

Go to page (83 pages in total): [First page] << < 34 35 36 37 38 39 40 41 42 43 44 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 3801 |
PF06076 (Orthopox_F14) |
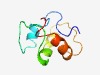 Estimated TM-score=0.55; hard target. |
>Orthopoxvirus F14 protein (Length=73) MKHRLYSEGLSISNDLNSIIGQQSTMDTDIEIDEDDIMELLNILTELGCDVDFDENFSDIADDILESLIEQDV |
| 3802 |
PF06074 (DUF935) |
 Estimated TM-score=0.55; easy target. |
>Protein of unknown function (DUF935) (Length=511) ILDIHGNPFRFEDRLQTENESRLAALQHHYSEHPASGLTPMKAARILRNAEQGDLVAQAELAEDMEEKDAHLQSELGKRR GAMLNVEWQIVPPANATPEEQRDAEMIEEILRDAVWFDDCIFDATDAILKGFSCQEIEWETHGNMTLIKGVHWRDPAWFM TPQFERNSLRLRDGSAHGVELQPFGWVKHIAKAKTGYLSRIGLVRTLVWPFIFKNYSLRDFAEFLEIYGLPLRLGKYPEG AGEKEKQTLLRAVMSIGHNAGGIIPRGMEIEFQKAADGSDATFMAMIDWAEKSMSKAILGGTLTSQADGATSTNALGNVH NDVRLEVRNADLKRLAATLTRDIVYPLYALNCKSFNDARRIPRLEFDIAESEDLNAFAEGLNKLVDIGFKIPKQWAHDKL QVPIAGENEEVLARNQPNPTAYLNSQSHQKMAVLSVQRDPDDLLDELEPSVEAYQAIIDPMLKPVVNALAEGGYEFAQEK LASLYADMDDSELEKILTRAIFVSDLLGRAN |
| 3803 |
PF06067 (DUF932) |
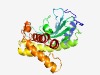 Estimated TM-score=0.55; very hard target. |
>Domain of unknown function (DUF932) (Length=226) PLSVVSQRYQVVQPREVLEFYRDLTERSGYELETAGVLKGGRKLWALARTGQSTALKGNDVVNGYLLLATSCDGTLATTA TPTTIRVVCNNTLTIAINGASQAIKVPHSTRFDPRTVKQQLGITVSQWDDFMYRMRTLAARPVKDHEAKAYLRSVLCEVQ TDKPEHTGLSNERALNKVLSMYEGHGRGAELESAKGTAWGLLNSVTEYVDHERRARSNEYRMDAAW |
| 3804 |
PF05987 (DUF898) |
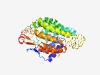 Estimated TM-score=0.55; hard target. |
>Bacterial protein of unknown function (DUF898) (Length=339) TPVEFTGKAGEYFGIWIVNLLLSIITLGIYSAWAKVRRKKYFYQNTLIDGVGFDYHASPIAILKGRIIAFVFFMAYIFSQ HISPFLPTVFALIILFLMPWLVIRTMTFNARNSSYRGLRFDFNGDMREAAIVFIGLPILIIFTLGLIVPYMVMRQSRYLI QNHKFGLTAFSMDAQAKDFYQIYGKALLLLLGIGAALSVAVTLTVDQNFLLNAVNDMRTVETFANFGIIIAVFFLFYFLV ILGIGAYIQSRISNLVWNSTTLGNLSFHSHLRARDMLWLYLSNLLIIICSFGLLTPLAHIRMMRYRASRLNLTGETDFER FIGDKKADVKATGAEMAEM |
| 3805 |
PF05886 (Orthopox_F8) |
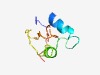 Estimated TM-score=0.55; very hard target. |
>Orthopoxvirus F8 protein (Length=65) MEGSKRKHDSRRLQQEQEQLRPRTPPSYEEIAKYGHSFNVKRFTNEEMCLKNDYPRIISYNPPPK |
| 3806 |
PF05846 (Chordopox_A15) |
 Estimated TM-score=0.55; hard target. |
>Chordopoxvirus A15 protein (Length=90) MFVDDKTFVVYKTWPTGLIQGTRKSVPFPTRHSCTFSSDVRIDNSYSSVVLSNPTFVQLLKMCVHIKRVQWKGKIILLFE CDNKPPPFRL |
| 3807 |
PF05832 (DUF846) |
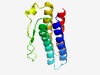 Estimated TM-score=0.55; hard target. |
>Eukaryotic protein of unknown function (DUF846) (Length=138) KPTIVLAHLSFKAASLFFYFFANFFTSSFIVQFLVILTLLSMDFWAVKNITGRLLVGLRWWNFVDADGNNHWKFESAKDM TRFPAIDRRVFWLGLVVGPAVWIFFVVTAFLTLKFEWMIVALLGALMNLANLWGYLRC |
| 3808 |
PF05732 (RepL) |
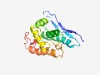 Estimated TM-score=0.55; hard target. |
>Firmicute plasmid replication protein (RepL) (Length=163) TTRKKVKVTGTETYINQSTGEIKEMQVIDIEERDFNFHKVWLEHIIHSMDLIGNQKTRLAFWLINNLNRDNVLIMTQRKI AEKTGISLETVRQTMKALMESGFLIKINSGAYCVNPDAVFKGGKTDRLNVLIQYRKSEQENTKTSEPTSEKDEPSLFNNE QLK |
| 3809 |
PF05710 (Coiled) |
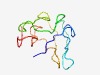 Estimated TM-score=0.55; very hard target. |
>Coiled coil (Length=88) MTILASISSIGNVKSISKSNNFSSLSNSSLQSSNSIQCGGCGGGSPLIGTVGNLVGGVLVGTGIIVGTVVGTVNGVVGGL LSGPNCGC |
| 3810 |
PF05696 (DUF826) |
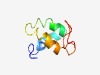 Estimated TM-score=0.55; hard target. |
>Protein of unknown function (DUF826) (Length=65) MPEIKGTVTEDLVKQALYSGEVNKLLKAQVRKDFEAQIDTYVDEVLAKLIGPAAAASGSDNEPKT |
