length=234) SKTWDDDFVRQVPSFQWIIDQSLEDEVEAASLQVQEPADGVAIDGSLASFKLAIAPLEIGGVFDPPFDRVRWGSICDTVQ QMVQQFTDRPLIPQAEMARMLYLDIPGSFVLEDEIDDWYPEDTSDGYGVSFAADEDHASDLKLASDSSNCEIEEVRVTGD TPKELTLGDRYMGIDEEFQTTNTDYDITLQIMNPIEHRVSRVIDTHCHPDNPDISTGPIYMERVSLARTEATSH |
| 5455 |
PF04425 (Bul1_N) |
 Estimated TM-score=0.48; hard target. |
>Bul1 N terminus (Length=439) KDLPVGESLDDLDEDDIVFDVLPSFEMYNALHRHIPQGNVNPDLHDFPPSYGEIQQGMATTAYIENNVDLTTYPLQQPPL AHHSNSGHDIHITPSYALNALHPLNTQHLTIQPTNDPSVDHIDTPFQDDLNESDNLFIDKLYTLPKIDTPVEIEIKITKH AGSPHTKPEEESILKEYTSGDIIHGYCIIENKSSKALPFEMFYVTLEAYTSVIDKTKGKRTVKRFLRMVDLSASWSYTNI FMGTGWKMTPGEIDFDNAVIGLRNSRILEPGTRYKKYFMFKLPNQLLDTTCKQEHFSHCLLPPSFGIDKYRNGCKYSGIK VNNVLGSGHLGTKGSPILTYDMVDDNLSVNYTVDTRVVGKDPKNKQRIVIMREREYNLRVIPFGFGSALVGERGCSTQLK DLIALIQERLSAVDKIFKRLEKNEPITSADIHGSDITGT |
| 5456 |
PF03752 (ALF) |
 Estimated TM-score=0.48; hard target. |
>Short repeats of unknown function (Length=44) RAEIQRLLADPSTGETVREAAQKALAAGTPAALRQFLATDLQSA |
| 5457 |
PF03580 (Herpes_UL14) |
 Estimated TM-score=0.48; hard target. |
>Herpesvirus UL14-like protein (Length=145) HRRNRVKLVEAHNRAGLFKERTLDLIRGGASVQDPAFVYAFTAAKEACADLNNQLRSAARIASVEQKIRDIQSKVEEQTS IQQILNTNRRYIAPDFIRGLDKTEDDNTDNIDRLEDAVGPNIEHENHTWFGEDDEALLTQWMLTT |
| 5458 |
PF03427 (CBM_19) |
 Estimated TM-score=0.48; easy target. |
>Carbohydrate binding domain (family 19) (Length=61) ITTTSLATASSALFPLLYAGRSCSSQSKVSCTSTGSYTICNYGKWVTAPCPPGVICLSSAQ |
| 5459 |
PF03216 (Rhabdo_ncap_2) |
 Estimated TM-score=0.48; hard target. |
>Rhabdovirus nucleoprotein (Length=307) WTNEMLKDRKVINLDAWNQVGTFDERIQIIREALTSLTTEVSVRSVGALMVCAWGLVSCENPNESVFGGLWESSKARELE EYTRLSDGILCKSSRWRSPPDDKDLILFTTTGNGLNWVRTACFYCAAVLRLATKEHDALVKAWSYLPEHYQSFYKAPLEF SLSLDHECLKCLRRLLQKSTTIRNSVAPFLLAFQELSGCSKNRAICKTLFESHLGFTGLHAYTLFISNASKLAVPHHVFR HVLRHHAAEEGLETIMEILKRYEDPSLDEGDRKSKCTWIYSRIFDSDMFGSLQTKRCVFLAALLAVI |
| 5460 |
PF03072 (DUF237) |
 Estimated TM-score=0.48; hard target. |
>Domain of unknown function (DUF237) (Length=138) LRWELINRANIQLQLQNVKVRENNFEVNGWGVPVSINWNDHNNGINYRATTPWTYGFEITMNYKGSYGLKGIYWTLANWG LGGIPPEWSGDMELKFQIDGKLANWITQKQDYPGSLFQFQNDKLLFTLHVVQRITVKD |
| 5461 |
PF02998 (Lentiviral_Tat) |
 Estimated TM-score=0.48; hard target. |
>Lentiviral Tat protein (Length=87) MEEAPRRYPGGNRDADGIFQYYEEWECWDWVSQRVPGDILQRWLAMLTNSQLRRKVVKEMQRWIWRNPRAPIIRNCGCRL CNPGWGS |
| 5462 |
PF02960 (K1) |
 Estimated TM-score=0.48; very hard target. |
>K1 glycoprotein (Length=120) GLSSRLSNRLCFSARCANITPETHTVSVSSTTGFRTFATAPTLFVMKEVKSTYLYIQEHLLVFMTLVALIGTMCGILGTI IFAHCQKQRDSNKTVPQQLQDYYSLHDLCTEDYTQPVDWY |
