

Go to page (83 pages in total): [First page] << < 40 41 42 43 44 45 46 47 48 49 50 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4401 |
PF13109 (AsmA_1) |
 Estimated TM-score=0.52; hard target. |
>AsmA-like C-terminal region (Length=214) DRVKIDGNYKNAMIMADLVHGALYLKAHNFSGDYINTILQKDFVEGGLFTLIGALEDQVFNGELKFQNTSLKNFALMQNM INLINTIPSLIVFRNPHLGANGYQIKNGSVVFGITKEYLGLEKIDLVGKTLDIAGNGIIELDKNKLDLNLEVSTIKALSN VLNKIPIVGYLILGKGGKITTNVNVKGTLDKPKTQVTLASDIIQAPFKILRRIF |
| 4402 |
PF13073 (DUF3937) |
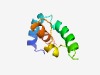 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3937) (Length=72) MFTNKKLIRFGLSLFVFLGIINFTISYFQTYLETAADIKWVIPEIWKTILLDVPQGILVLLGAIALYDFTKE |
| 4403 |
PF13066 (DUF3929) |
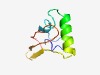 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3929) (Length=64) MVYHLEDGKTIQDIKEFYYRDHNKMLERVKHHVTDNQNVTAIDPQGTLISIACDDIVKVELNYL |
| 4404 |
PF13065 (DUF3928) |
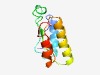 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3928) (Length=95) MYTLKIVSDREAVYQFASYVKVVQGVENVYVEVGEPLYEHPLMKFYVHIAIQATYDNEKALKEIARLVELGRFTYVHYRN EEIEQAFEAVKYESF |
| 4405 |
PF13007 (LZ_Tnp_IS66) |
 Estimated TM-score=0.52; hard target. |
>Transposase C of IS166 homeodomain (Length=72) KLKAQLAVLRRARFGASSEKIERSIAQLELALEDIEAAEAEIEVSARGVSGSREKAKPSRQPLPDHLPRQEV |
| 4406 |
PF12991 (DUF3875) |
 Estimated TM-score=0.52; hard target. |
>Domain of unknown function, B. Theta Gene description (DUF3875) (Length=52) ILKASTLESKFPIMAVEHDCIVSKDADITVAFRVTLPEVFSVSSAEYEAMHA |
| 4407 |
PF12982 (DUF3866) |
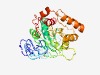 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3866) (Length=318) RAYNYPKLTGEVNIGDEVVLNTTAVELSLGTGGYHFVISNLNNMESNFTPGGHIMKLRYTPLQVKVDAVEEQESPYHDAI EEFESLENMPVVVGTLHSMLTPFVASYKRNNPSKKLVYIMTDGASLPIYLSKNVDTLKNKKLIDSTITIGNAFGGDYECI NIYTALITAKEILKADVVFVSMGPGIAGTGTKYGFTGIEQGPILDAVKKLKGMPVAIPRVSFADQRDRHMGISHHSLTVL NDIVNVEVNIPITTYEDEKLDVVKRQIETKELDKKHHIVYINNKNTEEDLNYFGLSVRSMGRNYQQDKEFFDAASSAA |
| 4408 |
PF12757 (Eisosome1) |
 Estimated TM-score=0.52; hard target. |
>Eisosome protein 1 (Length=124) RNALMAAAQRNVKARLQGMDEKVYNETGKVNPTLMSEWEVKAHQLANASHETRNENKGKIDIGGGKFMDPHEVDAIAARR MQPLLDDINEKAEIERERLATLKMEEEARKAEQEKQKAREREIK |
| 4409 |
PF12621 (PHM7_ext) |
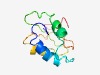 Estimated TM-score=0.52; hard target. |
>Extracellular tail, of 10TM putative phosphate transporter (Length=98) FLKPHIYADYATLRRLVPHDVAGSVSNYSPEVERDAYLPPSVKSETPLLWIPRDEMGISKQEIAHTAKVIPITDEGAALD EKGAVYWVAEKEEGARPP |
| 4410 |
PF12587 (DUF3761) |
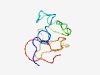 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3761) (Length=86) FVLFAVALLCNTAVARQHTQPSSQDSELIEQGDYTNSDGQEIHRPAHTKSGKAPDGASAQCRDGTYSFSTHRRGTCSGHG GVAAWL |
| 4411 |
PF12576 (DUF3754) |
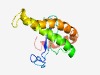 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3754) (Length=132) FEELILLYTKDASEKDEKSKDETRSSLQLEIFERIPIPDLPVIFPHKKLYFRVIDTVRLDIASILGLTAYFVNYKFENIS SSPSALFLDVIAVTALVIYATRVVLGYKQTWDRYQLLVNKTLYEKTLASGFG |
| 4412 |
PF12517 (DUF3720) |
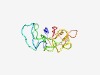 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF3720) (Length=97) KAVEAAEGKQELSGPDPPQDKSKVSSPDNKLGNNTQSTGAPGAGGGGVGGQPAELPPKPGGSGTEEQVGELEVMDTSGEE VAGDKKDHEDGTATGPQ |
| 4413 |
PF12497 (ERbeta_N) |
 Estimated TM-score=0.52; very hard target. |
>Estrogen receptor beta (Length=114) SPASYNCSQSVLPLEHGPIYIPSSYVDNRHEYSTMTFYSPAVMNYSIPGGASNLEGGTGRQTTSPNMLWPAPGHLSPLAI HCQSSLLYAEPQKSPWCEARSLEHTLPINRETLK |