| 3811 |
PF05599 (Deltaretro_Tax) |
 Estimated TM-score=0.55; very hard target. |
>Deltaretrovirus Tax protein (Length=87) MLIISPLPRVWTESSFRIPSLRVWRLCTRRLVPHLWGTMFGPPTSSRPTGHLSRASDHLGPHRWTRYRLSSTVPYPSTPL LPHPENL |
| 3812 |
PF05512 (AWPM-19) |
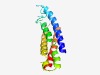 Estimated TM-score=0.55; hard target. |
>AWPM-19-like family (Length=141) LLNFCMYVIVLGIGGWAMNRAIDHGFIIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGAASAISGINHIRSWTA DSLPSAAAAATVAWTLTLLAMGFACKEIELRIRNARLRTLEAFLIILSATQLLYIAAIHSA |
| 3813 |
PF05078 (DUF679) |
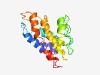 Estimated TM-score=0.55; very hard target. |
>Protein of unknown function (DUF679) (Length=168) TAANLAQLLPTGTVLTFQALAPSFTRGGACYTSNRCLISVLIALNAVFCVFFSITDSLVGADNRLYYGVATLRSLYVFNY NGADEEVRRRVLGDLEEYRLRPLDYVHAVFTVLVFLTIAFGDAMVQHCLFPDAGPNARELLVNLPLGAGVVSTLVFIVFP TSRKGIGY |
| 3814 |
PF04995 (CcmD) |
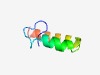 Estimated TM-score=0.55; easy target. |
>Heme exporter protein D (CcmD) (Length=44) GGYAFYVWLSVGMTLLALGCLIIHTLVQRKQLLKEIRQRESRER |
| 3815 |
PF04964 (Flp_Fap) |
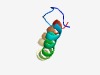 Estimated TM-score=0.55; hard target. |
>Flp/Fap pilin component (Length=44) FMSDESGATAIEYGLIAALIAVVIISAVTALGSTIKTKFNAVVT |
| 3816 |
PF04875 (DUF645) |
 Estimated TM-score=0.55; hard target. |
>Protein of unknown function, DUF645 (Length=59) LSDVQHGQFGFTKGSIIAVIWFSLSRTLNRGQLNLDRFEFWQPTSQLLVLDVCLFDAFA |
| 3817 |
PF04740 (LXG) |
 Estimated TM-score=0.55; hard target. |
>LXG domain of WXG superfamily (Length=201) KTLDVHALHHAIDQTLEQLKQQSDELSKVKKAVEGITSLDDALKGKGGDAIRAFYEECHTPFLQFYDTFIEEYSSTLKKM KSALNSLEPNHNGFISQSFLEHELEQGLNAADRTTKRLVSKTNATIAKVSHIVDLPDLNDSGFHEQNQKALKEINQTIEK LHAFDREQTSALKTAENDLETMQRYISRLEKMYTGPKIEIT |
| 3818 |
PF04714 (BCL_N) |
 Estimated TM-score=0.55; hard target. |
>BCL7, N-terminal conserver region (Length=49) SRSIRAETRSRAKDDIKKVMNVIDKVRHWEKKWVTIGDTTMKIYKWVPV |
| 3819 |
PF04654 (DUF599) |
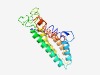 Estimated TM-score=0.55; hard target. |
>Protein of unknown function, DUF599 (Length=201) MVPLSFMISISYHLWLWHKVRTQPLSTVIGANAHGRRLWVSTIMKDNEKKNILAIQTIRNTIMGSTLMATTSIVLCCGLA AVVSSSYSVKKPLNDTIYGAQGQFMVALKYVTLLVLFLFSFMCHSLSIRFISQVNFLINCPQDSTLTSTYVSELLEKSFT LSMIGNRLFYSALPVMLWIFGPVLVLLCSSTMVPVLYNLDL |
| 3820 |
PF04550 (Phage_holin_3_2) |
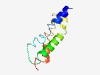 Estimated TM-score=0.55; hard target. |
>Phage holin family 2 (Length=86) MEEHEKTFVTLVLLGALIALGKMLTGDEPITLRLFIGRIILGSAVSVMAGVLLIWWPGISPIAVTGIGSALGIAGYQLIE VWLRKR |
| 3821 |
PF04448 (DUF551) |
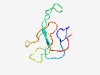 Estimated TM-score=0.55; very hard target. |
>Protein of unknown function (DUF551) (Length=68) GWISCSERMPEKGQNVLISVNFDSSLVEPLICSARYTGSTFRRGDATIKPGNAIEQATHWMPLPEPPQ |
| 3822 |
PF04336 (ACP_PD) |
 Estimated TM-score=0.55; hard target. |
>Acyl carrier protein phosphodiesterase (Length=104) YDHFLAKNWDNYSDTPLKDYVNNFYNSLEHHYDILPKRIQKMMPYMITDNWLLSYASIEGIAKVLAGMNRRTKNRSSMNE ATEELQEFYTEFEEEFTSFFNELI |
| 3823 |
PF04318 (DUF468) |
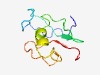 Estimated TM-score=0.55; very hard target. |
>Protein of unknown function (DUF468) (Length=84) MHGTCLSGLYPVPFTHKAHDYPHFNIYISFGGPKYCITALNTYVIPLFHHLLSTQFIYTYVNITKKSPLKSPKHKNILSF NDNT |
| 3824 |
PF04241 (DUF423) |
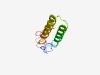 Estimated TM-score=0.55; hard target. |
>Protein of unknown function (DUF423) (Length=88) LGAFGAHILEGKLSDHYLSTWQTGVQYQMFHALALLAVGLLMSKFPASSLISWSGWLFFAGIVLFSGSLYVLSLTGVKVL GAITPIGG |
