

Go to page (83 pages in total): [First page] << < 32 33 34 35 36 37 38 39 40 41 42 > >> [Last page] [All entries in one page].
| # | Pfam ID | C-I-TASSER models ranked by estimated TM-score (click to view) |
Full name and representative sequence |
| 3601 |
PF06626 (DUF1152) |
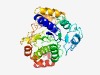 Estimated TM-score=0.56; hard target. |
>Protein of unknown function (DUF1152) (Length=298) FVFGIGGGGDIVSAIVVYTYLRKLGYNVLLGAVVWERYVEDPIPGPICFDDLRNAILVNQGLYKVNKDTYAIRGSRQVVP QLVRAIRALELEGAYGICNKSGAMGLYSSLKEFANTEKIDLIVGVDAGGDVLAKGCEETLGSPLIDFISLASLVKLEEDG YNVMLGVIGSGSDGELQYDYFLKRISEIASLNGLLDIKGYDRETATLVERVLNEVNTEASKIPFEAFKGLYGEIPIRNGT RNVFVTPVSAIMFFLDPIKVAETSPLYSLVKDSSSIDDANKNLNEVGIYTEYNFEIDL |
| 3602 |
PF06591 (Phage_T4_Ndd) |
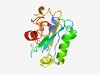 Estimated TM-score=0.56; very hard target. |
>T4-like phage nuclear disruption protein (Ndd) (Length=152) MKYMTVTDLNNAGATVIGTIKGGEWFLGTPHKDILSKPGFYFLVSKLDGRPFSNPCVSARFYVGNQRSKQGFSAVLSHIR QRRSQLARTIANNNVPYTVFYLPASKMKPLTTGFGKGQLALAFTRNHHSEYQTLEEMNRMLADNFKFVLQAY |
| 3603 |
PF06580 (His_kinase) |
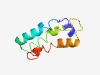 Estimated TM-score=0.56; hard target. |
>Histidine kinase (Length=80) ARLDALKLQLNPHFFFNSLASVRSLISEAPRRAKKMVARLARLLRKTLQASDEETVSLREELSTTQTYLQLEKVRFEDRL |
| 3604 |
PF06578 (YscK) |
 Estimated TM-score=0.56; hard target. |
>YOP proteins translocation protein K (YscK) (Length=205) ALTPYQFRFCPASYIHSDHLSPEWLTVLSSLPEWRHSPRLNGLLLTQFDLNVDYELPTGLGNIALLEQSCLEQLLTWLGA LLHGQAIRHCLMATELRHLHDSLGKEGHRFCLKYLDIIIGNWPTGWQRSLPPEINANYFRTSALQFWLTAMESPPIDFAK RLSLRLPSYENLAAWPVSQAERPLAQALCLKLAKQVNTECYHLLK |
| 3605 |
PF06515 (BDV_P10) |
 Estimated TM-score=0.56; very hard target. |
>Borna disease virus P10 protein (Length=87) MSSDLRLTLLELVRRLNGNATIESGRLPGGRRRSPDTTTGTIGVTKTTEDPKECIDPTSRPAPEGPQEEPLHDLRPRPAN RKGAAVE |
| 3606 |
PF06423 (GWT1) |
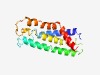 Estimated TM-score=0.56; hard target. |
>GWT1 (Length=161) QNREGIFSFIGYLSIFLAGQSLGSIALPRQPKPKPVPNPPSPTSPLHHLTRSTLGQLCLVSIAWTCLFYLCTSYHGLHLT VSRRLANLPYVVWVASFNSANIALCCAIESLLFPNLYHAKAGSRAEERRRARDATSTVLHAYNRNGLVVFLVANLLTGLV N |