| 4414 |
PF12491 (ApoB100_C) |
 Estimated TM-score=0.52; hard target. |
>Apolipoprotein B100 C terminal (Length=57) QFRYKLQDFSDQLSDYYEKFITESKRLIDLSTQTYHMFLRYITELLKELQSATVNNM |
| 4415 |
PF12466 (GDH_N) |
 Estimated TM-score=0.52; hard target. |
>Glutamate dehydrogenase N terminal (Length=95) MVSKKSLSTSVKGPAKDPAKQSLVGDVEPRCMVNLESVFAVLREQCPVSMQSDVQRLGAAFYCRMETDEFARRTPEQWAA LVMGMLEFARVRSSG |
| 4416 |
PF12430 (ABA_GPCR) |
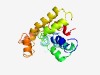 Estimated TM-score=0.52; very hard target. |
>Abscisic acid G-protein coupled receptor (Length=179) RSKTVLGRLLNVFSYGFSMYCLYRIGATTLTTFRRLHAPDTSFSNSDPINNVLALLAKHWDPALDRAAWSRQISFLLAGV MLLASFNSVLQTFLLFTRLAPSLLQHAQSNIALIVSQISATYVISSALLLRSNLPPEYRGVISEALGAPLEPGFVERWFE GWFLAACVVSGLGIWLGRK |
| 4417 |
PF12355 (Dscam_C) |
 Estimated TM-score=0.52; very hard target. |
>Down syndrome cell adhesion molecule C terminal (Length=118) SGPGPEYDDPANCAPEEDQYGSQYGGPYGQPYDHYGSRGSMGRRSIGSARNPGNGSPEPPPPPPRNHDMSNSSFNDSKES NEISEAECDRDHGPRGNYGAVKRSPQPKDQRTTEEMRK |
| 4418 |
PF12335 (SBF2) |
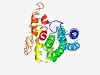 Estimated TM-score=0.52; hard target. |
>Myotubularin protein (Length=234) SIVNDPRHSVSNSARRLEVLRNCINCIFENKIADARKTFPAVLRALKSRAARLALCTELARHVQGNNAMLEHQQFDLVVR LMNCALQDGSSMDEYGVAAALLPLSTAFCRKLCTGVIQFAYTCIQEHAVWQNQQFWEAAYYQDVQRDIKALYLEPHNNPA HTPQSPRDGGREFVWRESRGRGNAHSRSQEPTALEIAAEQMRKWPSIDADKQKELTNSEESTLYSQAIHYANRM |
| 4419 |
PF12321 (DUF3634) |
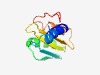 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3634) (Length=100) LYVILIAAIVIFWLIAVDRPVLKVSFDDGHLSKVKGHIPPSFKHNLQDIGEHDPFTGELKVYNQRSGMRLVFSKDVPKKV QQRIRNVFPHQGFKANKGKK |
| 4420 |
PF12267 (DUF3614) |
 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF3614) (Length=161) NFTARFPISAVRYENHYAATMTDDGIGWVPKGHVPDVEIVKRRRSRNRRLPLSHRVAEQSVPALPSSSVEVTSASSDRSS SSEKASTPSCDSVRSSDWVNVGYSRQNTTKVRESRFTGRGKIGNFTFPRGTVYNAPVDERAYKRVLKLRDTTACSFLRIL L |
| 4421 |
PF12168 (DNA_pol3_tau_4) |
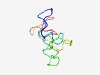 Estimated TM-score=0.52; very hard target. |
>DNA polymerase III subunits tau domain IV DnaB-binding (Length=77) LQQNTTQNGGTKKPKAASAKPESVIDRVAQLHGNSAQVSPNSTSKPTEPEVEEAYRWRPSKPVETKVNTKLTPTQLK |
| 4422 |
PF12015 (DUF3507) |
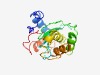 Estimated TM-score=0.52; very hard target. |
>Domain of unknown function (DUF3507) (Length=178) KSVVETTPLNGHSLFDDDKSLSDWTDNVFTQSVFYHGSDDLIWGKFFVCVYKSPNSNKLNAIIFDKLGTSCFESVDISSN SQYYPAIENLSPSDQESNVKKCIAVILLQRYPLLSPSDLSQILSNKSENCDYDPPYAGDLASSCQLITAVPPEDLGKRFF TSGLLQNRFVSSTLLDVI |
| 4423 |
PF11950 (DUF3467) |
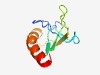 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3467) (Length=83) PEQPAPDANPALRARVPDHVAGGCFSTGAIVMTGPSEFIIDFLQTIGRPHRVATRVVIPHPVMPQFVDALQKNLDIYRSR FGD |
| 4424 |
PF11872 (DUF3392) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3392) (Length=104) LIEFLADSGQFIRPWLSEISTAMIACLLVVFGADINRLLRRQLSGTNFVIRTLVFILVNAFGYGFLIITVSPWLANQLSH IPSQWLICLVAATFIFIGTWAQRN |
| 4425 |
PF11862 (DUF3382) |
 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF3382) (Length=99) NLLNALISALVLLVLASFIMGLQLSLEGTRLVVHGADVVRWYWIAAGCAVVFVFQLFRPLLQKGLKKVSGPSFVLPSFDG STPKQKWLALLLIIAAVAW |
| 4426 |
PF11780 (DUF3318) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF3318) (Length=141) RRLLDLMPASGRMLTKIVSKPEQAKVIDATFPLPWNSERPIYINFDLWRRLTKPQRDLLLLRTVNWLTGVKWFKPDIYQG VALAGLLGGFVESAQADAVGVVVAGGLTTLAMLRIWRTNRSQQTELNADEAAIRVAQRRGY |
| 4427 |