| 3825 |
PF04144 (SCAMP) |
 Estimated TM-score=0.55; hard target. |
>SCAMP family (Length=172) EKNWPPFFPIIHNDIGNEIPVHLQRTQYVAFASLLGLVLCLFWNIICVTAAWIKGEGPKIWFLAVIYFILGCPGAYYLWY RPLYRAMRNESALKFGWFFLFYLVHIAFCVYAAVSPSILFVGKSLTGIFPAISLIGKTVIVGVFYFLGFAMFCMESLLSM WVIQRVYLYFRG |
| 3826 |
PF03804 (DUF325) |
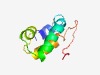 Estimated TM-score=0.55; hard target. |
>Viral proteins of unknown function (Length=73) LKYALRLTREYKENIIPHFDHLTRLRDLIDGMIKNEDVQRFNRTSRNDLISACMQINVHTYMPNATIDMRKQP |
| 3827 |
PF03788 (LrgA) |
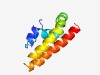 Estimated TM-score=0.55; easy target. |
>LrgA family (Length=93) QFIGELLSRFFHLPIPGNVMGMGLLLICLWRGWVKVQWIQEAAELLLSHLALFFVPAGVGVMVYFDLIAAEWLPIVVATV VSTFVVMAVTGRV |
| 3828 |
PF03777 (ChpA-C) |
 Estimated TM-score=0.55; hard target. |
>ChpA-C (Length=56) GYAHADSGASGSSSNSPGVLSGNTVQVPVHVPVNVCGNTVDVVGVLNPAAGNSCAN |
| 3829 |
PF03691 (UPF0167) |
 Estimated TM-score=0.55; very hard target. |
>Uncharacterised protein family (UPF0167) (Length=173) FPKFRYHPDPIGTGAFKKADKPQICECCGKKTGYVYESPFYSTEDVECLCPWCIADGSAAKKFDGEFQDAYSCEEIDDVS KLDELIHRTPGYCGWQQEVWLAHCNDYCAFVGYVGMAELEKMGLSDKLEDIYRKDEAMFDIGDIRECMTNGGSMQGYLFR CLHCGKYQLYADC |
| 3830 |
PF03683 (UPF0175) |
 Estimated TM-score=0.55; easy target. |
>Uncharacterised protein family (UPF0175) (Length=74) FPDSVFSALRKDPDEFAQEMRIAAAVKWYEMGEISQGKAAEIAGLNRAEFINILARYQVSPFQYTTQELAEELA |
| 3831 |
PF03682 (UPF0158) |
 Estimated TM-score=0.55; hard target. |
>Uncharacterised protein family (UPF0158) (Length=157) QNPLILRCHRLMEAFAKSDDERDFYIDRLEGFLIYIDLDKPQAELDALQEELEQNADRYCQIPKLSFYETKKIMEGFVNE KVYDIDTKEKLLDIIQSKEARENFLEFIYDHHAEQEKWQQFYQERSRIRIIEWLRLNHFHFVFEEDLDLPRPLVEKL |
| 3832 |
PF03631 (Virul_fac_BrkB) |
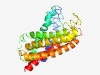 Estimated TM-score=0.55; hard target. |
>Virulence factor BrkB (Length=254) NDAMALGGQLAYFFLLFLFPFLMFLVSLGGLVVDDPEAILKTLVASTQGFLPQAAIEILSDHLDRTLRSPSSLAFLFSGL LTFAMGSVCALQIIGAANRAYGVPETRPFWKRWLIAILLILGFTLLVATMAFMVLSPHSGAYLQRTVGLPDVLVDLWGGL SWLIAFVNLTLAFAALYHMAPDTELSFRWVTPGGLMTTVLLLVSSQIFILWANNIFRSNQLFGQLGAGIVLLLWLFVIGL VVLAGIEINAELAR |
| 3833 |
PF03617 (IBV_3A) |
 Estimated TM-score=0.55; hard target. |
>IBV 3A protein (Length=57) MIQSPTSFLIVLILLWCKLVLSCFREFIIALQQLIQVLLQIINSNLQSRLTLWHSLD |
| 3834 |
PF03597 (FixS) |
 Estimated TM-score=0.55; easy target. |
>Cytochrome oxidase maturation protein cbb3-type (Length=41) LAYLIPVALFLGALGLGGFLWALRSGQYDDLDGAAERILID |
| 3835 |
PF03432 (Relaxase) |
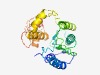 Estimated TM-score=0.55; hard target. |
>Relaxase/Mobilisation nuclease domain (Length=234) TKSTSRAINYAEKRAVEKSGLNCDVDYAKSSFKATRELYGKTDGNQGHVIIQSFQPGEVTPEQCNQLGLELAEKIAPNHQ VAVYTHADTDHVHNHIVINSIDLETGKKFNNNKQALRNVRDFNDEVCLEHGLSVPEKDTARLRYTQTEKAIADPNTKSTA QYSWKDEIREAIDQSQATNMDEFKDHLNQHGIEIERVTPKSITYRHLAEDKKVRGRRLGEDYNKGGIEDGFERQ |
| 3836 |
PF03245 (Phage_lysis) |
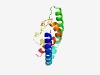 Estimated TM-score=0.55; hard target. |
>Bacteriophage Rz lysis protein (Length=126) AITYKEQRDTVTHKLTLANATITDMQTRQRDVAALDARYTKELADAKAENDALRDDVAAGRRRLLVNATCPAMPTGKSTS AARVDNAASPRLADSAQRDYFTLKERVTTMQKQLEGAQDYIRTQCL |
| 3837 |
PF03135 (CagE_TrbE_VirB) |
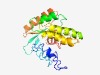 Estimated TM-score=0.55; very hard target. |
>CagE, TrbE, VirB family, component of type IV transporter system (Length=204) VLSTYEDKHGALCSQQLEFYNFLLSGEWQKVRVPSCPLDEYLGTGWVYAGTETIEIRTANATRYARGIDFKDYANHTEPG ILNGLMYSDYEYVITQSFSFMTKRDGKEFLTRQKQRLQNTEDGSASQIMEMDIAIDQLGRGDFVMGEYHYSLLVFAEDME TVRHNTSHAMNILQDNGFLATVIATATDAAFYAQLPCNWRYRPR |