| 5463 |
PF02697 (VAPB_antitox) |
 Estimated TM-score=0.48; easy target. |
>Putative antitoxin (Length=66) KTITIADDVYYELVKMKGNKSFSELLRELIGKKKKGNLDVLMIAFGTMTEEEAKEFEKEMKEVEEW |
| 5464 |
PF02646 (RmuC) |
 Estimated TM-score=0.48; hard target. |
>RmuC family (Length=298) EQMDQLLNPLGEKLEAFKKKVEETYDDENRQRATLKEQIKQMAELNKTMSEDAKNLTKALKGDSKTQGNWGEVILQRILE KSGLRKDEEYFVEQSVTMDDGRRIRPDVVVRLPDEKFLVIDSKVSLTAYEQFSSAEDEADQERALKEHINSIRTHVKGLS EKNYQHIHGDRSPDFVLLFIPIEPAFGAALQHDSNLYYEAFDKNIVIVSPSTLLATLATIDSVWKQEYQNKNAMEIAKRG GALYDKFVLFVESMNDIGQRIRQTQDSYDEAMGRLSTGAGNLVRQAEMIRKLGAKTSK |
| 5465 |
PF02398 (Corona_7) |
 Estimated TM-score=0.48; hard target. |
>Coronavirus protein 7 (Length=101) MLVFVHAVLVTALILLLIGRIQLLERLLLSHLLNLTTVSNVLGVPDSSLRVNCLQLLKPDCLDFNILHKVLAETRLLVVV LRVIFLVLLGFSCYTLLGALF |
| 5466 |
PF01837 (HcyBio) |
 Estimated TM-score=0.48; very hard target. |
>Homocysteine biosynthesis enzyme, sulfur-incorporation (Length=358) AVVVTAEEMIGIVEEKGYTKAAEEVDVVTTGTFGPMCSSGAFLNFGHSNPRIRMKRVWLNGVEAYSGIAAVDAYIGATQL PENDPENKVYPGKFVYGGGHVIHDLVARKMIRLEAVSYGTDCYPRKKIETMITLDDLNEAYLYNPRNAYQNYNCAVNLGN RTIYTYMGILKPNLGNASYSSAGQLSPLLNDPYYRTIGIGTRIFLGGGIGYVAWYGTQHHPNVPRAENGVPLGGAGTLAV IGDLKQMSPEWLVGASFIGYGASMYVGLGIPIPILDEEMVRYTAVKDEEILCPVVDYSEGYPYCKPIDLGYVNYRDLKSG KVTIRGKEVPTTPMSSYPRARKIAEILKEWIKSGRFEL |
| 5467 |
PF01623 (Carla_C4) |
 Estimated TM-score=0.48; hard target. |
>Carlavirus putative nucleic acid binding protein (Length=90) AVVKMLILRKFAEQGNVCPIHLCVDIYKRAFPRSVNKGRSSYARRRRALELGRCHRCYRVYPPLFPEISRCDNRTCVPGI SYNSKVKDYI |
| 5468 |
PF01526 (DDE_Tnp_Tn3) |
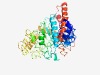 Estimated TM-score=0.48; hard target. |
>Tn3 transposase DDE domain (Length=384) EILDEVNCWTTFTRHFSHIKNDITRPDTRQLLTTILADGINLGLTKMAEACPGSTKSSLEDIQAWYTRDETYSAALAELV NAQGKRPLAAFWGDGTTSSSDGQNFRTGSSGRYAGQVNPKYGQEPGRQFYTHISDQYSPFYTCIISRVRDSTHVLDGLLY HESDLEIREHYTDTAGFTDHVFALMHLLGFAFCPRIRDLHDKKLFIKGKADQYPALQSLISTTSLNLKEIEIHWREVLRL ATSIKQGTVTASLMLKKLASYPKQNGLAKALREIGRIERTLFMLDWFRDPALRRRVQAGLNKGEARNALARAVFLHRLGE IRDRKPENQSYRASGLTLLTAAISLWNTIYMERAVDALKRKGMKINERLLSHLSPLGWEHINLT |
| 5469 |
PF01254 (TP2) |
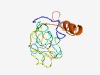 Estimated TM-score=0.48; hard target. |
>Nuclear transition protein 2 (Length=137) MDTKTQSLPNAHTQPHSNSGPQSHACNQCSCSHHCQNCSQSCDRSQSCSRSRSSSQSPTGHRSLPGHQSQSLSPSPSPRH RKRAMHSHRCPSRPGTRSCSHSKKRKNVEGKANKRKGIKRSQQVYKTKRRSSGRKYN |
| 5470 |
PF01203 (T2SSN) |
 Estimated TM-score=0.48; very hard target. |
