

Go to page (83 pages in total): [First page] << < 38 39 40 41 42 43 44 45 46 47 48 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 4201 |
PF12278 (SDP_N) |
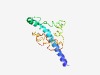 Estimated TM-score=0.53; very hard target. |
>Sex determination protein N terminal (Length=138) QRRRREWQRQQERERQHEKLKQQKILEYERNRAQALKYAEEITSHHSQSKSGSESPSHVRYRDRSTSTASKSGSLHEELD GSTSGTVPLFRGPQNAQIDTSELRRIKVDIHRNIPVKGPVTELERDILNPEDVIVKRR |
| 4202 |
PF12069 (DUF3549) |
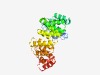 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF3549) (Length=338) TIHTLSELLKNSGCQYQVFDLGRRIQAIDTAQFEQVELGHQPYPYPLKRTAHLAIVYWNESNQPWIWFLKFTLDERGLLA QADVGQFIKYVIEAMGTRLNQEMSDEQQQKLANNPYTFKPADDKMAVFHSQIRAQLGLPCSQYYEHAQHYFTGDLGWENW QTVGLQGITDICARLGQEQNGVLLRKALNHLPPQPLYALLGALEHIALPEKLAARLQEHAEAQIAAHEPDLFLLSALVRA LSGAPAAIQNQVLDAVLNSPRLSHPEVLIGIAGRSWSMLADSGRAQRFLLRLAQSGNQSLFNQLFADLVMLPELRLVLLP LLHTSPSPELAQALMNLQ |
| 4203 |
PF12067 (Sox17_18_mid) |
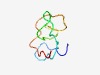 Estimated TM-score=0.53; very hard target. |
>Sox 17/18 central domain (Length=49) YREAQAMGAHFDSYSLPTPDTSPLDIVENDPVFFPSHSQEDCHMMSYSY |
| 4204 |
PF11914 (DUF3432) |
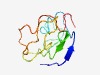 Estimated TM-score=0.53; very hard target. |
>Domain of unknown function (DUF3432) (Length=89) GSTVSVTSISIPSPVSSYPSPITSYPSPVSSYPSPVPSCYSSPVHTSYPSPSIATTYSTGTSMFQTQVATSFPSSVNIYN SPVTTPLPD |
| 4205 |
PF11888 (DUF3408) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF3408) (Length=127) KIDEDFMKELISQGVPAKRESQPDDAPQPGGETGTAQEGQQTETVRVEKTTPRKRKGGSGDYRETYFQKVELADRQPLYV SRTTHEKLMRIVTVIGGRKVTVSSYVENILLHHFEQYQDEINTLYES |
| 4206 |
PF11815 (DUF3336) |
 Estimated TM-score=0.53; hard target. |
>Domain of unknown function (DUF3336) (Length=137) SAWRGRRGQLRKQLRAAKTYEEWKQVALELDEYMRFNQWKTTDEDPFYDWRLVRKVRRSLKTLREKDDARGILGVLETCI RNNFAGIESPRLYSETFYGTKNLIEDYINEQEKALRYVLDTSHLSKEEKRRFFKTVN |
| 4207 |
PF11801 (Tom37_C) |
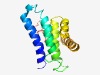 Estimated TM-score=0.53; hard target. |
>Tom37 C-terminal domain (Length=140) SKALESASQLAQSKTFKITHENKVKNKQELQQVKYNLQFDNRLQNCIKHWLEVREKLDESMVLPSDILFHANIYVQLNLP DGNRIRSKLEQTFGDEFINNTSNKMEEIAQSSDEDLGQRDPHFKEQGNVVMSLYNLACKY |
| 4208 |
PF11763 (DIPSY) |
 Estimated TM-score=0.53; very hard target. |
>Cell-wall adhesin ligand-binding C-terminal (Length=126) GDSINPSYVYKNTNVSDPDEGNMYTSTEGNTEAVNVFYYDPTVERILTCDCVRPVYTIYTDDPDSSFDIIKNNNGTFTFV ESSSGEIQTLHVSQYGALWITSPEYDGETGGMDDVGFRADDVILVA |
| 4209 |
PF11747 (RebB) |
 Estimated TM-score=0.53; hard target. |
>Killing trait (Length=68) ITDAVTQANVKVLGEAPAMAMGNLYQAMAHSTGILFQNAVSAQNQQNILAQAATTQGVMQIYSIDTAA |
| 4210 |
PF11742 (DUF3302) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF3302) (Length=76) LDYFALGLLVFVALVIFYGIIVIHDIPYEIAKHRNHPHQDAIHVAGWVSLFTLHVLWPFLWIWATLYRPDRGWGFN |
| 4211 |
PF11688 (DUF3285) |
 Estimated TM-score=0.53; easy target. |
>Protein of unknown function (DUF3285) (Length=44) EPKPSYVKLAMRNMVRKRGTSLKHFFLTTVGLLGVLIGLAYLTR |
| 4212 |
PF11374 (DUF3176) |
 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF3176) (Length=104) AIYLRINGSSLSSSGTALTFANILGKISSAALIIPTSEALGQLKWNWFNNSKAMWDFEIFDKASRGPWGAALLLFRTKGR SLAALGALLIVLLLAIDTFFQQII |
| 4213 |
PF11337 (DUF3139) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF3139) (Length=78) YKKWWITIGIVLILCVIGYVYWFAIPKHTANKAVDNYLAEQKIKSSQIETRVIKKDWKMGGYLTTIVFKDDPNLKYEY |
| 4214 |
PF11271 (PorA) |