| 3838 |
PF02705 (K_trans) |
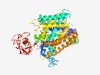 Estimated TM-score=0.55; easy target. |
>K+ potassium transporter (Length=571) YQSLGVVYGDLGTSPLYVFYNTFPHGAKDREDVIGALSLIIYSLTLVPLLKYVFIVLRANDNGQGGTFALYSLLCRHANL KIIPNQHRTDEELTTYSRATILERSFAAKTKRWLETHAFNKNSILMLVLVGTCMVIGDGILTPAISVLSAAGGIKVNRPD VDSGVVVLVAVVILVGLFSLQHYGTDRVGWLFAPIVLLWFLLIGGIGMYNIWNYDSSVLKAFSPIYIYRYLRRGGREGWT SLGGILLSITGTEALFADLAHFPVSSVQIAFTLVVFPCLLLAYSGQAAYLLLNLDHTKDAFYRSIPEKIYWPVFVVATAA AIVASQATITATFSIIKQALAHGCFPRVKVVHTSKNFLGQIYIPDMNWILMILCIAVTAGFKNQNQIGNAYGTAVVLVML VTTLLMILIMLLVWHCHWILVVIFTLTSLVVECTYFSAVLFKVDQGGWAPLVIAGVFFIIMYVWHYGTLKRYEFEMHSKV SMAWVLGLGPSLGLVRVPGVGLVYTELASGVPHIFSHFITNLPAIHSVVVFVCVKYLPVYTVPEEERFLVKRIGPKNFHM FRCVARYGYKD |
| 3839 |
PF02582 (DUF155) |
 Estimated TM-score=0.55; hard target. |
>Uncharacterised ACR, YagE family COG1723 (Length=183) VFLFSYGVVVFWNFTENQEKDILADLTFAENDSGQSLLTRPLNEEDFETEEFHFEYSENIKRPRIFNDMITLLPRSDHMI KLSISHAIAQSTKLCLFEERMSETMLDAQHVPKRLALTGELNMTRTEILKILGRLFKSRVDINLSSNILDVPNFFWDSEP TLNPLYVAIREYLEIDPRIKVLN |
| 3840 |
PF02553 (CbiN) |
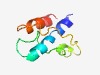 Estimated TM-score=0.55; hard target. |
>Cobalt transport protein component CbiN (Length=67) NLIIIALVILLAVLPLFLNKGAEFGGADGQAEELITEINPDYTPWFSSIWEPPSGEIESLLFALQAA |
| 3841 |
PF02505 (MCR_D) |
 Estimated TM-score=0.55; hard target. |
>Methyl-coenzyme M reductase operon protein D (Length=148) EIEIFPHRYLKASTTEKFLNKLYDLKTVERVVIHGQPLPKVVTYGPARGLPVNHSERKIINVKGVPIELTVMAGRFIVTL SDDSELEKIKKICEEMFPFGYDIRVGKFLKDRPTVTDYLKYGEDGVYLINEMDRRLIGMVDPRSKLSS |
| 3842 |
PF02079 (TP1) |
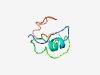 Estimated TM-score=0.55; hard target. |
>Nuclear transition protein 1 (Length=51) STSRKLKSHGMRRGKNRAPHKGVKRGGSKRKYRKGSLKSRKRCDDANRNYR |
| 3843 |
PF01998 (DUF131) |
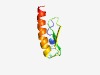 Estimated TM-score=0.55; hard target. |
>Protein of unknown function DUF131 (Length=59) GIIKSSKKERNENQREETKTEYGGVIFIGPIPIVFGSNKDMAKMALIIGVIMVVLIIAF |
| 3844 |
PF01710 (HTH_Tnp_IS630) |
 Estimated TM-score=0.55; easy target. |
>Transposase (Length=118) MAYDLDLRIKVIDFIESGGGITKASKIFKVGRATIYRWLGREELGATKVETRQRKINRKELEEDVKNNPDMLLKERAKKF GVTPASLCYQFKKMKVTKKKNSYFIKREMPKSEQNIRK |
| 3845 |
PF01384 (PHO4) |
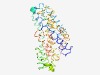 Estimated TM-score=0.55; hard target. |
>Phosphate transporter family (Length=332) IGANDVANTFGTSVGAKVLTLRAACTLASICELAGSILLGGNVSSTIKDGIVDTGRFNNTLDGPRLLMQGACIWMLIATF FRLPVSGTHSIVGATAGYGLVHFGLKGIKWMGLLKIVVSWLISPILSGAVSFFIFLFIKRFVLSKEKPLEPALRSLPFIF ACTIIFAESAIKTDRPEEAKVFSYAQILTATFGSFVHGGNDVSNAIGPLIGLWIVSTTGSVTIDKNTPIPLLIYGGAGIS VGLWVWGRRVIQTIGEDLTSMTPSSGVSIELGSAVTALVASKAGLPVSTTHCQVGSVVAVGLARSRKDVNWKLIISIFVA WVVTLPVSAGIS |
| 3846 |
PF18870 (HEPN_RES_NTD1) |
 Estimated TM-score=0.54; hard target. |
>HEPN/RES N-terminal domain 1 (Length=126) NYQDGYCDYCGKELKVVPLESLMEFIMDGISNFYEDAANFMSYNSREGGYLGEIYTPDELIQEHIELNAEPFEVIEDVVN SIEDIAWAQPDLYYDNIKDDLEFQWNYFKEIIKHKSRYLFSSGDSS |
| 3847 |
PF18860 (AbiJ_NTD3) |
 Estimated TM-score=0.54; hard target. |
>AbiJ N-terminal domain 3 (Length=164) VSEVTRRNILDELYSRGKITGKYDLIEFLNRIWNLEQMPSSDSRFKTASGDIWQHMINNSDWDEVYLFERYLELVTSADE TFIKFLEQVVHPLIREQDEQDDYINFINKHLSNDGYKLFLADNISGYPVYKVMKLEVGVKGKVKNLIFAAIGHKPEIIFS DSIN |
| 3848 |
PF18809 (PBECR1) |
 Estimated TM-score=0.54; hard target. |