| 3607 |
PF06139 (BphX) |
 Estimated TM-score=0.56; hard target. |
>BphX-like (Length=132) MKNARLFLIAIGVFYIINLIGTLPFSTLGLFGRMYPGVELHVGAPIFTLLQDAWAVVGLQLGAIGAVALWGARDPGRYRA VIPVVIATEVVDGLWDFYSIVWSHEALWFGLVTLVIHVLWIGWGLHAWRALA |
| 3608 |
PF06096 (Baculo_8kDa) |
 Estimated TM-score=0.56; hard target. |
>Baculoviridae 8.2 KDa protein (Length=63) DNYSVQNFYNNDRKPLKPTTLHDGNIKKSVYEDVTYIRKLMCKEIMPGEHDHKFYNYGYNKEN |
| 3609 |
PF05919 (Mitovir_RNA_pol) |
 Estimated TM-score=0.56; easy target. |
>Mitovirus RNA-dependent RNA polymerase (Length=479) EFDDSNHFVSVKSSPTGPSTLTALWGLWNHSYESFNWLFKITSLSGVDYLTKIMNWTWLKDLGNKNNNKWSKQFLGSLSL IYDPECKVRIVAMLDYTTQLFLRPIHNDLFKLLKKLPQDRTFTQNPLNDWENNEHSFWSIDLTAATDRFPISLQRRLLLY IYSDPEIANSWQNLLVHREYARNGLNPIKYSVGQPMGAYSSWPAFTLSHHLVVHWCAHLCHINKFKDYIILGDDIVIHND KVAKKYIEIMGKLGVGLSDSKTHVSKDTYEFAKRWIHKGQEISPLPITGIVNNITNPYIVLMNLYDFFKIKRNQYNFSGD ILTMLLKLYQGLKIYKGDKSKKHYYLSLNRFKNLDIFLFALNNTFGYSTYDHKRSFLISKEYSKDLPGPKIMQDLIXYVI STGLSNLTVESVNKSLKFVDTIIDNKSLLNIEDPNDLSNLPLFKGFQAYISNLRKTVEGWNNKSISLIEASRGLVQMDI |
| 3610 |
PF05813 (Orthopox_F7) |
 Estimated TM-score=0.56; very hard target. |
>Orthopoxvirus F7 protein (Length=80) MTLVMGSCCGRFCDAKNKNKKEDVEEGREGCYNYKNLNDLDESEARVEFGPLYMINEEKSDINTLDIKRRYRHTIESVYF |
| 3611 |
PF05751 (FixH) |
 Estimated TM-score=0.56; hard target. |
>FixH (Length=140) KQFWPWFLIILPMCAVIASLTTLKIALDNSDSLVAEDYYKQGKAINMDLRKIKYAQQIGMKFLVTVDDQELTITQHGGPE YRAALNVRFYHPTIEAKDFSLNATADANYVYHIPLDAAITGPWEVRIEGFDAKWRIQQRI |
| 3612 |
PF05628 (Borrelia_P13) |
 Estimated TM-score=0.56; very hard target. |
>Borrelia membrane protein P13 (Length=143) LLVYETSKQDPIVPFLLNLFLGFGIGSFAQGDILGGSLILGFDAVGIGLILAGAYLDIKALDGITKKAAFQWTWGKGVML AGVVTMAVTRLTEIILPFTFANSYNRKLKNSLNVALGGFEPSFDVAMGQSSALGFELSFKKSY |
| 3613 |
PF05595 (DUF771) |
 Estimated TM-score=0.56; hard target. |
>Domain of unknown function (DUF771) (Length=90) EDMVLITRVEYEMLKQKELSGLYWNMKDLEKRINKKSEWIKENILYPGRFRKILDINKGGFVYYPETKGQHWSFQATKMA EFLDNNFGNI |
| 3614 |
PF05398 (PufQ) |
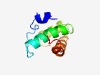 Estimated TM-score=0.56; hard target. |