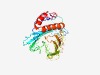 Estimated TM-score=0.53; hard target. |
>Porin PorA (Length=313) LACILVGLGAFLLAVAILIPTYTVGKLEKTPLDLEVTTIAEGSGSVLNSQALLAGKAEVNTNVPLVSQRYVTTEDPANAD VVTLQAGQTLRRTDKQGDTGLLSAIVDRNTVDRKTSMPTNDPVGTIQTQPNQPAEEVPRDGLQYKFPFNSEKQSYPYFDL NARATQDIDFVEETEINGLKVYHYNQTIEPVDLSKVVSSPTNKLTLPAEAWGVPGGTLPVTMTRWYTNVRDLWVEPETGV VVKGQEQLHQYYARNKDKPEIDVLKVELPFNEQTIEYQVQQAKDGMDKLSTFGRTVPIIAGILGVIALIAGLV |
| 4215 |
PF11247 (DUF2675) |
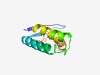 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2675) (Length=99) MAMTKKFKVSFDVTAKMSSDVQAILEKDMLHLCKQVGSGAIVPNGKQKEMIVQFLTHGMEGLMTFVVRTSFREAIKDMHE EYADKDSFKQSPATVREVF |
| 4216 |
PF11226 (DUF3022) |
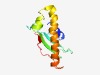 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF3022) (Length=105) IDMLAHVICDLFPEQTQFAERRDEAGRTSLVIHYVAMRFGATARRITIDVRFDPAALARYRAMPPRMHARSYAVLRAYVE ATLGSLEETYANGEAVPREVEIEMG |
| 4217 |
PF11207 (DUF2989) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2989) (Length=202) LFGCDNAANTATICKNNPELCDDLHRDSWCRYERADLIQKRYTLKNTPSPTGKQIYQHLINLENYSKCIELAAGVQHIMH PERTNDRVRAFGISAQNLAQLRETTKDNPDLYLSYYHWIRLNDQAALSRVIKAQQAGSIKDLEILASLASYYQKSDLDKA KALYLQLLDSADPDSFNPDWLLGLASIYHQQNDLEKTYLFSK |
| 4218 |
PF11203 (EccE) |
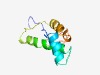 Estimated TM-score=0.53; hard target. |
>Putative type VII ESX secretion system translocon, EccE (Length=93) AVAIRTVWVVLRFDPGANVDAVFRRGGGRRGAERCAVIATARVRRALAAAHCASTILQAGQIHRISAEMLRQYAGAPMHE DWSGLTLIDSYNV |
| 4219 |
PF11169 (DUF2956) |
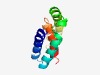 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2956) (Length=101) SPETQQEAMKIAKATQKPGQTKEQTKLIAQGIEKGIALYKKQQKAKARDRDKERKKQLKQKHVDTVSTNDVTETVEPMST SRNLLPWGLLIVSWVGFAAYI |
| 4220 |
PF11126 (Phage_DsbA) |
 Estimated TM-score=0.53; easy target. |
>Transcriptional regulator DsbA (Length=67) IKEASDHKLKISGYNELIKDIRIRAKDELGVDGKMFNRLLALYHKDNRDVFEAETEEVVELYDTVFS |
| 4221 |
PF11026 (DUF2721) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2721) (Length=117) TTPALLFPAISLLLLAYTNRFLAIATLIRNLHEKHENEPTKNLIEQIKNLRKRVNLIRNMQFLGVSSLLLCVVCMFVLFW GQIELGHWLFGISLILLMLSLSFSVIEIQISVKALNI |
| 4222 |
PF10953 (DUF2754) |
 Estimated TM-score=0.53; easy target. |
>Protein of unknown function (DUF2754) (Length=70) MQLSAKIRRDWHYYAVAIGLIFIMNGVIGLLGFEAKGWQTYAVGLVTWVICFWIAGFIIRRRPEESNAAE |
| 4223 |
PF10931 (DUF2735) |
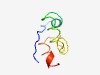 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF2735) (Length=52) TATIYQFPTRISRKAEERRSEAQRNADLASSDVCAAAIDGCWYHEEAVREET |
| 4224 |
PF10912 (DUF2700) |
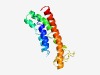 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2700) (Length=145) PARPLVCVLAFIGCARGVASLIFSGNWGERVGDIIFLLLNLMLLFGAAKNNECALKWSQRVVFIAVILAVIQFLIWPVMF ASYTASGLSDKNNTMAEIEDVLYKTEDEKKKMFARGMITGYALEIGTIVLVGAEVLKYVLVNRLW |
| 4225 |
PF10898 (DUF2716) |
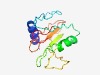 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF2716) (Length=130) LNDPEYRTLWDTVHERLAFRPHPRCFPGITEPPGAVTWNLGAGEADTLQEVLERGLRSVGEPGERLHWLDWNHIGYGFDP DLCGGEGQPEWPGAVYPDGDYYLYLPSDLRFGTFGHPWEATLCVWGTGLV |
| 4226 |
PF10896 (DUF2714) |
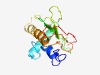 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2714) (Length=144) YNDARAEDSFIKYDKLIASVLLKNNISFNSEIYIKFVEKMTMAINKHYDLLFRDFVITFNVNGRFGNDLLVPMIANFESS NNEAINFREALTNDTKASQFLYDLNNEIARLLNQKSYVEIFPNIILYISPNTEHLKLLFSRETV |
| 4227 |
PF10888 (DUF2742) |