>phage-Barnase-EndoU-ColicinE5/D-RelE like nuclease1 (Length=111) AFYREDLGYIDLVWGSVEGKGKEVKGLGLSKIIEKHIEDFKDFEGATPQEKLASGLNEIIKNGEFKAQENTSATIKYTKD GHLFRVGLRQNWMGEPTRNKWVITAYKDERE |
| 3849 |
PF18718 (CxC5) |
 Estimated TM-score=0.54; very hard target. |
>CxC5 like cysteine cluster associated with KDZ transposases (Length=117) PRSIYPLQHHCINPNCQRTQKGLALKKAEQRQAVLYMLDSGPIPVWSVHLYCEKCKVNYHSNFHVHQGKRVYYGGIPDVI QVGEHQFVEHKVVELWVTLMVVSWTSATNCTRFYNTA |
| 3850 |
PF18399 (CLIP_SPH_Scar) |
 Estimated TM-score=0.54; hard target. |
>Clip-domain serine protease homolog Scarface (Length=67) GCAAAMKCVTEEFCSVDGVMVNTPVRLSPYEKEHLRVPLMECTNSETNQVGFCCRDPLYEDPWPADM |
| 3851 |
PF18153 (S_2TMBeta) |
 Estimated TM-score=0.54; hard target. |
>SMODS-associating 2TM, beta-strand rich effector domain (Length=180) LAVLSILLAWLLSSILSYLSISIPWMIDAPSVVGFYGITYVLFDRYLWKMPFFRTIRLVNIPDLSGTWQGYLVSSYDEHA KQHETTLLISQTWSKISIILRTKHSSSRSMMASIEGGDPNVIRLMYGYLNEPNPESASTMVIHLGTACLKMHNNSPDQSL DGEYYAGRGRMNYGGMHFKR |
| 3852 |
PF18095 (PAS_12) |
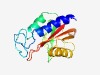 Estimated TM-score=0.54; easy target. |
>UPF0242 C-terminal PAS-like domain (Length=152) IPEATILLKKCITMAQKLTGANYYSNEATRYREFSSSNYAIDQRRLFDGLRNEEGGLIVVYSQKEHKLLFANHQSKALLG WSPDKFTSDFSSIIQEGLADWKRGLNTLSSATESQIRLLAKNKNGQEILLSCHLGIVPSGLFRNYIIGVLYP |
| 3853 |
PF17712 (DUF5557) |
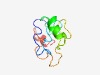 Estimated TM-score=0.54; very hard target. |
>Family of unknown function (DUF5557) (Length=90) MGAERVCTKAPEITQDEAEIYSLTNMEGSIGIKGCEFKSWLFKFYQARSQVLLCGEVKNLYLLTSNKTTVKEQNACLTHP DRSAMAGLLL |
| 3854 |
PF17647 (DUF5518) |
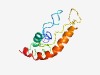 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5518) (Length=116) WKYAILGGVASLPFTAASYWQTGSRVSLVSVFGGGLLAGYFQARDTGTRDHVGARAGLIGGLPILVMLFDMLRFTFESSN PLWFSVAAVLMTLAFAVLSFGLAALTGELGAIVGNK |
| 3855 |
PF17597 (DUF5493) |
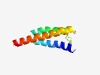 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5493) (Length=82) MSALGDVIYILGFLFPALGLISRNYLVNLMAFIIGTVAFLVFVQGYTDIAFSSSTFYLGVLPLLLGLVNLGYFFNWLREE RI |
| 3856 |
PF17521 (Secapin) |
 Estimated TM-score=0.54; hard target. |
>Honey bee peptides (Length=45) VSNNMQPLEARSADLVQEPRYIINVPPRCPPGSKFIKNRCRVIVP |
| 3857 |
PF17491 (m_DGTX_Dc1a_b_c) |
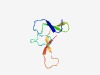 Estimated TM-score=0.54; trivial target. |
>Spider Toxins mu-diguetoxin-1 a, b and c (Length=55) AKDGDVEGPAGCKKYDVECDSGECCQKQYLWYKWRPLDCRCLKSGFFSSKCVCRD |
| 3858 |
PF17472 (DUF5425) |
 Estimated TM-score=0.54; very hard target. |
>Family of unknown function (DUF5425) (Length=76) CDLSINNDRNKIDGASHFKKKYMDNLNYQCLSKKESEAKNSQIKLDENNNKNHFYSSRVSNVSNYYDRTHISCKKN |
| 3859 |
PF17452 (YnfE) |
 Estimated TM-score=0.54; hard target. |
>Uncharacterized YnfE-like (Length=78) EILKQYMVLYKKMSNMINGPDYPGKEKDIQHQKDQIEVYEKQLQQGFSTDYDYDVFADSVIKCAYGDITLEDLEAVYY |
| 3860 |
PF17393 (DUF5402) |
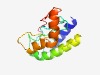 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5402) (Length=120) AEKLLEDRKKLEERLRRLTGGANVFVSEADLFAMSCGCIGIISYIQGMQFEDVEIYHEELMNIIEELSLDLGIYPSVSYA QMKPGTFDLERLQAHDLCDNCQKEYAGVGGKPWPDILIFK |
| 3861 |
PF17356 (PBSX_XtrA) |
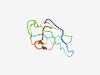 Estimated TM-score=0.54; hard target. |
>Phage-like element PBSX protein XtrA (Length=66) MRLKELKINPSTMRLEIDIMEQKGSFAIVVCDGKAKLTELPPHGETKIITHQGKVKRVKFDEGEEF |
| 3862 |
PF17347 (DUF5377) |
 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5377) (Length=95) MTVQTEVLFSNNWNVRISDPGEEGAHSHFFETIYITLVAHIDGSNISYEFTRKVEEQVKIHRTFTDLSELFKFLGDYLDP VSMGFLGIKIGNLGV |
| 3863 |
PF17345 (DUF5375) |