>Type II secretion system (T2SS), protein N (Length=212) GLSIEGVEGTVWQGRANNIAWQRVNYGSVQWDFQFSKLFQAKAELAVRFGRNSDMNLSGKGRVGYSMSGAYAENLVASMP ASNVMKYAPAIPVPVSIAGQVELTIKHAVHAQPWCQSGEGTLAWSGAAVDSPVGSLDLGPVIADITCEDNTIAAKGAQKS EQVDSEFSASVTPNQSYTSAAWFKPGAEFPPAMRSQLKWLGNPDSQGKYQFT |
| 5471 |
PF00524 (PPV_E1_N) |
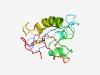 Estimated TM-score=0.48; very hard target. |
>E1 Protein, N terminal domain (Length=121) MAARKGTDSETEDGGWVLIEADCSEVDSADETSENASNVSDLVDNASIAETQGLSLQLFQQQELTECEEQLQQLKRKFVQ SPQSRDLCSLSPQLASISLTPRTSKKVKKQLFATDSGIQSS |
| 5472 |
PF18842 (LPD26) |
 Estimated TM-score=0.47; hard target. |
>Large polyvalent protein associated domain 26 (Length=57) DGEMRRAWGGKLPEEFTQRDTLAFMAETDLQLQGEVSAETLSAIHAAGYDYKGGAVI |
| 5473 |
PF18721 (CxC6) |
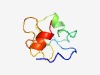 Estimated TM-score=0.47; hard target. |
>CxC6 like cysteine cluster associated with KDZ transposases (Length=65) VTDGVTIGHPCCAVHNCHEPLPTIAARYCSTHDHLNHICAVRGCTAPIQPGGRTCNTPDHIQLQE |
| 5474 |
PF17742 (DUF5579) |
 Estimated TM-score=0.47; hard target. |
>Family of unknown function (DUF5579) (Length=202) TPPGWEPAPDAPWCPYKVLSEGPEAGGGRLCFRSPVRGFRCQTPGCVTLASAGGSLRAHVLRNRSVLLQWRLAPAEARRV RVFALNCSWRGTYTRFPCDRVLLGASCRDYLLPDVHDSVRYRLCLQPLPLPLRAELAAAPPELAECVEFTAEPAAMQEIV VAMTAVGGSICVMLVVICLLVAYITENLMHPTFRRPSLRRQP |
| 5475 |
PF17702 (DUF5548) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5548) (Length=177) MQVEVQSLSLEECPWRLPGPQCECEALLPSGARRRIDLRLSGRAVAVWVHVRDGPGQLNLSYATGRHKKPNPHQNMNRGM EFIAPVSAPTKSGAPWHFLSQGPTDAQRAVRIRPGTRMGLSSDPVVGTLSSSYLDLLTLSYKPGRRVTSSYLNVRGHEVR KLQNSVEATRISRTGSS |
| 5476 |
PF17700 (DUF5546) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5546) (Length=135) MTQPRNCMRLSRSCSLTWETPRWYMAGRVATSTSGCRCWMSRRDLTPLPHPSEPAVLDCLGPCRQPASPGSPCWVLGLHF SLHPPSASASHALTITSLPPGLLPFVGVELTAHPQALMGGFPSGMGAAGRHLCFL |
| 5477 |
PF17695 (DUF5541) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5541) (Length=116) TIAFKRAEAVEPDGCVQSWCCRLPCGLGQASCFIHATVCSAIRWRSCKGQRNFAERHILPAELEEQSNHAGMGPIPPAMP VVDGNHFQHPAGDCHPYGILCLQAHSASVTARQVLQ |
| 5478 |
PF17691 (Croc_4) |
 Estimated TM-score=0.47; very hard target. |
>Contingent replication of cDNA 4 (Length=138) HPSRHSFSPGAGGFCTTLPPSFLRVDDRATSSTTDSSRAPSSPRPPGSTSHCGISTRCTERCLCVLPLRTSQVPDVMAPQ HDQEKFHDLAYSCLGKSFSMSNQDLYGYSTSSLALGLAWLSWETKKKNVLHLVGLDSL |
| 5479 |
PF17670 (DUF5530) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5530) (Length=140) MTAIRLREFIERRPLIPPSIFIAHQGRDLQGYYPGQLARLHFDHGAKRAPRPLIDLTIPPKRKYHYQPQFDQQTLIRYIC FRRHSKPAEPWYKETTYQRDYSLPFYEIDWNQKLATVSLNPRPLNSLPELYCCEERGGFE |
| 5480 |
PF17635 (DUF5515) |
 Estimated TM-score=0.47; hard target. |