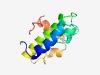 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2742) (Length=96) ASRAVSWWSVHEYVAPVLDAAGSWPMAGTPAWRQLDDADPRKWAAICDAARHWALRVETCQEAMAQASRDVSAAADWPGI AREIVRRRGVYIPRAG |
| 4228 |
PF10825 (DUF2752) |
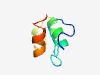 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2752) (Length=46) CPFKLLTGYDCPGCGSQRAVHALLHGDIQQAFRYNPLFVIAIPYVV |
| 4229 |
PF10803 (GerPB) |
 Estimated TM-score=0.53; very hard target. |
>Spore germination GerPB (Length=52) MNFYINQTIQINNLRVDSVSNSSVLQIGSAGMIRSLSNLYNTGGFTEPAPEA |
| 4230 |
PF10726 (DUF2518) |
 Estimated TM-score=0.53; very hard target. |
>Protein of function (DUF2518) (Length=146) MPTTADFLQYTQWSGIATIAFAVLTILAFILKWGIRFRLVGTTGFMIVLTAGLFSLSLVPLSRTVIPGAVKYTLVYDNGS NQAVIATTPEITPEELEATLRQAASNLYSYGRLGAGGNNQLTIRARTIIHPEPGISIPVYLGKVER |
| 4231 |
PF10721 (DUF2514) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF2514) (Length=160) WKVLAIVAVLISCWVWGFNHGSNKVEADWQQKWDRELKTQSQAVASTTTEYRTEEQRRQKAANQVANDARQEQAVAIADA VVADASGDRLRSEAGKLAASASCVPSDPGIADRGKNAARAAMVLSDLLSRADSRAGELARHADRLTVALQACNASYIGVT |
| 4232 |
PF10718 (Ycf34) |
 Estimated TM-score=0.53; very hard target. |
>Hypothetical chloroplast protein Ycf34 (Length=76) MCICINCYYVDRCNTYHAVEAQHQQPHLTENPSFEPESPSINVNIRPQEDYIEMEWDVVGCASFTEEKGKWAKLRP |
| 4233 |
PF10628 (CotE) |
 Estimated TM-score=0.53; very hard target. |
>Outer spore coat protein E (CotE) (Length=172) YREIITKAVVGKGRKFTQSSHTLAPKNRPTSILGCWVINHEYKAKKSGSNVEVDGRYDINIWYSYNNNTKTEVWTETVSY KDNIKLRYKDEDSIGDDYEVIVRVLQQPNCLECTISPNGNKTIVQVERELLAEVIGETKVCVAVNPKGCDDEDEFDLDVD DDEFEDLDPDFI |
| 4234 |
PF10612 (Spore-coat_CotZ) |
 Estimated TM-score=0.53; very hard target. |
>Spore coat protein Z (Length=167) MGCGKPTNPIGPIHSAGCVCDVVRAILDIQNQAVRDECSPCTANCFLEPLGGIVSPARSTADTRVFMLITKDGTPFKAFF SSPSADPCKCTSVFFRVEDMFDECCATLRVLEPLCTYDSTESTVDLLSRDGCCVNMKKICKVDDWNSTDSCITVDLSCFC AVQCIAD |
| 4235 |
PF10611 (DUF2469) |
 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF2469) (Length=100) MSAEDLENYETEMELQLYREYRDVVGLFSYVVETERRFYLANQVDLKVRSADGEVFFEVTMSDAWVWDVYRPARFVKSVR VVTFKDVNVEELAKSDLELP |
| 4236 |
PF10495 (PACT_coil_coil) |
 Estimated TM-score=0.53; hard target. |
>Pericentrin-AKAP-450 domain of centrosomal targeting protein (Length=88) RFQREASLRADAAYAKKFLQLQLDVANACNKAQLRELEDIRTNLLGNKKALSLPSHTTAATSNTKPTLKTFLVMARFIAR ARLSANNW |
| 4237 |
PF10269 (Tmemb_185A) |
 Estimated TM-score=0.53; hard target. |
>Transmembrane Fragile-X-F protein (Length=237) LKLDHHISCSWWIIFLPLWMFHGIVARGRFSLPAPSAPRNRNWAPCHAVVAMPLLIAFELLLCIYLESLDVRGSAAVDLK IVFLPLLTFEIIILIDNFRMCRALMPGDEENMSDEAIWETLPHFWVAISMVFFIAATVFTLLKLSGDVGALGWWDLFINF VIAECFAFLVCTKCGLVVSSEENQRQGRMCSLQDIGGHFMKVPIIVFQVLLVMHLEGTPACAVYIPLPVLFSPIFLL |
| 4238 |
PF10051 (DUF2286) |
 Estimated TM-score=0.53; very hard target. |
>Uncharacterized protein conserved in archaea (DUF2286) (Length=138) KVLVIRAEEGKIVSSDILDGDFAKIAKDVTARALTEWNPEESDLTALRTKLELRYPKPLPPEIADKILELSLKYSLEGND VIVNLPVLVVSFDNRWIDETTYLDKKIYVIALYLDDNAKKQLEEYAAEATKAPKSVEA |
| 4239 |
PF10041 (DUF2277) |
 Estimated TM-score=0.53; hard target. |
>Uncharacterized conserved protein (DUF2277) (Length=78) MCRNIRTLHNFEPPATDEEVRAAALQYVRKIAGTRDPSQANREAFDQAVEEITAATQRLLDGLTTSAPPKDREEEAAK |
| 4240 |
PF09977 (Tad_C) |
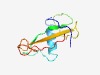 Estimated TM-score=0.53; very hard target. |
>Putative Tad-like Flp pilus-assembly (Length=88) AGAQRMDDQCAQPTATATANASANGFNSQASGYSLSIVCGRWDLAAYPTPPYFGAAASQLNAVQVTVTKPVPYFFLGPMR TVSATSIA |