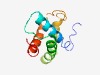 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5375) (Length=106) KECHLTPALKAALYRRAVACAWLTLCHEQKRYASLTLARLEHAIETELEGFYLRQHGRQRGQEIACALLADLMAAGPLKA APCLSFLGQVVMDELCGRLKDVPVLH |
| 3864 |
PF17339 (PH_15) |
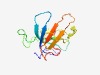 Estimated TM-score=0.54; easy target. |
>PH domain (Length=105) SMDELCREPGAMCGYVEKALALSNWERRLAVITRDGKLVIYKYYLLDICPGEPLMGKTYQLNKVKYLHLERFENGDIISR IKTMDGKRVKLKLTGEYATAWAAKI |
| 3865 |
PF17301 (LpqV) |
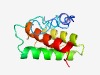 Estimated TM-score=0.54; hard target. |
>Putative lipoprotein LpqV (Length=113) PPADKTSAETSSEETPTVAPGSVAVSPGGVTTAVGAPADSTEDEYFKACTSARQWMDEQGGDPKSQIEPYLGMLQTSDSA GPGTYDTPWTQLPPPRQAAVIVAVQAAADGLCG |
| 3866 |
PF17240 (DUF5313) |
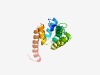 Estimated TM-score=0.54; hard target. |
>Family of unknown function (DUF5313) (Length=123) DRPNLLQYIAYSYGRRLPDSMREWVAHDLADHGAVRRHMIRMAIPPALVLAPFWLLPASLYVHIEMTVPIYIWSLLMALA LNKVWRRHRLAQHGLDPNLVDEIRYKKQAHIHEDYIRRFGPRP |
| 3867 |
PF17114 (Nod1) |
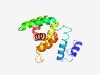 Estimated TM-score=0.54; easy target. |
>Gef2-related medial cortical node protein Nod1 (Length=145) MVCLSTSIAENPYQSLSLESVPSKILFGSFLFYVKDRIPEKAGYIIEEQFCKRLQQIDYDAYSFSPKECNEKLKNIFSEF SQMKLKMLSVFFSIVQILLPKLSGDYQEHVQFFASISAVVAPRGYVFEIYHAVEHLSLEAREVFQ |
| 3868 |
PF16974 (NAR2) |
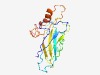 Estimated TM-score=0.54; hard target. |
>High-affinity nitrate transporter accessory (Length=169) KQTLLINSSPSQGQVLQAGEAQVTITWSLNNSYPTGTDTNYKTVNVKLCYAPVSQLDRGWRKSNNNLKKDKTCQINLVTM PYKPSNNNFTWTIHKDVPTATYFIRAYVYDSTGEVVAYGQTTDAHKKTNLFKITAITGRHITIDVCAACFSVFSIISLVG FYFVEKRKA |
| 3869 |
PF16949 (ABC_tran_2) |
 Estimated TM-score=0.54; hard target. |
>Putative ATP-binding cassette (Length=545) LLSGSLLRCLMVLACSAVFWTVLFGIFFEGFMFVGMYVDLKNTIIEHLFSMFFLSLLIMLFVSTGIIVYAGLFQSREAAF LLTTPASTDRIFVYKFFEAIVFSSWGFLLLVSPMMVAFGITVIAKWSFYAIFLVYLLSFVMIPGSLGAVAAILVANFFPR RQKTVLILGGGLILIGAAFVVTQITTTPGSALSTDWLGGLLGRLAFSQHPLLPSRWMSAGLLASARGNWSDGLFYLMVLT AHAGLFYLVATVVARDLYRQGYSRVQGGRSSRRRSGLYWIDALFHRLFFFIPHPVRLLILKDLRTFRRDPAQWSQFLIFF GLLAFYFLNIRRLSYDVQSPYWRNLVSFLNLAVTALILSTFTSRFIFPLLSLEGRNFWVLGLLPLKREAILWGKFAFASG ISLVATEVLVVLSDLMLRMSTAMIALHMGMVAILCLGLSGISVGLGARLPNLKETDPSKIAAGFGGTLNLLVSLVFISAI VTALALPCHLYFAGQDASEFRTFVLSEERFRFWMAIAVVTSLVIGAVGTFLPLRIGIKAFKRMEF |
| 3870 |
PF16825 (DUF5075) |
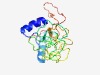 Estimated TM-score=0.54; easy target. |
>IGP family C-type lectin domain (Length=173) YFVSYKNAEAYCVDNGAVLPADQTEAAHQSLQSAIKRVITGSDAFSYLGGDAVYSSGEAIEADKRCKPGDIASSRYCVYR WNKGLFAKALPDGQGVAFWRGSYIKYDGAASMNGYPSFWGGEYPKRGQLYTLSWYHTGDDVTTWYDGKDMSGRIKWASDN PDTKTENYVILCE |
| 3871 |
PF16633 (APC_u9) |
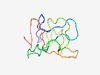 Estimated TM-score=0.54; very hard target. |
>Unstructured region on APC between 1st two creatine-rich regions (Length=88) SAEDEIGRDQSTRVTDANNTLQIAELKENSGALSTEGAVSEITSTSQHIRTKSNRLQTSSLSPSDSSRHKAVEFSSGAKS PSKSGAQT |
| 3872 |
PF16632 (Caskin-tail) |
 Estimated TM-score=0.54; easy target. |
>C-terminal region of Caskin (Length=63) RQKLEETSACLAAALQAVEEKIRQEDAQGSRSSAAEEESTGSILDDIGSMFDDLADQLDAMLE |
| 3873 |
PF16631 (TUTF7_u4) |
 Estimated TM-score=0.54; hard target. |
>Unstructured region 4 on terminal uridylyltransferase 7 (Length=88) RRKVRRRRDQEDTLNQRYPENKEKRSKEDKEIQNKYTEREVSTKEDKPMQCTPQKAKPVRAAVDLGREKILRPPVEKWKR QDDKDLRE |
| 3874 |
PF16624 (zf-C2H2_assoc2) |
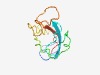 Estimated TM-score=0.54; very hard target. |