>Family of unknown function (DUF5515) (Length=70) MLPPCYNFLKEQHCQKASTQREAEAAVKPLLAPHHVVAVIQEIQLLAAVGEILLLEWLAEVVKLPSRYCC |
| 5481 |
PF17634 (GP67) |
 Estimated TM-score=0.47; hard target. |
>Gene product 67 (Length=80) MEGLIEAIKSNDLVAARKLFAEAMAARTIDLIKEEKIAIARNFLIEGEEPEDEDEDEDDEDSDDKDDKKDEDSDEDEDDE |
| 5482 |
PF17631 (DUF5512) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5512) (Length=139) GCGGSSVPYKKDDTYKDSKDNFITITAENEWRVKGSQNSEANYKVESTEYKGDSHSVVAISVKEKINGSDPLLVVNDYHY YILSPRENGFSLTPIGTSHPNSAQWKEFQEGFKGANDKEEFLKKEVANVNKQNIFKKTN |
| 5483 |
PF17615 (C166) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (Length=158) VYVLLFDVFLSLVVGAYSGIAPPSIPPVPTYASAQLTASLITWTVGWPPITLWPQITLIPPFSILGANFPGLTIPSLTIP GVTLFSISFSWLAPIIYIANWIIWVFQTVASVLSYLLNIFTGSVGLLSSVPVLGPFLTAFVLIVNFVLVWELIKLIRG |
| 5484 |
PF17497 (DUF5432) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5432) (Length=74) MLHEFHTWRLLKFRVESCSSIHLLSLHPLYVIMSPMDIIGFNKYFRYRNKILFVDVFDYHLLCFWRTFLCMIFT |
| 5485 |
PF17494 (DUF5429) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5429) (Length=76) MCMMSHNKAFFLSLQHAAVSGVAVCLSVRRGAGSVPAGNRGKKTIITEYRITGTRALARCPTKPVTSMWNSSWTSR |
| 5486 |
PF17460 (RP854) |
 Estimated TM-score=0.47; easy target. |
>Uncharacterized protein RP854 (Length=211) MFEKYIKYLKNLIFFQFIIYFFFISLTIWIIKNCQQEYSKSILDKQVLQENLTEEVLKLYSVINSKEEILESYKKYVDLS VPSSVSCLNYQELIPKIKSLRNKYDLVEPIDVFINSVFFKNNVQAIEGEGEAIHVNNYSINIKYACADFLTFLKIFSEIY SYMPPNTLISFVRVRNEEVLTPKNIYKLSVNHAPNLIYAKLILYIRELASK |
| 5487 |
PF17457 (DUF5420) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5420) (Length=184) PEFRYFKCALSVEPIKSLDEQWRKDRELRDKKLDAIFDTIPFYECWRGSERNIFGIVCSLDSDEFAKIKEDKTYKFEIVE NEKVVITGNGRTKAGKAFNDKIQSVRDILNQYPSFNDFMLRKLKLNCWVLGARTSYISVCGVASDYFIVSIPVKSDGFGG DGFPAIPECLTEIKQSEFLMLQGK |
| 5488 |
PF17424 (DUF5411) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5411) (Length=135) MKESYWGYWLMVFGVFIIVVLLLIQSFTSTNTQDYYLIKEISEASMVDAIDYGYYRTYGELKMNKEKFYEVFLRRFAEEA SLTTNYTIEFTEVYEAPPKIGVKVSSKSATFQVAGDTTTFDIVNKVDAILESKVD |
| 5489 |
PF17398 (NolB) |
 Estimated TM-score=0.47; very hard target. |
>Nodulation protein NolB (Length=150) SPSFGEQAQFEHALAQAAASMKNDTASGPATSAPIPPQLDVQRATAQSSPLGDRVLQTISSMYRDNAVTSAALDHQVGLA KDGLPGPAQKLPVEGGAAGAQMSGVPQAGHDFESMIADLRDLYNGATQVALISKGVSGITSSVNKLLKEG |
| 5490 |
PF17366 (AGA2) |
 Estimated TM-score=0.47; hard target. |
>A-agglutinin-binding subunit Aga2 (Length=60) IPSATLESTPYSVSTTAILANGKSMQGVFEYYRSVTYVSNCDPALTATKKDSPINTQYVF |
| 5491 |
PF17281 (DUF5346) |
 Estimated TM-score=0.47; very hard target. |