| 4241 |
PF09932 (DUF2164) |
 Estimated TM-score=0.53; hard target. |
>Uncharacterized conserved protein (DUF2164) (Length=74) IEFSKEEKDILVKNIQAYFREELEQDIGNFDAQFLLDFFSEKIGPYFYNRGLYDAQAVLEKRLDSIAEAIYEIE |
| 4242 |
PF09919 (DUF2149) |
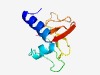 Estimated TM-score=0.53; very hard target. |
>Uncharacterized conserved protein (DUF2149) (Length=92) MSSVANLFDVAMVFAVALLVAMVMFYNMPELVSPNENITIVKNPGEPDMQIIIKEGQEIEVLNMTDKLSGGQGVKMGVAY RLKDGRVIYVPE |
| 4243 |
PF09893 (DUF2120) |
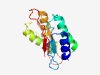 Estimated TM-score=0.53; very hard target. |
>Uncharacterized protein conserved in archaea (DUF2120) (Length=136) EIAGHIMGKLDAFEGSKAAMDTTELLIVRGRSRLRIPAEELESTIDQILKELGARKLDMFSDEAADIITIMDEQIRAQVP IQEETDIYGIYRLKESFEHMNCHADYGMGVVDDIAIFIILWKDKSGMGPLFVELVV |
| 4244 |
PF09886 (DUF2113) |
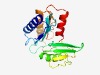 Estimated TM-score=0.53; very hard target. |
>Uncharacterized protein conserved in archaea (DUF2113) (Length=182) MKVECYDKEGEKVYDMIVRQILQDVQVIRSINDLRVFVDPREPLFIIVIKFEKAASPIMLEDLAEYEYDKQANEVFIRIK DETYLPELLKKLWELEGRNKIHQPSRYEVIIDDPKVDLNKLVVHDPQEDMKKKIYDAMFRIIPEGFRVIKHKSEDDIIAM TCSDEIIKDEWLEKTDKIIEEV |
| 4245 |
PF09831 (DUF2058) |
 Estimated TM-score=0.53; very hard target. |
>Uncharacterized protein conserved in bacteria (DUF2058) (Length=176) SLQEQLLKAGLVDQKKAKQLKQEKRKQAKQTPKGHQVEDETKLRAQQAREEKAQRDREMNRLRQEEADRKALKAQVKQLI EMNRINRNKGEVGYQFADEGKVKKIYVTQELQDGLASGRFAIARMGDAYEVISRKVAEKIRERAPEFVVVLNDNKNAEPD EDDPYAAYQIPDDLMW |
| 4246 |
PF09743 (E3_UFM1_ligase) |
 Estimated TM-score=0.53; easy target. |
>E3 UFM1-protein ligase 1 (Length=261) QVLLSERNCIEIVTKLIAEKQLEVVHTLDGKEYVTPAQISREIRDELHVCGGRVNIVDLQQVINVDLLHIENRANDIVKS EKTIQLVLGQLINQSYLDQLAEEINDKLQETGQVTISELCKAYDLPGDFLIQALSRRLGRIIHGQLDQENRGVIFTEAFV SRHRARIRGLFTAITRPTPVSNLITRYGFQEHLLYSVLEELVNSGRLKGTVVGGRQDKAVFVPDIYARTQSNWVDSFFKQ NGYLEFDALYRLGIPDPAGYI |
| 4247 |
PF09721 (Exosortase_EpsH) |
 Estimated TM-score=0.53; hard target. |
>Transmembrane exosortase (Exosortase_EpsH) (Length=254) IGIVLLYLPLLPELVGDWWNDPNYSHGFLIPLVSGYLCWERRDTLRRLPLGSDRGGLLLLVGGLFLFLAGVVAAEEFTQR FSLIVVLAGVIRYNLGRAVFREVRFPLAYLAFMIPLPYLLYDQIAFPLKLFAARCATFSLHLLGIPVFREGNIIELAAMR LEVADACSGIRSLISLFALSVIYAYLSQPETWKRIVLVVATIPIAIVANGARVSGTGIIAEYAGIEAAEGFFHSFSGWLI FVVATLLLVGLGKL |
| 4248 |
PF09436 (DUF2016) |
 Estimated TM-score=0.53; very hard target. |
>Domain of unknown function (DUF2016) (Length=72) ADAALQQSFPSVMVPRFGALAPMERSGERLLIAANGVFLEIVRPWLSVVRHLGAFQHRTAIPYGEAAETTDL |
| 4249 |
PF09430 (DUF2012) |
 Estimated TM-score=0.53; easy target. |
>Protein of unknown function (DUF2012) (Length=111) QQNWAADSRVLLNHGEYIGFVREDGSFIIDNVASGSYIVQVENVHFVFEPIRVDITAKGKIRARKLSVLQPNVVNQLPYP LKLSAREPTKYFRKREEWRITDILMNPMVLM |
| 4250 |
PF09362 (DUF1996) |
 Estimated TM-score=0.53; very hard target. |
>Domain of unknown function (DUF1996) (Length=231) DPIVSPGQISGHVHTIMGGNGFGFDMTYQQARNSQCSSCPITKDMSNYWVPTLYYHAQNGSFISVPQVGGATVYYEQRGG PNNDQLLAFPEGFRMLAGNPFLHTYNDTFEQQAVSYACLNYNGPATAETHAFPNNNCPDGLRAQVYFPSCWDGVNLDSPD HKSHVAYPESVAYNDGPCPASHPKHLISLFYEVIWDVNQFANDWYGDEQPFVWSSGDATGYGYHGDFVNGW |
| 4251 |
PF09331 (DUF1985) |
 Estimated TM-score=0.53; hard target. |
>Domain of unknown function (DUF1985) (Length=136) ISRQLKVAKKHEVWFIFAGKPIRFSLREFAIVTGLSCGRFRKRSKKRSNRNISEKPYWGELFGTLKEIPVSSVIRMLKKK TVRGDLRLKYAYLALLSSVILPTTHTPKMSQDYAEMIKDLDTFFAYPWGRVSFDML |
| 4252 |
PF09133 (SANTA) |