PF11727 (ISG65-75) |
 Estimated TM-score=0.52; easy target. |
>Invariant surface glycoprotein (Length=280) HILLVFSAVASASEESYKIDRYKADRKLDKEGAIALCKLRDVLTNISEETTDYLVEEVQKMGESLKMDMDAINYYVKRVR SLKESGREDAKINESDSNEIDSLCKEAKAHANKKSRDQVDQTVAKIRRVSGIVKDAATKTLGDENELEDHDNADGIFKVF NWHCGGDRIGEVHAGSQSCDVIGFRKNFSAGQRNVISCKKWKDEYGETIPHENVTVTQMESYLSRWFEARPKRVGEDICK DGEYHSPHSCTVWEGWVTGYEETMEKMEELRNNIKKLNRI |
| 4428 |
PF11594 (Med28) |
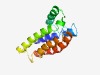 Estimated TM-score=0.52; hard target. |
>Mediator complex subunit 28 (Length=134) RVRSYLEQLSKSFVLLDVIREKKVWVIEKTNELSTSVYDFKTTLENEAEQYKLAPEQMLMKHIQSSSESLATQLKTLREK GQNVFGDGTRIDSTIDYLEKLKTNLSEAENVYKVRDEVLNEARQRIAELTTWTT |
| 4429 |
PF11189 (DUF2973) |
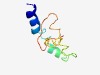 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2973) (Length=68) MLHLLYILAFTILAFIAVGNLIRNLIMFSFERERSYPTNYSSKNQFIPHPELLDNAGNLIKEPLLVMR |
| 4430 |
PF11140 (DUF2913) |
 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF2913) (Length=211) NYHLEIQNVVNTALAELEAEHKAGKLANAPVANNHFLVHWVTKALKAQRFHRCVGDDLTQWQKAGRSKGTESQLLPTFQR ISAYYAHFFAEQEHTPITDKQIEAFLDEMEQAGWEVSTSEPLVNAGKVQIFTDGQNSLALCSVQCEACFDGEQLVKPMNW FVRGHHAGFIEKAFAAGFMVHKQTDYKSNVKYHGEYLIFPANQGTQLAEIP |
| 4431 |
PF10971 (DUF2773) |
 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF2773) (Length=81) ALNNRHTPPAVLAAEIDPEWWIVAMNNPRFPVDVLKARLKRDPLLALELVNPELDLVRQLALNGKTRAIREQAMRKLDEL Y |
| 4432 |
PF10970 (GerPE) |
 Estimated TM-score=0.52; very hard target. |
>Spore germination protein GerPE (Length=125) TSQVKFSKVNSVGISSVVQIGDTQELCPKVKVLAVQRTVSLFYGNEAENLDAKEFEVYYEPIPLMLPERSVRTAFYHEKP VIKVSSIHVEGVSSSSVFQIGSTCGMNGVARVKHIRELFSNDESP |
| 4433 |
PF10969 (DUF2771) |
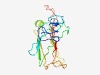 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2771) (Length=159) ILALIIAVVIVVVATVLVQNWWNNRPGPEPEEVTVTASVGDESIEVSPYLVCEPGVECPEGEVPNLPVGPDETLKLELPE PIHNHDWQLLMIYDDPAANNQQLHGPHDTTEVEIPGSVDPVGDSTERPRLVVVEVSSVMIGHDDEGVETPYTTIWSLST |
| 4434 |
PF10961 (SelK_SelG) |
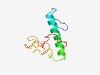 Estimated TM-score=0.52; very hard target. |
>Selenoprotein SelK_SelG (Length=89) YISNDGQVLESRSPWRLSVIPEMIWGLINFIVLFFQTLINPSKNSKGDRYTSEYRRPGGGGPGGGGPRRRMGGVGGRGGA PGPPPMAGG |
| 4435 |
PF10959 (DUF2761) |
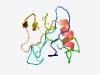 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF2761) (Length=94) PKQPGYVCPMTGRVAVLVKDYADSDLNGDAPAYWFNPEAEGWGMDPWKLVEGVDPHAQGSSMDVCFADGSSKTVGPLMTF FLTKTDAARLAAVR |
| 4436 |
PF10944 (DUF2630) |
 Estimated TM-score=0.52; easy target. |
>Protein of unknown function (DUF2630) (Length=80) TDNNIFTKIDALVAEEHELRSKHAAGEISDGEEHDRLKALEVELDQCWDLLRQRRAKREFGENPDTAEARPASTVEGYLN |
| 4437 |
PF10899 (AbiGi) |
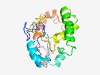 Estimated TM-score=0.52; very hard target. |
>Putative abortive phage resistance protein AbiGi, antitoxin (Length=189) QTFVPRYCFEEIKLSDELERPGFESAIPMVCFCDISLSQMANHIKMYGGYGLGMTKEWGIRKKLNPIIYVNPNSNISESV SQMAENIYQALDKHCSEITGNIFDEYMNLLNFLKPYDGDFKRGNKTHKKVRFYNEREWRYVPKIEFDSEVKNNLTKEEFL NPTILENENKKLFEYRLGFKPSDIKYIFV |
| 4438 |
PF10870 (DUF2729) |
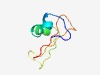 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2729) (Length=57) KLLTYCKLKLVKGFSKKFASLFCRCVVSHLSDEDDSDGDRYYQYNNNCNFIYINIVK |
| 4439 |
PF10849 (DUF2654) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2654) (Length=70) AERKANKLLSKNKRELNRLYKHAQIAAENNNFAQYEYAIKKSRDILKQPYNDELISILWKTTRSQIEDMI |
| 4440 |
PF10810 (DUF2545) |