>PufQ cytochrome subunit (Length=74) MTDMTSNTPVEARSHKPPKTEFMVYFAIIFVATLPLATLTWALAAIKSGSFTEKGPVARAWSQARIITPMIFTA |
| 3615 |
PF05385 (Adeno_E4) |
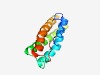 Estimated TM-score=0.56; hard target. |
>Mastadenovirus early E4 13 kDa protein (Length=108) MVLPALPAPPVCDSQNECVGWLGVAYSAVVDVIRAAAHEGVYIEPEARGRLDALREWIYYNYYTERAKRRDRRRRSVCHA RTWFCFRKYDYVRRSIWHDTTTNTISVV |
| 3616 |
PF05295 (Luciferase_N) |
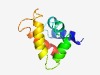 Estimated TM-score=0.56; hard target. |
>Luciferase/LBP N-terminal domain (Length=85) MASRGQLVQFLANEGKVDQKVIAYMTNQMKLTSVSDFANYFTSSEFEKGVQDDIVAQVSPFNADVSKPDAKIQTARLRAA WKAAQ |
| 3617 |
PF04885 (Stig1) |
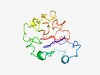 Estimated TM-score=0.56; hard target. |
>Stigma-specific protein, Stig1 (Length=130) AITLTITFTGSNHEPTPITEPSSSVAKPFPKRVSRFLAESKNPRATDHCYKDDAICYILEGKNSTCCNXKCMDLSQDKXN CGACKNKCKFTSSCCRGECVNLAYDKRHCGSCGNKCMPGGYCIYGLCSYA |
| 3618 |
PF04883 (HK97-gp10_like) |
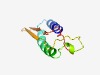 Estimated TM-score=0.56; hard target. |
>Bacteriophage HK97-gp10, putative tail-component (Length=85) KKMAKSFQKALDERVIERWIREFLLEMAFRAERKIKKRTPVGVYPNKTGGYLRRNWQVGNVIKQGDAYIVEIFNNTEYAS YVEYG |
| 3619 |
PF04770 (ZF-HD_dimer) |
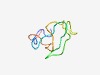 Estimated TM-score=0.56; hard target. |
>ZF-HD protein dimerisation region (Length=54) NVRYCECQKNQAVGVGGHAVDGCMEFMPSGAEGTDAALACAACGCHKNFHRRVV |
| 3620 |
PF04554 (Extensin_2) |
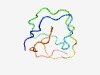 Estimated TM-score=0.56; hard target. |
>Extensin-like region (Length=59) YVYKSPPPPPYIYKSPPPPPYVYKSPPPPPYIYKSPPPPPYVYKSPPPPPYIYKSPPPP |
| 3621 |
PF04259 (SASP_gamma) |
 Estimated TM-score=0.56; hard target. |
>Small, acid-soluble spore protein, gamma-type (Length=78) GTDVQHVQKQNSGAASGQYGTEFASETNVQEVKQQNQQAEAGKSYGAQGSGQYGTEFASETNVQEVKQQNQQAEAGKS |
| 3622 |
PF04143 (Sulf_transp) |
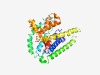 Estimated TM-score=0.56; hard target. |
>Sulphur transport (Length=311) RGRFCLTGAYRDLYLTKDNRMFIAVILAIAVQSIGVFLLYSFGLIQFKEGNFMWLATIIGAFIFGIGIVLAGGCATGTWY RAGEGLIGSWIALIAYMVSSAMMKGGVLQSLNDSLQSYTVNHVTIYDSFGVSPWVFVILLSVITGYFVWKQLRKPSVKIP SLKQKKTGIAHVLFEKRWHPFVTAILVGCIAILAWPLSEATGRMYGLGVTTPTANILHFLVSGDVSYINWGVFLVLGIFL GSLIAAKGSGEFRWRTPDAKTTIFSFIGGILMGIGASLAGGCSIGNGLVGTAIFTWQGWIALPFMIFGTWT |