 Estimated TM-score=0.53; very hard target. |
>SANTA (SANT Associated) (Length=90) LLFEWWLMKAERDYNGRRLTVGGFTKRGEAVGIFRSAPIIKRHDHCTLETADGINIKIQGTINKTRTIDNGFPLEACNAF LFGFPTAWDK |
| 4253 |
PF08897 (DUF1841) |
 Estimated TM-score=0.53; hard target. |
>Domain of unknown function (DUF1841) (Length=135) SREEVRRFFCDAWQKQIAGGVLTPLEAIAVDWIGEHPEYHALLRDTEGALAQDYTPEQGQTNPFLHLSMHLSISEQVSVD QPRGIRQAYEALARRLDSPHEAQHQVMECLGEMLWQAQRTGQPPDGDHYVDCVKR |
| 4254 |
PF08620 (RPAP1_C) |
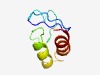 Estimated TM-score=0.53; hard target. |
>RPAP1-like, C-terminal (Length=66) QARFDFNGNLLSKTDSIPEYIGLHHHGNEPSAPGYTLEELFVLARSTFQQQRVFALQTLARIIKQA |
| 4255 |
PF08508 (DUF1746) |
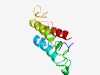 Estimated TM-score=0.53; hard target. |
>Fungal domain of unknown function (DUF1746) (Length=116) NLDALVYAEICALYYMDCSFFRLLMRVLVQSFFLSPKSDEFVLLMPTHRPHVVAVLAPNLLCMLLHLLTSLPTAGESTRG YLHGGVIIDFVGEKAPTSKFKLLLLDMLVLALQCTM |
| 4256 |
PF08285 (DPM3) |
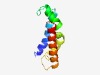 Estimated TM-score=0.53; hard target. |
>Dolichol-phosphate mannosyltransferase subunit 3 (DPM3) (Length=92) MSKGTRFVTFAVPAVIAYFLLLLHVLPTPFLAQETADQILPVIPWWLLVAFGAYSLSSLGLGLLRFHDTPEAYQSLLGEI QQAKNELRDQGV |
| 4257 |
PF08268 (FBA_3) |
 Estimated TM-score=0.53; easy target. |
>F-box associated domain (Length=124) ICINGILYYLALDSDGRYFMVVCFDVRSEKFKLVDIECRFDQLVNYKGKLCGIDLKNAYYGGFPLELRMWVLEDVQKREW SKYVYTLRDDNKVVKVNYNLSVSGMTATGDIVLLLNNASNPYYV |
| 4258 |
PF08165 (FerA) |
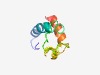 Estimated TM-score=0.53; hard target. |
>FerA (NUC095) domain (Length=65) LEANAPTAETASLLIALLDQLINDCRRPLPKIDSSTPGVNELDLKIQDTRILEKLAIMEEATKLR |
| 4259 |
PF08151 (FerI) |
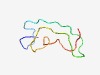 Estimated TM-score=0.53; very hard target. |
>FerI (NUC094) domain (Length=51) SDPEDFSAGAKGYLKVSACVLGPGDEAPLEKKEVSEDKEDIEANLLRPTGV |
| 4260 |
PF08140 (Cuticle_1) |
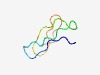 Estimated TM-score=0.53; hard target. |
>Crustacean cuticle protein repeat (Length=41) GPSGIISPDGVNTQFSHDFANDIVLVGPAGIVTKSGANRQL |
| 4261 |
PF07790 (Pilin_N) |
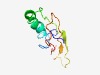 Estimated TM-score=0.53; easy target. |
>Archaeal Type IV pilin, N-terminal (Length=80) DAVSPVIGVILMVAITVILAAVIASFVLGLGDQAQQATPQASFSWDYEQDDPSHTTGNVTITHDGGDSIEAQQLYIRGDF |
| 4262 |
PF07335 (Glyco_hydro_75) |
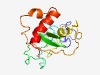 Estimated TM-score=0.53; very hard target. |
>Fungal chitosanase of glycosyl hydrolase group 75 (Length=161) MDIDCDGANNSAGGCANDPSGQGQTSFKDTVKTYGIPDLDANIHPYVVFGNEGASPSFSPQKNGMKPLSIMAVVCNNQLF YGVWGDTNGFTSTGEASLAMGKLCFPNEGLSGDNGHDPKDVLYIGFTGSDTVPGKSGANWKAKKTTEFEDSIKALGDKLV A |
| 4263 |
PF07226 (DUF1422) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1422) (Length=112) KPGRENGTLLLALIAGLSINGSFAALFSSVVPFSIFPLIALVLAVYCLHQRYLNRAMPDGMPKLAAACFLLGVLLYSAIV RAEYPQIGSNFLPSVLCVALVLWIGMKLKARK |
| 4264 |
PF07131 (DUF1382) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1382) (Length=60) MHKASPVELRTSIEMAHSLAQIGIRFVPIPVETDEEFHTLAASLSQKLEMMVAKAEADER |
| 4265 |
PF07127 (Nodulin_late) |
 Estimated TM-score=0.53; hard target. |
>Late nodulin protein (Length=52) MAQILKFVYALILFLSLFFILINGDRIPCATDADCPPKILPIIHKCINNFCK |
| 4266 |
PF07114 (TMEM126) |
 Estimated TM-score=0.53; hard target. |
>Transmembrane protein 126 (Length=177) ISNIISRKVKQLPESERNLLEYGSAYVGLNAGFSGLIANSLFRRILHVTQARLASSLPMAVMPFLTATLTYVNFLSYPLS TGDLNCETCAMTRGALVGFGMGGVYPILLAIPMNGGLAARYESSPLPRRGNILNYWITVSKPVCRKMVFPILLQTVFAAY LGSRQYRLLIKALQLPE |