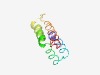 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2545) (Length=80) MIYLWMFLALCIVCVSGYIGQVLNVVSAVSSFFGMVILAALIYYFTMWLTGGNELVTGIFMFLAPACGLIIRFMVGYGRR |
| 4441 |
PF10753 (Toxin_GhoT_OrtT) |
 Estimated TM-score=0.52; hard target. |
>Toxin GhoT_OrtT (Length=54) YQHMLVFYAVMAGIAFLITWFLSHDKKRIRLLSAFLVGATWPMSFPVALLFSLF |
| 4442 |
PF10746 (Phage_holin_2_2) |
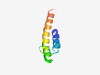 Estimated TM-score=0.52; hard target. |
>Phage holin T7 family, holin superfamily II (Length=56) SELVSGGMKLAPSALVSGGYFLGISWDNWVLIATFIYTVLQIGDWFYSKYSLWKEK |
| 4443 |
PF10734 (DUF2523) |
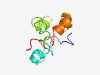 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF2523) (Length=80) IAEWLAKISWPLVSRVLVSLGFGYTTYEGADTAITNAISSIQGAFAGLGAEVLQLLAMAGFFDAMSITSGGIVSGLAWMV |
| 4444 |
PF10705 (Ycf15) |
 Estimated TM-score=0.52; very hard target. |
>Chloroplast protein precursor Ycf15 putative (Length=86) ETLVSSIFWTLAPWKNMLLLKHGRIEILDQNTMYGWYELPKQEFLNSKQPVQIFTTKKYWILFRIGPERRRKAGMPIGVY YIEFTR |
| 4445 |
PF10561 (UPF0565) |
 Estimated TM-score=0.52; hard target. |
>Uncharacterised protein family UPF0565 (Length=305) RINGVKGHQNRTNSIIYCPPLLRTNPQKDKCSAIIYFGGDVQDIPEKMESNRDNKNYIKWNLENTALLLRESFPQSHIIV VRPMRMEYSTFSCFDNFVRGNNAGIPDHTPMHFSLQHLEELLINLTKKLTKPILDQEFLHKLLAASSSKTLAGDVSCKQN NVDILQSDIYNSTSGDSNEGDLTDLLWWRENLNLDKASLKLIGFSKGCVVLNQFIYEFHYYKTLTPDDSTMMRLVSRITD MYWLDGGHGGGKNTWITSRSLLETLCRLCINVHVHVTPYQIQDDHRPWIRKEEKAFTDLLKRLGA |
| 4446 |
PF10554 (Phage_ASH) |
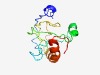 Estimated TM-score=0.52; very hard target. |
>Ash protein family (Length=112) YASRAFAKSKAECGNSKEHQANNSTPLTRAFFVRSSRTPKECQNKACSSMVACSGQGFALDCLPDVAVFHPVTRYRPTLW KMLAIAPKNQHQELSKMKAFAYSFLCAIRTPE |
| 4447 |
PF10490 (CENP-F_C_Rb_bdg) |
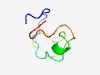 Estimated TM-score=0.52; very hard target. |
>Rb-binding domain of kinetochore protein Cenp-F/LEK1 (Length=45) RAEQDDEEFRPEGLPDLVQKGFADIPLGEASPYIIRRTTGRRCSP |
| 4448 |
PF10367 (Vps39_2) |
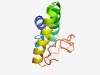 Estimated TM-score=0.52; hard target. |
>Vacuolar sorting protein 39 domain 2 (Length=100) LDLLSRRWDRINGAQALRLLPRETKLQNLLPFLGPLLKKSSEAYRNFSVIKSLRQSENLQMKDELYNQRKAVVKISSDSM CSLCNKKLGTSVFAVYPNGT |
| 4449 |
PF10347 (Fmp27_GFWDK) |
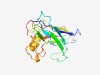 Estimated TM-score=0.52; very hard target. |
>RNA pol II promoter Fmp27 protein domain (Length=161) DYPLPLIHIPGLKPGQPARQSAMSLRTNFVIAEEYRGKKSTRHIKVNIIPPRTLESDASQEGGFAIDVRRTIGPIKSFSD MRIDINTALPTRITWGGPCYQPAIQDMMMVIESFTKPQVDPSERVGFWDKIRLNFHSRIHIAWRGDGDLHLALKGTRDPY C |
| 4450 |
PF10261 (Scs3p) |
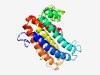 Estimated TM-score=0.52; hard target. |
>Inositol phospholipid synthesis and fat-storage-inducing TM (Length=245) PSYFARKDNLVNVLFVKRGWAWVSAAFAAFVLTHPHHTKNTTTGRGGGQQGGSSSRRLRAVLRWALVTGWWVLVTQWCFG PPLIDRGFRYTGGKCEEAEQKVRGADNYAASPVDIFSAAACRTAGGRWRGGHDISGHVFLLVLGSFFLLQEAGWVAVRAW EGASGGGGRRDSRAVVMPDGAVKGVDVEADGRREDPSELGFGGRFVAGVVGLSMWMLLMTAIYFHTWFEKLTGLLVALIG LYGVY |
| 4451 |
PF10206 (WRW) |
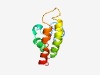 Estimated TM-score=0.52; easy target. |
>Mitochondrial F1F0-ATP synthase, subunit f (Length=97) FGDYPAEYNPKVHGPYDPARFYGKPDTPFGQVKLGELGSWFGRRNKSPQALAGAVSRAFWRWQHKYVQPKRTGVAPFFQL TVSAMIFFYCINYGKIT |
| 4452 |
PF10066 (DUF2304) |
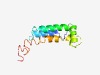 Estimated TM-score=0.52; hard target. |
>Uncharacterized conserved protein (DUF2304) (Length=105) FRITLIIVSVFTLLYMCKKVKQEKLQIEYSIFWTLFAIMLVLFSIFPQIADWLSELVGVYSTTNFIFLLIIFILLVKCFQ LTMDISKLDNKVKELVQKIAIDEKE |