| 3623 |
PF03935 (SKN1) |
 Estimated TM-score=0.56; easy target. |
>Beta-glucan synthesis-associated protein (SKN1) (Length=504) FPLHIDEKEPDDYLHNPDPVQDALYDKHRFWNDLKNADRRSAWGLVGFIILITAAVGVFIILPVLTFTGVEVHRTPQEYE VLTDYQYPMLSAIRQTLVDPDTPEDALTITAKDGSQWVLVFSDEFNAEGRTFYEGDDQFFTAPDLNYAATRDLEWYDPDA ATTADGTLQLRMDAFNNHDLFYRSGMLQSWNKFCFTQGRIVMSARLPNYGTVSGLWPGMWTMGNLGRPGYLASTEGVWPY SYDTCDAGITPNQSSTDGISYLPGQRLNSCTCKGESHPNPGVGRGAPEIDVIEAAIGTDPTGKTPNIGVASQSLQLAPFD IWYMPDYSYIEIYNSSVTTMNTYAGGPFQQALSGTTTLNSSWYEFGPFEHNFQRYGFEYLNDDDKGYLTWFVGDDPTFTV YPQSLHPNGNVGWRHLSKEPMSIIMNLGISNSWAYIDWPSITFPVTFRIDYVRVYQPKDQINVGCDPPDYPTYDYIQQHL NIFENVNLTLFEMGGYTFPKNKLT |
| 3624 |
PF03855 (M-factor) |
 Estimated TM-score=0.56; very hard target. |
>M-factor (Length=44) MDSIATNTHSSSIVNAYNNNPTDVVKTQNIKNYTPKVPYMCVIA |
| 3625 |
PF03690 (UPF0160) |
 Estimated TM-score=0.56; hard target. |
>Uncharacterised protein family (UPF0160) (Length=320) KIGTHNGTFHCDEVLACFFLRQLPEYSDGEIVRTRDPAQLAECDIVVDVGGEFDPKRHRYDHHQRTFSDTFNSLCPEKPW VTKLSSAGLVYLHFGRRVLSQLTQLKEDDRQLEVLYDKLYENFVEEVDAVDNGISQYDGESRYSISTTLSARVSHLNPRW NSTDQDTEEGFKKALKMVGEEFLDRLDFYKSSWLPAREVVEEAVKKRHQIDPSGEVLLFSQGGCPWKEHLFALEKELHVE TPIKFVLYPDQNGQWRVQCVPAGLNTFQNRLSLPEEWRGVRDEALSELSGIKGCIFVHAGGFIGGNKTQEGALEMARRTL |
| 3626 |
PF03295 (Pox_TAA1) |
 Estimated TM-score=0.56; hard target. |
>Poxvirus trans-activator protein A1 C-terminal (Length=63) AMIKTHVALREEPKITLLPLVFYNKPEEVINIINSLRNKDGVYGSCFYKENDQSVQISLRSLL |
| 3627 |
PF03181 (BURP) |
 Estimated TM-score=0.56; very hard target. |
>BURP domain (Length=213) FFRESMLKTGTVMPMPDIRDKMPQRSFLPRAIVSKLPFSSSNVNVLKEIFHASENSSMESIIKDALSECERQPSKGETKR CIGSAEDMIDFATSVLGNSVTLRTTQNVQGSKQNIMIGSVKGINGGKVTQSVSCHQSLFPYLLYYCHSVPKVRVYEADLL DPKTKAKINHGVAICHIDTSAWSQTHGAFLALGSGPGRIEVCHWIFENDLTWT |
| 3628 |
PF03124 (EXS) |
 Estimated TM-score=0.56; hard target. |