| 4267 |
PF06892 (Phage_CP76) |
 Estimated TM-score=0.53; hard target. |
>Phage regulatory protein CII (CP76) (Length=157) FQISKHPHYDEACRAFAQRHNMAKLAERAGMNVQTLRNKLNPEQPHQFTPPELWLLTDLTEDSTLVDGFLAQIHCLPCVP VNELAKEKLQSYVMRAMSELGELASGAVSNERMTSDRKHNMVESVNAGIRMLSLSALALQARLQANPAMSSVVDTMT |
| 4268 |
PF06880 (DUF1262) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1262) (Length=104) EGPSSGYLVRKDEGSDGGGSACCWGLCEGTRVRDLPFPQNRILTVQYDEEHARENTTYSAVVFFIPVLDKPLSSNHYYVV SGKGKDKGKIYTCSKEEDMSTCCF |
| 4269 |
PF06712 (DUF1199) |
 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF1199) (Length=52) MLHRNSGFVPASIYRNNNIYKSNQCSGSMETSTISSPSRRIRNNFLGLLGTR |
| 4270 |
PF06674 (DUF1176) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1176) (Length=316) SFSHKDWELACDNTLTCRAAGYSTEEGAGGSVLLTRQAGENTVVSGEVVLADAMDEDPKPVAELSLWIDDTALGALQKAD NDEWRLTEAQTQAVIRAVKGNGTVEFRGGEEPFELSDNGAFATLLKMDDAQGRTGTPGALVRKGDKPESSVMVAVAAPVI QQVKVIQAQPRPLTAQELSSVKPQLIDSLTDDDSCDNLQPSDDQPEEGAEPITLTPVDKSHVLISALCWRGAYNEGYGYW LMDKRLQQKPELITDSGSDYHDGVISLAQKGRGIGDCWSTAAWVWDGKDFQQSSEATTGMCRAIRAGGPWQLPTLV |
| 4271 |
PF06614 (Neuromodulin) |
 Estimated TM-score=0.53; very hard target. |
>Neuromodulin (Length=172) KKKDEAPVADGVEKKGEGPTTTEAAPATGSKPDETGKAGETPSEEKKGEGDAATEQAAPQAPASSEEKAGSAETESATKA STDNSPSSKAEDAPAKEEPKQADVPAAVTAAAATTPAAEDAAAKATAQPPTETGESSQAEEKIEAVDETKPKESARQDEG KEEEPEADQEHA |
| 4272 |
PF06396 (AGTRAP) |
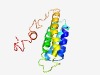 Estimated TM-score=0.53; hard target. |
>Angiotensin II, type I receptor-associated protein (AGTRAP) (Length=145) KMILLVHWLLTTWGCLVFLGPYAWANFTILALGVWAVAQRDSIDAISMFLIGLAVTIFLDIVHISIFYPRQNLTDTGRFG AGMAILSLLLKPFSCCLVYHMYRERGGELQLHAGFLGPSQDRSAYQTIDSPEAPSDPFSGPEGKN |
| 4273 |
PF06341 (DUF1056) |
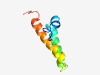 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1056) (Length=63) MIFRRIIGYLWQLSDVLLFISAMIVLDYTAFRINATLGWFVISLILFVLGWLVEVISERKRGD |
| 4274 |
PF06260 (DUF1024) |
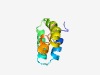 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF1024) (Length=82) MNNREQIEQSVISASAYNGNDTEGLLKEIEDVYKKAQAFDEILEGMTNAIQHSVKEGIELDEAIGIMVSQVIYEYKEELE NE |
| 4275 |
PF06209 (COBRA1) |
 Estimated TM-score=0.53; easy target. |
>Cofactor of BRCA1 (COBRA1) (Length=465) MGLILEEPWVPEKKLKLVMADKELYRACAVEVKRQIWQDNQALFGDEVSPLLKQYILEKENALFSTELSVLHNFFSPSPK TRRQGEVVQKLTQMVGKNVKLYDMVLQFLRTLFLRTRNVHYCTLRAELLMSLHDLDVGDICTVDPCHKFTWCLDACIRER FVDSKRARELQGFLDGVKKGQEQVLGDLSMILCDPFAINTLSLSTIRHLQELVSQETLPRDSPDLLLLLRLLALGQGAWD LIDSQVFKEPKMEVELITKFLPMLMSFVVDDYTFNVDQKLPSEEKAPVTYPNTLPESFTKFLQEQRMACEVGLYYVLHIT KQRNKNALLRLLPGLVETFGDLAFSDIFLHLLTGSLALLADEFALEDFCSSLFEGFFLTASPRKENVHRHVLRLLLHLHA RVAPSKLEALQKALEPTGQSGEAVKELYSQLGEKLEQLDHRKPSPAQAAETPTLELPLPSMPAAA |
| 4276 |
PF06191 (DUF995) |
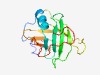 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF995) (Length=138) IIAGASFAEAAAARAERKTTPMTHAELYQLYGQKSWIWKEGAGYFSVQARRFIAWSKENGKPSYGVGRWFITGPGKLCFR AEWHARDGSSPATTCFSHAKRGGVIYQRREPDGAWYVFKNAQVRPDDEFAKLRRGNYV |
| 4277 |
PF06146 (PsiE) |
 Estimated TM-score=0.53; hard target. |
>Phosphate-starvation-inducible E (Length=68) FGAFLNILIALELLENITAYLKKHVIHLELVVVTALIAVSRKIIIFDFKEYSSWHLMSLGVAILCLAA |
| 4278 |
PF06025 (DUF913) |
 Estimated TM-score=0.53; easy target. |