| 4453 |
PF10032 (Pho88) |
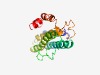 Estimated TM-score=0.52; very hard target. |
>Phosphate transport (Pho88) (Length=185) LNPQITNLLIILGMMQVSKRVPFDDPNVLNSVRAIYVTSNIIIALVYFYVQSKINAKNDMTTLKYVEPAPMGSTEEPKLV TTTIKDYDLAQLKTAWRGQLTGVAMMAFMHLYMKYTNPLLIQSIIPVKSLFESNLVKIHLYGQPASGDLKRPFKAAAGLM GAMQGGATQSDKKAVEAAEKAGRGG |
| 4454 |
PF09998 (DUF2239) |
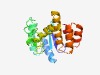 Estimated TM-score=0.52; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2239) (Length=185) SYTAFDGHRRLVSGPLATVALAVRQAAGDATPGTILIFDDATGRAIDLDLRGTADEIRARYAAAPADASAPASADEPRGR GRPKLGVVSREVTLLPRHWDWLATQPGGASVALRKLVEDARRTHAAADRRRDAQARAYHFMSAIAGDLPGFEEAARALYA NDLARLAELIAGWPDDVRDHALALA |
| 4455 |
PF09971 (DUF2206) |
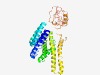 Estimated TM-score=0.52; hard target. |
>Predicted membrane protein (DUF2206) (Length=375) RAILSVIFAISLIVSHYGLSYIYLMSLVMVYIILIFVKSSFFKKIVRKFRPNVDPDSNNWNQTILTSTYVFLFIVFALTW YMYLSSSSIIYKIFNIGNHIASSIFTDFLNPEAAQGAAAITFKTTSLLHEVAKYLNLLFQFSIFIGILALLMGRLKNYRF KKEYVVFAMVNFIILIISIVVPYFASALNITRLYHITLFLLAPFSVIGGIILIRFFLTKISNKSWTKDRDIQLKIIYMGV VVLFLFYSGFIYEISGDNPTSFSLSNIDYPVFNDQEVQGLQWLSTNREHGYVYADGYRLSLLSRSFTVSESKDIPQNFSQ LKTDSYIFLGNYNTKTGTILIRNKNTTTKNDFYMDYSSLANTKSQIYTNGNVTIY |
| 4456 |
PF09946 (DUF2178) |
 Estimated TM-score=0.52; hard target. |
>Predicted membrane protein (DUF2178) (Length=104) MAIIVSMVALVGWFTAAGNALLVAISITAGVVLLYLCKSKIKEVVEDERDVKVSEKASKLAIEIFAATNILIGVVLIALR YRYSEYTNIGFTLAFSASSLMILY |
| 4457 |
PF09877 (DUF2104) |
 Estimated TM-score=0.52; hard target. |
>Predicted membrane protein (DUF2104) (Length=98) LYLLYIISFVVGSIAGLVLSYKKYSAPFVAQNVDIVALVLAIVGWVLALNSQLITAVPSYISITVGIFLIALVLGMRPGY GRYETFTGFVVAGLIWIL |
| 4458 |
PF09684 (Tail_P2_I) |
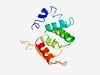 Estimated TM-score=0.52; hard target. |
>Phage tail protein (Tail_P2_I) (Length=129) TPPLERAIEAATDEVAEVPLRVLYNPDTCPAHLLHQLAWAWSVDRWDEAWPEAIKRSVIRSAFYVHAHKGTIGALRRVVE PLGYLIEVXEWWETVPQGVPGTFALQVGVLETGITEEMYQELTRLIDDA |
| 4459 |
PF09617 (Cas_GSU0053) |
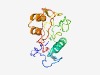 Estimated TM-score=0.52; very hard target. |
>CRISPR-associated protein GSU0053 (Cas_GSU0053) (Length=170) LRCVTEYQPAGGPGDKIFPPTYEGGKYATEERVIDGERVPCVLVDSVQSQANRMELALLEAWEREQLPLPVITVDFHDKD VLKPLRVTSLEAPHRIADAILRDSTLGGKPFRKCEPGNSLDLVDNGYATPLFELCPTALIFGMWDSTGPRGGLGAKFARA MVSEIIGLHA |
| 4460 |
PF09580 (Spore_YhcN_YlaJ) |
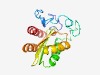 Estimated TM-score=0.52; hard target. |
>Sporulation lipoprotein YhcN/YlaJ (Spore_YhcN_YlaJ) (Length=160) NHQRYRMQGANKQQRQGMMQGSNQQQRQGMMQGQQHQRSQQGQGHQGQQGQHRGYYDGEDGHLAQRIANRVEDMDKVEDC RVIVNGDDIVIGVNTLDGDDQRVQQKIHRMTSDLDDHKDIYVSSDREQVDRIRGMDDRLRAGEPFDEVGATFNDMVQDLG |
| 4461 |
PF09550 (Phage_TAC_6) |
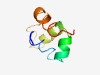 Estimated TM-score=0.52; hard target. |
>Phage tail assembly chaperone protein, TAC (Length=59) FPWEEAIGFGLGVLRLPPEQFWRMTPRELACAVAAVRGSAGAPMDRAGLDALMQAFPDR |
| 4462 |
PF09521 (RE_NgoPII) |
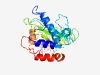 Estimated TM-score=0.52; hard target. |
>NgoPII restriction endonuclease (Length=264) TNILSAFRTIISNYQTNVQHVTNGSNRANNMGDGLEAFIKDAFANTFNETNEQTKLEVFSRIYSYSGNKNNPPDLILQNS DAIEIKKLESASTAIALNSSYPKAKLFSNSTMITTACRTCEAWSEKDMLYAIGNVPKTTNQLKSLWLVYGDCFCADKEVY ERIQQTISSGITSIPNVEFTQTNELGKVKKVDPLGITDLRIRGMWHIENPTKIFNYLYHYDETATFQLICLMKKEKYEAL PLEDRNAIENLSNLHVTLSDIRIK |
| 4463 |
PF09482 (OrgA_MxiK) |