>Unstructured region upstream of a zinc-finger (Length=95) SKREKGPNNTANSSEIKVKVEPADSVESSSPSITHSPQNELKGTNHSNEKKNTPATQKNKVKQDSESPKSASPSAAGGQQ KARKPKLSVGFDFKQ |
| 3875 |
PF16611 (RGS12_us2) |
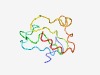 Estimated TM-score=0.54; very hard target. |
>Unstructured region between RBD and GoLoco (Length=72) PSRGKDKQKSAPVKQNTAVNSSSRNHSATGEERTLGKSNSIKIKGENGKNARDPRLSKREESIAKIGKKKYQ |
| 3876 |
PF16605 (LSM_int_assoc) |
 Estimated TM-score=0.54; hard target. |
>LSM-interacting associated unstructured (Length=61) NPPRRSVPDKPGSSRSTGDMMPRQVYGARGKGRTQLSMLPRSLHRQNAPVGKVENGTAAEQ |
| 3877 |
PF16594 (ATP-synt_Z) |
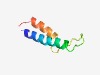 Estimated TM-score=0.54; hard target. |
>Putative AtpZ or ATP-synthase-associated (Length=53) ICVWSFLVSMAGGLLLLWWDYEYHPTNSQLWLVPFGLILFVTPVIACFAVVVS |
| 3878 |
PF16476 (DUF5053) |
 Estimated TM-score=0.54; hard target. |
>Domain of unknown function (DUF5053) (Length=59) QEVAEIVSLSYLAKKYFNKSRSWLYQRLNGNLVNGKPARFTQEELQTFNNALQDISKKI |
| 3879 |
PF16428 (DUF5025) |
 Estimated TM-score=0.54; very hard target. |
>Domain of unknown function (DUF5025) (Length=160) VQFIKMQIAGQSLEIKGSIDQNRNIFSGSWTGIGYGDGTQKEMYTVNVTVPKTFLNTTTDSKFQVRIFDIEKKEYLLSDS DPYKQSFASSIYLVTNLGSADSKVYTTNETKPPFKVQITRYEKPKDSGVPFVGGKLNGTLYNIKNLQDSVVIKDGVFDVR |
| 3880 |
PF16410 (DUF5018) |
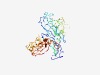 Estimated TM-score=0.54; easy target. |
>Domain of unknown function (DUF5018) (Length=351) YLLLGVLALMSSCQDPEYVLPTAERQGITSLTALFTSGPYVDKEAVVYTIADASVDKYVIPIPWFYPEDSENETAEYMKT MRVQAKLAPNCTINPVLTILDLTKENYFTYTDAQGYKKQICITGERVKSNKCQLLSFSIPAEDISGIIDEEHKTVSLISA EDLSSCLAEYSLSSHATMSPDPKTEALDFNSPVELTVIAHDGVTKQTYTVQKAVPDKIPYGYRKGSETELFKLDMGVIGL PWTSANSPSLAVNGNNLVVCLGDGSTAPTYYNASTGSKIGNMTLGSVGVASLGCVTSDSKGNILLATKAANGKSFSIYKT SSVTTVPTLLISYTNNTSLDMGTKVSVQGDI |
| 3881 |
PF16386 (DUF4995) |
 Estimated TM-score=0.54; hard target. |
>Domain of unknown function (Length=72) MKKKLVLFASVAIALASCQTTPKEDYSWIKKGLDVASAQLQLTAEEIDGTGKLPRSIRTGYDMDFLCRQLER |
| 3882 |
PF16372 (DUF4984) |
 Estimated TM-score=0.54; very hard target. |
>Domain of unknown function (DUF4984) (Length=164) LYKDYTHVVMQKACPFDIHNFSGWCMVTSTFYSQYLNNVTNRLIKTEVVEGEENTVLLKDVYYKGYDLKLKFKTGNLLEP KVEMDDQIAGETGEAFGTIYGDGKLRISQPSLYTSYYNTCQNYVLQYVNMSVNKKDGSLYGNVGTFITMFEWVSDAEAEK LKEQ |
| 3883 |
PF16301 (DUF4943) |
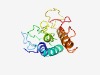 Estimated TM-score=0.54; very hard target. |
>Domain of unknown function (DUF4943) (Length=150) DYNNPDVDLFVRQLKAGNYNTKSPKGFVEVPKFTEKDIPTLLNYAEDLTLITSFPLPPVSAYYSGKVRLGECMLWVVETI RLGHYASFGCKMVRANAENYEGIYFLTDEELLDAAARYRRWWENRQYPRTAWTIDACFDEPLCGSGYRWW |
| 3884 |
PF16270 (DUF4923) |
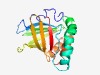 Estimated TM-score=0.54; hard target. |
>Lipocalin-like domain (DUF4923) (Length=173) LKDLFNKENVEKVVSAVTGKNTVDMTGTWSYTGAAIEFESDNLLMKAGGAVAATTAEAKLNEQLSKVGIKPGQMSFTFNA DSTFNAKLGTKTLNGTYMYNATEKHVTMKFVRLINMNAKVNCTSGSMDLLFESDKLLKLITFLSSKSSNATLKTISSLAN SYDGMMLGFSLEK |
| 3885 |
PF16200 (Band_7_C) |
 Estimated TM-score=0.54; hard target. |
>C-terminal region of band_7 (Length=59) IVSQAIQESGGMEAVSLRIAEQYIEAFGKLAKEGTTVLLPSNAAEPANMVAQALTIYKN |
| 3886 |
PF16098 (FXMR_C2) |
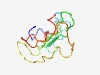 Estimated TM-score=0.54; very hard target. |
>Fragile X-related mental retardation protein C-terminal region 2 (Length=90) GYKGNEEIQWNESRARNSRDPKPRQQEDSLQIRIDSNNERSVHTKALQSSFADSGTSNSSSRPRPGKDRGPKKEKQDSLD APRVLVNGVS |
| 3887 |