>Family of unknown function (DUF5346) (Length=96) PAVAPTGQCPGGPSLPIECDPKRPWPQCPPQSYCYATNSVDVGPYFCCPIWSTYGAAWRPATPFYNYVPPMPANWPDVAK ITANWPAAAIGLPVKA |
| 5492 |
PF17237 (DUF5310) |
 Estimated TM-score=0.47; hard target. |
>Family of unknown function (DUF5310) (Length=44) LPNLRRIFASFRTEKEERGYSRTAFFNLIGFLGTCVVFSLIAQK |
| 5493 |
PF17029 (DUF5100) |
 Estimated TM-score=0.47; very hard target. |
>Domain of unknown function (DUF5100) (Length=126) MSNESKLQKILSILNSPIDLEKYIEMLESIDLVLNVNDDEKDYFICPEEANLQLPNVFLRQTKLLECKQFIKSKASSCMG EHFYENCREILDFLESVELQEPEFMESRKKSDIDVDLLRELVKKYI |
| 5494 |
PF17026 (zf-RRPl_C4) |
 Estimated TM-score=0.47; hard target. |
>Putative ribonucleoprotein zinc-finger pf C4 type (Length=107) MTICSYCRKSKLIKSDEYIYCPTCESYFTQTNVKQKHAYVKIEDTDQITLYGNLCDYCEKKYNTEKYITCQTFNDYLYGL NFCTKCRIRNLKFLKNSYNKSFLLYKA |
| 5495 |
PF17020 (DUF5097) |
 Estimated TM-score=0.47; very hard target. |
>Domain of unknown function (DUF5097) (Length=120) MIESVVIRKYIIQYFLDNGHLDLWAMSKDLNEYFSSHFLSKEYICIKKLKCTGRIVNFGKSECMVAVNVDNGNKMQKINT EDITRRDELTINDIKNFLETVTQENLPFGRMLKPNILESL |
| 5496 |
PF17015 (DUF5094) |
 Estimated TM-score=0.47; easy target. |
>Domain of unknown function (DUF5094) (Length=189) MKKGVSRTPKKQSRVVSKSKNRKKKENSKEQENLQIQEPAHCEEKEINEKAHTKTVVMKEYMENETQQNGVMCSECRNYT DVKLELVTQLQEENRMLREEIQHLLSTMNMLGITSHASGDTQVWGFHREGSKGNIGLEFEIKDSGNNYTVQITSTHNCDV PEYMYKGIEFKKKSFILFFYKLMQIICEN |
| 5497 |
PF17012 (DUF5091) |
 Estimated TM-score=0.47; very hard target. |
>Domain of unknown function (DUF5091) (Length=145) LIFKLNNAIQKPTLEAFLNIFEHDGYVRLNMVYISPISLKHLRQKLESFDLIEMFFTTNGLAFIFKTFRLNFFGNMNVVV KNKIYCIEINFDPTIDENCGIPIMSEIEHYLKGCDALFFMTYFVDIDKEILHNSLSESVDFLYKR |
| 5498 |
PF16981 (Chi-conotoxin) |
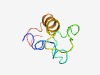 Estimated TM-score=0.47; hard target. |
>chi-Conotoxin or t superfamily (Length=59) MRCLPVFVILLLLIASTPSVDALLKTKDDMPLASFRDDVKRTLQTLLNKRFCCPYFECC |
| 5499 |
PF16935 (Hol_Tox) |
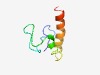 Estimated TM-score=0.47; hard target. |
>Putative Holin-like Toxin (Hol-Tox) (Length=60) MVAILEGGGAYGFKTVNPDPRKGEASCPVSDALQLMLAFGGFILGILTFVVLLIEKISKK |
| 5500 |
PF16882 (DUF5079) |
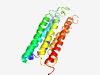 Estimated TM-score=0.47; hard target. |
>Domain of unknown function (DUF5079) (Length=223) KENIDTLRKPGAQALSLASLFMILFSCLTFFFGLDYERFPNYLKITTIIELIIIIISLLQWIRFIDFEKESAQKYKKIYA RFLVIINVLTTITAVFATCNLYYFVAVQNHYDLFNYWLMGTISIIISYLLLVIGGMFTLLKLPKVTKRWGGKTKTHFGLL LTALSSFIYIEKIIEYILVPNVVESKFIIIVSMLVIAGAQFVAFQFIMQYSRFYIFELNTEDD |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218