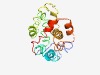 Estimated TM-score=0.52; very hard target. |
>Bacterial type III secretion apparatus protein (OrgA_MxiK) (Length=184) VMRVMYHPVSYLHASRFPSGWHREDQAAHPLLNHWILRHYHCPSDDGADRGDPFYRFLLDNWQRLGDAALLIGAHLLRNR LLSEGRFILLSAPVQSFLALPLPPLTPGALPQEQEEPATTLLQDPDPAAWGAYCLLSMLKRLSPAVYRRACLLFPVEQPL PQQATSLSPSSINLIKMALHHAQY |
| 4464 |
PF09139 (Tam41_Mmp37) |
 Estimated TM-score=0.52; very hard target. |
>Phosphatidate cytidylyltransferase, mitochondrial (Length=341) LRQLLWEFKAPIRYAFAYGSGVFSQGTASGKDKPMIDLIFGVTYTQHWHSLNLTEHRDHYSFLGRMGSGVVAKVQDGFGA GVYFNTYVEVNGMMIKYGVVNLDTFCKDLADWETLYLAGRMHKPVKVLRDDARVRLANQANLLSALRVALLLLPEEFTEQ QLYHTIAGISYLGDPRMNLFTENPHKVANIVSNQLPNFRRLYSPLVDTLPNLVFKNDPKTNIPKDTILLQDMDIVRRGNM VRRLPKAFRKRLYFQYQRKWQIPQLEYEKLVGTSEEREMMEGRQGGEFEQRIAADMENIRKEVKNVIKGTINWPSTSQSF KGFFTTGFVKTWRYLGEKIKK |
| 4465 |
PF08907 (DUF1853) |
 Estimated TM-score=0.52; hard target. |
>Domain of unknown function (DUF1853) (Length=286) RDLAWVILAPPMLSDTPWPQRHPLAGSDWVQAPERLQHWLRQLDRDSYGLLHWLAQARTRRLGLYYERLWQFAIQHAPGI ELIAANMPIRREGHTLGELDMLLRDRDGVHHLELAIKLYLGPQNGDGHDAAQWLGPGCHDRLDRKLAHLCQHQLPISARP ESREVLEALDIREFSAHLWLGGYLLYPWPGKATPPRGAHPQHLRGRWLHQKDWAEFAAQSPPGRWQPLPRHAWLAPAHYP AEQTWSAEQLDAWLSDLEPQAPAQLLVRLVKNAQGDWEEAERLFLV |
| 4466 |
PF08591 (RNR_inhib) |
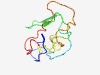 Estimated TM-score=0.52; very hard target. |
>Ribonucleotide reductase inhibitor (Length=101) NLLSVGMRVRKSVPEGYKTGTYSSFKLWSDQSTVSRPTASTKKRSTAASSQRELMPFCGINKIGGLSAQPVGSSDDEDYE RDIPGLDDIPGLSSSQESVES |
| 4467 |
PF08589 (DUF1770) |
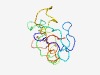 Estimated TM-score=0.52; very hard target. |
>Fungal protein of unknown function (DUF1770) (Length=107) AETVQTAHINRHPSPRHDLNPSTAADKAEPVSLREKPDGDDLDSLDDEAEDVQGEDGDDDDIPYSVLRPTPRRPAHQLPP LPDLRFEQSYLRSIAKADTWWKVAWIT |
| 4468 |
PF08499 (PDEase_I_N) |
 Estimated TM-score=0.52; hard target. |
>3'5'-cyclic nucleotide phosphodiesterase N-terminal (Length=61) QMLDTEDELREMRSDAVPSEVRDWLAATFTQQARAKGRRAEEKPKFRSIVHAVQAGIFVER |
| 4469 |
PF08488 (WAK) |
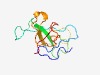 Estimated TM-score=0.52; very hard target. |
>Wall-associated kinase (Length=102) DKICGGYRCCQAKIQADRPQVIGVNLESSGGNTTQGGDCKVAFLTNETYSPGNVTEPEQFYTNGFTVIELGWYFDTSDSR LTKPVGCVNLTETRIYTSAPSC |
| 4470 |
PF08310 (LGFP) |
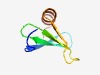 Estimated TM-score=0.52; hard target. |
>LGFP repeat (Length=50) LGEPVGEEQVVSDSIRYQQYDNARIYWSEGQGVHSVSGAVLEAYLNAGGH |
| 4471 |
PF08195 (TRI9) |
 Estimated TM-score=0.52; hard target. |
>TRI9 protein (Length=43) MLAAAKLIDSYEMDPDVSWLEVFAYSGVSAALCATIWVAAKAC |
| 4472 |
PF08039 (Mit_proteolip) |
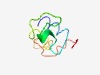 Estimated TM-score=0.52; very hard target. |
>Mitochondrial proteolipid (Length=58) MLQSIIKNVWIPMKPYYTQVYQEIWIGMGLMGFIVYKIRSADKRSKALKAPAPARGHH |
| 4473 |
PF07989 (Cnn_1N) |
 Estimated TM-score=0.52; easy target. |
>Centrosomin N-terminal motif 1 (Length=72) SLREQENVIDRIEKENFGLKLKIHFLEEALRKAGPGFSEAALKENTELKVDKVTMQRDLQKYKKHLTTAEKD |
| 4474 |
PF07954 (DUF1689) |
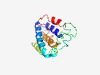 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1689) (Length=162) YNSALKFYEKDSALDSKDRLELSRTFVSIARAELWGGWAAFALVFGAPFGYRLYKTGAVRGVKVPRNFVLGLFAMFTTTQ ACGRYAFNSKLAQLQNDGNFTPRGSFGDDDDQSRPETNQQRQYEMMKLVGYGMAPKWGNYFYTTFHNPEKRLPNPKVKLQ EM |
| 4475 |
PF07935 (SSV1_ORF_D-335) |
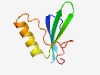 Estimated TM-score=0.52; hard target. |