>Domain of Unknown Function (DUF913) (Length=385) TRAVRVVDLITNLDMAAFQSHSGLSIFIYRLEHEVDLCRKECPFVIKPKIQRPSTAHEGEEMETDTDGVQCIPQRAALLK SMLNFLKKAIQDPAFSDGIRHVMDGSLPTSLKHIISNAEYYGPSLFLLATEVVTVFVFQEPSLLSSLQDNGLTDVMLHAL LIKDVPATREVLGSLPNVFSALCLNARGLQSFVQCQPFERLFKVLLSPDYLPAMRRRRSSDPLGDTASNLGSAVDELMRH QPTLKTDATTAIIKLLEEICNLGRDPKYICQKPSIQKADGTSTAPPQRANHAAEEASSEDEEEEEVQAMQSFNPPAQSDT DSNQQVIRTEERVPIPLMDYILNVMKFVESILSNNTTDDHCQEFVNQKGLLPLVTILGLPNLPVD |
| 4279 |
PF05957 (DUF883) |
 Estimated TM-score=0.53; hard target. |
>Bacterial protein of unknown function (DUF883) (Length=93) DLLADFQTLVNDTEKLLQDTASLAGEQADELRVQIHESLKRARDTLRDTEQNLRERGQAAVQATEDYVQEHPWQSVGLAA GIGFLLGLLAARR |
| 4280 |
PF05915 (DUF872) |
 Estimated TM-score=0.53; hard target. |
>Eukaryotic protein of unknown function (DUF872) (Length=117) INTRDSSRTSIRSSQWSFSTISNTTQRSYNACCSWTQHPLIQKNCRVVLASFLLLLLGMVLILVGVGLEVAPSPGVSSAI FFVPGFLLLIPGVYHVVFIYCAVKGHRGFQFFYLPYF |
| 4281 |
PF05725 (FNIP) |
 Estimated TM-score=0.53; trivial target. |
>FNIP Repeat (Length=44) FSKVLPPTTLPNTITTLTFGNDFNQVVRPGTLPNSLTTLKFGYD |
| 4282 |
PF05422 (SIN1) |
 Estimated TM-score=0.53; very hard target. |
>Stress-activated map kinase interacting protein 1 (SIN1) (Length=147) FISTDDTGMCETVMLSDDMPKHYLRKFGNSGAGGDHYHWRRAHKTPPTGGGTTPERNTRHPDAPLQEVDFICYPGLDLSD DEEDMSTHSFDIQMYPEVGAHRFRSNTAQKLEKLDIAKRRAARIKSVNYQEEVQPPESDDFFKRKEL |
| 4283 |
PF05396 (Phage_T7_Capsid) |
 Estimated TM-score=0.53; hard target. |
>Phage T7 capsid assembly protein (Length=122) EELSAESYAKLAEIGYTKAFIDSYIRGQEALVEQYVNSVIEYAGGRERFDALYNHLETHNPEAAQSLDNALTNRDLATVK AIINLAGESRAKAFGRKPTRSVTNRAIPAKPQATKHEGFADR |
| 4284 |
PF05338 (DUF717) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF717) (Length=55) CISEEDFKDCQAFFAKPLPILVAEISKSLANIDFVDTASQQVDQLGIFLDLIGTE |
| 4285 |
PF05155 (Phage_X) |
 Estimated TM-score=0.53; hard target. |
>Phage X family (Length=87) LLNNGAVDTQRKANTTMNYFTLWQNGVDMREILSSSQYYEHKARLKKIGIDIGQQFDVSRMCPTLRRSEEIEVQSLQIPS WYQLPKV |
| 4286 |
PF05119 (Terminase_4) |
 Estimated TM-score=0.53; hard target. |
>Phage terminase, small subunit (Length=100) AKKEFNRIAKELQEIGLLTNVDIDMLAAYCDAYTEYQKCTKIIEEEGLMVEYTNKAAETNKVPHPLLTKKKQLFEQMKSI AGEFGLTPSARAKLAIPKQE |
| 4287 |
PF05106 (Phage_holin_3_1) |
 Estimated TM-score=0.53; hard target. |
>Phage holin family (Lysis protein S) (Length=98) ENPDVWAELLNGLQNSWPQISGSVLAIAICYGRLIYDGVERKNRWVEALLCGALSWSVSSGLEMFGVPSSFAPAIGGVIG FIGVEKIREFAIRAINKR |
| 4288 |
PF04947 (Pox_VLTF3) |
 Estimated TM-score=0.53; hard target. |
>Poxvirus Late Transcription Factor VLTF3 like (Length=167) HTTYMYKRQNHFKTWLKRTQGKETTTVDKEVVDSVRLELKKQRIDDIKTISHTKVREILKKLRLNKHYNNCVQITAMITG ITPPQMTSEQETALLQMFDAIQTPFQKIIQSEPRQNMLSYSFLIHKFLELMSWDEYLPYFPLLRSVDKIQYQDWIWRKLC AEVGFEY |
| 4289 |
PF04931 (DNA_pol_phi) |
 Estimated TM-score=0.53; easy target. |
>DNA polymerase phi (Length=785) ALQRLFRGLCSSRKAARVGFSIALTEILSQIFSPSQTLGSEWSISKALDIWESQSDASGGASGQEERDHHFGRLFGAEAI IKSSVLFHAKASFQDWTRVLTVIFDLAKKKPWLREECGWIMYRCISDLAAQKLDTKYVEAILDQLYANDLARTPEGVAIW LAAKDSFSKAKFPQNVWKYEDPLDSRDKSTLAKIMKESSDVEAQSEGGGAKASGVWNSKLHFAWDAVLSRIFVSPQKKEK SKDKASKSSRTSFVDFWVEVVDNGLFAAASSEERKYWGFLLFIKTLNEAPQSFASLVFTKNLIRCLMNQLAVEDRYLHRM AVKAAKTIQGRVSKEPEFAAAAINGLMGPTGTVNFDQITKTKTVEKIVAEADEESLKQIVVLFERLIAAPGTDDIKAAAS SRQNLANLLVSIVRSRASVKAEEGDFHDAIEAILSIFVRFAYFVDKGENTQPPLTQATQELFRNRINSCLNSLIAARKDP ASLPYAVVRKIRDAAESAEYGKFIIKMDETVREAIDGAFKSLKKLASKEKHSEKEISGVQAFKLLYSMTILQVYNGDGDA VSMLEELQFCYDKFMGKKSKDDTSDASDALVEILLSFASKPSQLFRRMSEQVFGAFADQVTADGLQSLISILEVKETLAG QQEMFEQQDDDEDEDMADVDDEEEEDSDVEVVDAKDEGSSDEGSDEGSGSEGSDEEDEDDDELAVFDAKLAEALGTHRAD QDLKEDSEESDADMDDEEMEALDDQIVKVFRARNKATSKKKDKKDAKENMVNFKNRVLDLLEIYV |