PF16082 (Phage_holin_2_4) |
 Estimated TM-score=0.54; hard target. |
>Bacteriophage holin family, superfamily II-like (Length=76) MKTETIDALATAGGRTSGGGAVVGFLGWLASSQAIGLFGVCIALLGALVSWYYKREANRRLAAEHALRQQERQMRI |
| 3888 |
PF16044 (DUF4796) |
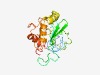 Estimated TM-score=0.54; hard target. |
>Domain of unknown function (DUF4796) (Length=190) GILCFQHCDKKVFCFELPNLCPVCKKNLSTEQFKLLPIRIPYPFVRAAQHPCSILIKPTSGDFLNDYFNSVDLHIGVTDS TGAVVEYDKNGLQRQKNNSWNQCLVLDGMDEPWADQWDEALKAVIKEGHWTPQMYNENSYNCYTFVLAFLKKLKHDLLSN AAYSSTYFCEKFILPRTTAAGKYISLYRKL |
| 3889 |
PF16025 (CALM_bind) |
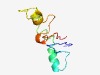 Estimated TM-score=0.54; hard target. |
>Calcium-dependent calmodulin binding (Length=78) LNSKSRFHLDGQPILAPLMTASKVNEMQMARQMAMQIEARLTATAEADATDDDDEDNSSMEGRRSFPKLEQTKTYIYD |
| 3890 |
PF15990 (UPF0767) |
 Estimated TM-score=0.54; very hard target. |
>UPF0767 family (Length=83) MLPVIFTFLRTYAPYVTLPVAAVVGFIGYNLEAILSDKYTPYQKSIEEKREDRLMEENLSKDVTKVDSLKERKFVPRSVF EKN |
| 3891 |
PF15980 (ComGF) |
 Estimated TM-score=0.54; hard target. |
>Putative Competence protein ComGF (Length=96) NVKQVKQQENQDWLLFVQQMEAELEGCQFVKVEGNKLYVKQDNQDLAFGLSSATDFRKTNADGRGYQPMLFNVKSSTISQ NNQIVTIQVTLKNGLQ |
| 3892 |
PF15845 (NICE-1) |
 Estimated TM-score=0.54; hard target. |
>Cysteine-rich C-terminal 1 family (Length=90) SAKGFSKGSSQGPAPCPAPAPTPAPASSSSCCGSGRGCCGDSGCCGSSSTSCCCFPRRRRRQRSSGCCCCGGGSQRSQRS NNRSSGCCSG |
| 3893 |
PF15825 (FAM25) |
 Estimated TM-score=0.54; hard target. |
>FAM25 family (Length=65) AVHAVEEVVKEVVDHAKQVGEKAIADALKKAQESGDKVVKEVTETVTNTVTNAVTHAAEGLGKLG |
| 3894 |
PF15749 (MRNIP) |
 Estimated TM-score=0.54; hard target. |
>MRN-interacting protein (Length=104) VLRCFSCETFQVHQVKKSNKWNCKLCGAKQSVRKEYGRGSGADCRRHVQKLNMLRATMDSAPSTCEDTDVSEQDPEWQQD TEDATDPQYNYTDDSQKSSKWEKF |
| 3895 |
PF15689 (NRIP1_repr_3) |
 Estimated TM-score=0.54; very hard target. |
>Nuclear receptor-interacting protein 1 repression 3 (Length=88) VKIKSEPCDDLHIHNTDVHLSHDAKGAPFLGMAPPVQRSAAALPAPEDFKSEPISPQDFSFSKNGLLSRLLRQNQESYLA DDLDNSHR |
| 3896 |
PF15615 (TerB_C) |
 Estimated TM-score=0.54; hard target. |
>TerB-C domain (Length=155) PATAVKGATQPANGGFTLDHERIAQLQRETAAVSALLAQVFTEDQTDDHFSDSPISSAPSNDSESAACIEAGRVDEQVTS SDIPGLDLEHSAFLRLLVSRTEWSRSELEEAASDMELMLDGALEQINDMGFDRFNMPVTEGDDPVEINADILEEL |
| 3897 |
PF15531 (Ntox27) |
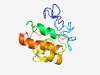 Estimated TM-score=0.54; very hard target. |
>Bacterial toxin 27 (Length=130) LGINPHSDDLTKLAGGPGGRTFNNQDYGIQLPKSMGLGERPIWTVGVEQAVANRNVRISVSLDGVPGATNADEALNALLK RGEGITPGDWETIRQRGMGTAWEMTKLRSAVRMEDRGWKSIDWWMTNSKG |
| 3898 |
PF15448 (NTS_2) |
 Estimated TM-score=0.54; hard target. |
>N-terminal segments of P. falciparum erythrocyte membrane protein (Length=50) MDKSSIANKIEEYLKEKSNDSKIDQSLKADPSEVQYYGSGGDGYYLKNNI |
| 3899 |
PF15446 (zf-PHD-like) |
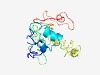 Estimated TM-score=0.54; hard target. |
>PHD/FYVE-zinc-finger like domain (Length=164) CHTCGGSRQRGQLLYCQGCCLAYHKNCIGYRSNREHLATKVGEDDFVLQCRFCLNIYSKKDSSAPRHSACQQCKKDGRSC TPFSQKKTARQEEKLRAENGGVDPVTPVSPQLINNPDNVLFRCSMCHRGWHVEHLPPTGTDAIETDVKSERLKEYSVDWR CTDC |
| 3900 |
PF15382 (DUF4609) |
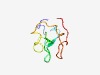 Estimated TM-score=0.54; very hard target. |
>Domain of unknown function (DUF4609) (Length=68) EKPDVKQKSNKKKFVIPQIIITRASNETLISYCSTGNEEQRTIREQADWGPYHRHRNPSTVAAYDLHT |
[an error occurred while processing this directive]
zhanglab![]() zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218