>ORF D-335-like protein (Length=68) RYKYGDIIIRERKGRYYVYKLEMINGKVRETYVGPLIDVVESFIKLKEIGGVGISPTTDPPGFEPGTT |
| 4476 |
PF07889 (DUF1664) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1664) (Length=124) GGLSSYILPAAAVGAMGYCYMWWKGLSLSDVMFVTKRNMANAVQSMSKQLEQVSSALAATKRHLTQRLENLDGKMDEQVE VSKTIRNEVNDVKDDLSQIGFDIEAIQQMVAGLEGKIELLENKQ |
| 4477 |
PF07812 (TfuA) |
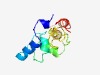 Estimated TM-score=0.52; hard target. |
>TfuA-like protein (Length=120) DGYFETVPTVWHKEILWAMSQGIHVYGAASIGALRAAELADFGMKGVGHIYRQYRTGELTDDDEVAVLHGPAEVDYVQVT EAMVNVRATLEHALQSGIVEPDVASALVEVAKSLFYKDRT |
| 4478 |
PF07800 (DUF1644) |
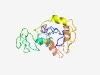 Estimated TM-score=0.52; easy target. |
>Protein of unknown function (DUF1644) (Length=159) AAKCSVCLEFPHNAVLLLCSSYNKGCRPYMCATSKRFSNCLEQYKKAYTKVTSADSMDNSSLTSGVEQPTEKMEVPELLC PLCRGQVKGWTVVEPVRKYLNREKRSCMQDKCSFVGTYKELKKHVRTKHPLARPRAVDPVREEKWKKLENERERNDVIS |
| 4479 |
PF07778 (CENP-I) |
 Estimated TM-score=0.52; easy target. |
>Mis6 (Length=516) SQKRKRVSRGRRSTQTDLAAWRLGERTSGSKPVNLKTGQKQREEDFTSDDDQAQDGERDSLRKALRYFEKGQISVPLRKN PTLQKHLETVENVAWRTGLPPESIDTLLNVALSGRFAEAVNTRLLKCMIPTTLILETSIVAAVSWLCVGKCSGSNKILFF RWLIAMFDFVDHKEQINSLYGFIFAFLQDENLCPYICHLLYLLTRRENVKPFRVRKLLDLQAKMGMQPHLQALLSLYKFF CPELVSISLSTNRKTYFKDSECLWKTALSAVRQRNQGPSPGSPKLILGTTSIQSRRRKWNTSLVVPLCSSQDSNPQSGNR ELKLVHFLKTDESFPPEQLHTFSQLLQNAHCLELPSQMGSVLKNSLLLHYLNCIKDESVLLRLHYWINNTLKEDCIWHPT GDHLDNEEFKEFLNLIFRTECFLQEGFSSCEKFLYKSLPSWDGFGFRSQILPLVTWIPLRNFSEIKALLCEPLAQLFFTS SIYFKCSILQTLRELLQNWLLSHATDMDEEPIINSP |
| 4480 |
PF07762 (DUF1618) |
 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF1618) (Length=136) WVDLWRNIIFCDVLAERPKLSYLNLPPALRPRPNMGAGDARSHRNIAVLGNTIKYVEMLPHPDRSSSKPASHRWEVAAWS IQKANRSWPKDWHTECNLDSTHIKVDSGTTAAALPTLSTLHVGLPTVSLQNDAIIY |
| 4481 |
PF07734 (FBA_1) |
 Estimated TM-score=0.52; easy target. |
>F-box associated (Length=156) GLSLKGNTYWYAIDYETEDCFLHYFDFTRERFENRLPLPQPPSSYSYAQVNISLSSVKEEKLAVLFSPRGRFVIDIWVTN KTEPDAVSWSYFFKVETTQHLSGFLFENFLIDEEKKVIVIFDIYGGANNAYTISGETGHFRKVDLRQSPYKRPYRL |
| 4482 |
PF07583 (PSCyt2) |
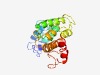 Estimated TM-score=0.52; very hard target. |
>Protein of unknown function (DUF1549) (Length=207) PIDAFILHRLEAQQLKPSPEADRTTLIRRLSLDLTGLPPSVKQVQEFLADTKPGAYERLVDRLLASPHYGERWARHWLDV ARYADSNGFTIDGPRSIWKYRDWVIEAINANMPFDQFVTEQLAGDLLPKPTTEQLIATGFHRNTLINQEGGTNPEQFRVE AVVDRVNTTGAAFLGLTVGCAQCHKHKYDPITQRDFYQLYAIFNSTA |
| 4483 |
PF07292 (NID) |
 Estimated TM-score=0.52; hard target. |
>Nmi/IFP 35 domain (NID) (Length=88) LLTLEDEEVAHKLTQMGKHTVNLDGKTADLKVVPFTVGMGVKFELHVTISGKKINVSEIPELPIPDEWMRDKLELNFYKS KHGGGEVE |
| 4484 |
PF07259 (ProSAAS) |
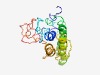 Estimated TM-score=0.52; very hard target. |
>ProSAAS precursor (Length=188) MARSPLLCGPQAGGVGLSVLLLLGLLRPVPALGTRPVKEPRGLGAPSPPMAETGASRRFRRAVPGGEAAGAVQELARALA HLLEAERQERARAEAQQAEDQQARILAQLLRLWGSPRPSDLSLGQDDDPDAPATQLARALLLTRLDPAALAAQLLPVPAS VLRTRPSAYDDRPTGQGAEDAGDWTPDV |
| 4485 |
PF07227 (PHD_Oberon) |
 Estimated TM-score=0.52; hard target. |
>PHD - plant homeodomain finger protein (Length=119) CKNSACRAVLSVADTFCKRCSCCICHLFDDNKDPSLWLECTSDSSHGESCGLSCHIECALQQGKVGVVNLGQLMQLDGSY CCASCGKVSGILGYWKKQLTIAKDARRVDSLCYRIYLSF |
| 4486 |
PF06887 (DUF1265) |
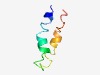 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1265) (Length=47) EELQKNYEDIMYVCNMLIIAEDAHFTNVQSCCIATLVFYHVRDFIRI |
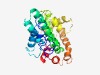
| 4487 |
PF06798 (PrkA) |
 Estimated TM-score=0.52; hard target. |