| 4290 |
PF04776 (protein_MS5) |
 Estimated TM-score=0.53; very hard target. |
>Protein MS5 (Length=120) LPEWPGENAFDDKKHYYVVKKSELRENDWIRVYLELAFLTAHLSIRSLDLSKVVITKVAVYTSDEDMALPNEKLNARNAI FYIKYKYCPNENEDHGCKPTRDGVAIVRRNMDKISGDITL |
| 4291 |
PF04767 (Pox_F17) |
 Estimated TM-score=0.53; hard target. |
>DNA-binding 11 kDa phosphoprotein (Length=91) KHTPFYIDTKEGKYLVLKAIKVCDIRKVECNSGSASCVLKVDKPTPSCERPMSPCERGRSPTRRNDVVPFMRTNMLEDIQ NSNRNVVSRIL |
| 4292 |
PF04573 (SPC22) |
 Estimated TM-score=0.53; very hard target. |
>Signal peptidase subunit (Length=168) LTRANAILAYTLSVSACLTFCCFLSTVFIDYRANATLNTVKVVVKNVPDYSASREKNDLGFLTFDLQTDLSPLFNWNVKQ LFLYLTAEYRTETNEINQVVLWDKIVLRGDNAVLDFKNMNTKYYFWDDGNGLRGNKNVTLTLSWNIIPNAGFLPSVSAHG SHTFSFPA |
| 4293 |
PF04556 (DpnII) |
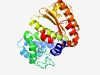 Estimated TM-score=0.53; hard target. |
>DpnII restriction endonuclease (Length=279) NFNEWLSKFRSSIANYGYYIDFDKVYRNVNDIKVELNILNSLIGSTDIENDFKNIITKYPETLKCIPILLAVRANEIYAI DGDGEFSYNFKKAKYSPEQYAVFMRKTGLFELLEKHIIHTLVDYVTGVETGLDSNGRKNRGGHLMEDLVESYIVKAGFEK NKTYFKEMYIHEITEKWGIDLSAISNSGKMEKRFDFVIKTDKAIYGIETNFYASGGSKLNETARSYKQIAQEVATINGFE FVWFTDGSGWTSARHNLEETFDVMEHIYSIDDLEKGILS |
| 4294 |
PF04510 (DUF577) |
 Estimated TM-score=0.53; easy target. |
>Family of unknown function (DUF577) (Length=171) LHEIKPLVIECLTMQETKESDMKFLGRIVSLITYNVVICDNGGWYELSDCILWLAENEPVKAFHVFIDLPSLYQSFIDKF MKVIVEKAEKVLFNPEKDRVDDWSLALETVVKMGIQTLDTEMRVDLIRILLNILVNSVKMLVQKGMEQFLVQGLKHLEKF VLRDKNLYNYT |
| 4295 |
PF04467 (DUF483) |
 Estimated TM-score=0.53; hard target. |
>Protein of unknown function (DUF483) (Length=123) LKFQIELVEKYKPKVRPAIDPMVSTELGIYRRLDDFEIGKLLDYPECCIKSFVEDVRVAIDREHLKEVEEMKEELKNKGI YAIVLPSGFIPCSLKCEEAIKRGFIGYLTKEEFDKILELEKEL |
| 4296 |
PF04385 (FAINT) |
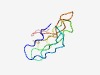 Estimated TM-score=0.53; hard target. |
>Domain of unknown function, DUF529 (Length=73) LDLNTSKSTNEFDYTDQNGVVTFKPNSGYVFSKVSQGTKMVWESTNNVHVTLVRTKSEDNKKYLIVLLTNDMF |
| 4297 |
PF04334 (DUF478) |
 Estimated TM-score=0.53; very hard target. |
>Protein of unknown function (DUF478) (Length=68) MGVQKYTYTNPVLFWGIFEVRGTSKGVGVILTRFFWVFSGRERVLKRSSYIVILNLWLLRGFLEASDY |
| 4298 |
PF04295 (GD_AH_C) |
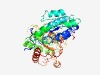 Estimated TM-score=0.53; very hard target. |
>D-galactarate dehydratase / Altronate hydrolase, C terminus (Length=389) KTFQGYRRKDGRVGVRNEIWIIPTVSCVNRAAQLIARQAMEELASAKNIDGIYEFTHPYGCSQLGQDQLNTQKILADLVN HPNAGGVLVLGLGCENNNIAEFKKVLGEYDPQRVRFLVAQEVEDEIQEGLKLIKELVEYASQFKREEVPVSELTVGLKCG GSDGFSGITANPLVGSFSDKLISCGGSTVLTEVPEMFGAETILMNRAKNQEIFEKTVKLINDFKEYFMGYNQPIYENPSP GNKKGGITTLEEKSLGCTQKGGTSNVVDVLEYAETVKQKGLNLLSAPGNDMVASTALAASGCQIILFTTGRGTPLGTAVP TVKIATNSDIFKRKSNWMDFDAGRLLNGESMEKLTDELMDCVLKVASGMATKSENMGFREIAIFKNGVT |
| 4299 |
PF04217 (DUF412) |
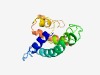 Estimated TM-score=0.53; hard target. |
>Protein of unknown function, DUF412 (Length=139) WLQLLQRGQHYMKTWPAEKQLAPIFPENRVVRATRFGIRIMPPLAVFTLTWQIALGGQLGPAIATALFACSLPLQGLWWL GRRSVTPLPPTLAQWFHEIRHKLLESGQALAPLEEAPTYQTLADVLKRAFNQLDKTFLD |
| 4300 |
PF04195 (Transposase_28) |
 Estimated TM-score=0.53; hard target. |
>Putative gypsy type transposon (Length=67) VCLYADALEAGMRVPLHGFFCDVLAHFGIAPTQLAPNGWRILAGFLVLCHSAGVPPSLPVFRHFFLL |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218