>PrkA serine protein kinase C-terminal domain (Length=250) IKKMKLYDGESVEGFKQKDVAELQAEAEGEGMSGVDPRYVINRISSALITKDTKCINALDILRALKEGLDQHPSITRDER EKLLNFIVLARKEYDEVAKKEVQRAFVYSYEESARTLFDNYLDNVEAYCNGVKVKDPISGEELDPDEKLMRSIEEQIGVT ENAKKSFREEILIRLSSYARKGKKFDYNSHERLREAIERKLFADLKDVVKITTSTKTPDPEQLKRINAVIDRLISNHGYC PICANELLKY |
| 4488 |
PF06796 (NapE) |
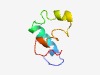 Estimated TM-score=0.52; hard target. |
>Periplasmic nitrate reductase protein NapE (Length=52) TDQKDQSTRGREWKAFLFITVFLFPILSVMVVGGYGFIVWMLQLFVFGPPGH |
| 4489 |
PF06763 (Minor_tail_Z) |
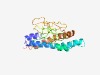 Estimated TM-score=0.52; very hard target. |
>Prophage minor tail protein Z (GPZ) (Length=192) MKGLENAIRNLNSLDRQMVPRASIWALNRVAQKAVSVATRKVARETVAGDNQVRGIPLKLVRQRVRLFKAGTDGKRSARI RINRGNLPAIKLGAAQVRMSKRRGKLLYRGSVLKIGPYLFRDAFIQQLANGRWHVMRRVNGKNRYPIDVVKIPLSGPLTQ AFESATQSLIDEEMPKQLGYALKQQLRLYLSR |
| 4490 |
PF06756 (S19) |
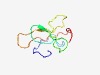 Estimated TM-score=0.52; very hard target. |
>Chorion protein S19 C-terminal (Length=71) GNHGGNYGGHQRGYGRPRWSVQPAGATLLYPGQNSYRRYASPPEYTKVVLPVRAAPPVAKLYLPENNYGSH |
| 4491 |
PF06749 (DUF1218) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1218) (Length=96) AAVLLLLAQVTVSAIGGCGCCCGNGKPRGIPSSKTNRVVGIVFAVASWIAAVIAVVLFVEGAAWNANVARDTAPVCYFLK DGVFAAAAVLALAATA |
| 4492 |
PF06741 (LsmAD) |
 Estimated TM-score=0.52; hard target. |
>LsmAD domain (Length=71) FGVKSTFNEELYTTKLDRGPQMRELEKEALRLAREIEGEDTQDLHLAEERGIHFHDKFDLDEETKYSSVTR |
| 4493 |
PF06738 (ThrE) |
 Estimated TM-score=0.52; very hard target. |
>Putative threonine/serine exporter (Length=238) VCLLAGKMMMQNGGETYRVEDTMMRIAAAYGMKDSQSFVTPTGILFALDGSSPTTRLIRISERSTDLNKVTLVNDISRRI TRGELTPDEAHRLLKQAEAAPAAYPPWLQIVLAALASGCFLLMFQGRMADFIPAVLAGGLGFAALIWFHRLVPIKLFAEL VASVVVGMLSVLLVQAGLGQELDRIIIGSVMPLVPGLLLTNAVRDLIAGHLVSGLSKGAEAFLTAFAIGTGVAFVLSV |
| 4494 |
PF06709 (DUF1196) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1196) (Length=51) MTVPLEAFVMCVFLMPTLPFKGVAKGIYAKQHSIKNYHIHKTKMLHVDIFR |
| 4495 |
PF06673 (L_lactis_ph-MCP) |
 Estimated TM-score=0.52; easy target. |
>Lactococcus lactis bacteriophage major capsid protein (Length=347) MGVNETQEMMKQAIEAGVKVRELETKVEELNKEREELKKEREANIPSEKPQDVERKFMRELGDKMTEMPEQGFLREFANA SALNVVNSLGSITSKYARKSGIYDGAMKARFQGLTLAEDGVDDTFIEGTFKAGTDKNKAQTASKRSLRPQMAEAYLQMDK ATVRGVNDSGALSEYVMSEMVNRVIQKVEHNMILGSADGSNGFYGLKTATDGWTKQIEYTDLFDGITDAVAECSISDAIT IVMSPQTFAELRKAKGTDGHSRFNELATKEQIAQSFGAVNLETRVWMPKDEVAVYNHDEYVLIGDLNVENYNDFDLRYNV EQWLSETLVGGSIRGKNRSAYLTKKAS |
| 4496 |
PF06667 (PspB) |
 Estimated TM-score=0.52; hard target. |
>Phage shock protein B (Length=71) AMLVMPLVIFMIFVAPIWLILHYRSKKQVNQGLSAEEQASLQSLAEQAEKMSDRIQTLEAILDSEAPEWRN |
| 4497 |
PF06634 (DUF1156) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1156) (Length=72) ALPLEAINKAAAREKSIRHGHPSTLHLWWARRPLAAARAVIFAQMVDDPSAHPDLFPTKEKQEKERNRLFKI |
| 4498 |
PF06633 (DUF1155) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1155) (Length=42) MSATLSTTLSFEPPLSLLAEPGTWFADTMDFRKKHSVRWYES |
| 4499 |
PF06592 (DUF1138) |
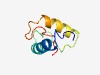 Estimated TM-score=0.52; easy target. |
>Protein of unknown function (DUF1138) (Length=66) GKYIIGSLVGSLGIAYVCDAIVSDKKIFGGTVCKTATDKEWFQATDAKFQAWPRVARPPVVMNPTS |
| 4500 |
PF06493 (DUF1096) |
 Estimated TM-score=0.52; hard target. |
>Protein of unknown function (DUF1096) (Length=51) GTTLREKRQSCGCAPAVQPSCSCQRTTYTQPQQYSCSCQNTAPVQKSCSCA |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218