>EXS family (Length=326) LQIWGGFLLMILMAWLFSLNCLVWAKFKVSYKFIFEFNEHDALDFKQYMVIPSLLLFIGGLTAWFSFEDFWPNHFSGYDF PWVFLAAAVAILFCPFDVFFLSARLWLLSTLTRLLLSGLYPVEFRDFFMGDILCSLTYSISNVSMFFCIYAQHWNDCSNC GSAKSRLLGFLQCVPSIWRFLQCFRRYADTGDWFPHLANMGKYTVSMMYYMSLSLYRIDTVNHYRALLIFWATTNSIYSI VWDVVMDWSLFQLDSKYFLLRDEITFKEPMIYYAAIIADVVLRFQWVFYALFPKQIQQSAITSFGIAVAEVVRRFIWVFF RMENEH |
| 3629 |
PF02519 (Auxin_inducible) |
 Estimated TM-score=0.56; hard target. |
>Auxin responsive protein (Length=91) KKWQRMAALGRKRLTMKAKENEECCTSVAGKGHCVMYTADGSRFEVPLAYLGTAVFSELLRMSQEEFGFTSDGRIMLPCD AVVMDYAMCLL |
| 3630 |
PF02049 (FliE) |
 Estimated TM-score=0.56; easy target. |
>Flagellar hook-basal body complex protein FliE (Length=85) TNNTNTVTPHKTQSTFSNFLKDAINEVNETQINSDQLTEKLALGENVNLHDVMIASQKATITLQTTMEVRNKVVEAYQEI MRMQV |
| 3631 |
PF18868 (zf-C2H2_3rep) |
 Estimated TM-score=0.55; easy target. |
>Zinc finger C2H2-type, 3 repeats (Length=126) CAICLDSFTNKKVLETHVQERHHVQFVEQCMLLQCIPCGSHFGNTEELWLHVLSVHPVNFRQSKAGQQHNLSAGEDSPLK LELYNTAPMENNSENSGGFRKFICRFCGLKFDLLPDLGRHHQAAHM |
| 3632 |
PF18813 (PBECR4) |
 Estimated TM-score=0.55; very hard target. |
>phage-Barnase-EndoU-ColicinE5/D-RelE like nuclease4 (Length=172) IIVHAAKEYEEKLNNKHFLIVYQKKSLTKTVEIGFRDMNFLHLTGVKTKLSAQQFYAACLESKLAERDFEIDNKGKVQQK LMVLPCLSKLMSHHCLVGEFINSGVYIRADYFVGDTKAILSVGFRNGNRVDYPVTLYNEDVKRLSNPTNKVLAIFSKSYK EKCYNKCTYLSK |
| 3633 |
PF18803 (CxC2) |
 Estimated TM-score=0.55; very hard target. |
>CxC2 like cysteine cluster associated with KDZ transposases (Length=108) LKSLGLRIQLGHAPGQTCPNPHRAFNDDFVIIDSHGVHEVSLDFCDCTQAETHVQQLLRMSLFPSTTADPKTAATFRLLE EFHLLSFESKVSAYEFYNALTRRSDNTG |
| 3634 |
PF18745 (SNAD2) |
 Estimated TM-score=0.55; hard target. |
>Secreted Novel AID/APOBEC-like Deaminase 2 (Length=204) LYNIINFLRNNYGGDYQYALGINVPPRYCEQVAPLDPPDQNFLPSDNNAQRVKDDMAGDDKIYKGDQLIGARPKPIPHSQ NNNYHSEYLLLINSTSSKLEPKPLMETLLDSNPDGCAVFFTRNSPCVRTCSTPNGLYSIIPALRSQFQNHKGLKAFVFNR VWPRDENDPAWRNNIKTINDIIPVYRCDGNCTLCVNNNDVDRCC |
| 3635 |
PF18730 (HEPN_Cthe2314) |
 Estimated TM-score=0.55; hard target. |
>Cthe_2314-like HEPN (Length=178) WIRGLISSLDELEQSLFAASFFRKSVKAGYMDDMSVSEQGDYARYVYFYKNGFIRVFSLLDKLGTVLNTLYDLNTVRVKA HFSYFTVLRRFRSTKKHRELSGRLDEIKESYRDPMQKLRNRRNAEIHYMNSEMQDDLWQRHQGLHDKIRLEDLDEHLEDL RQGMEMVCKTLVEVFTYS |
| 3636 |
PF18717 (CxC4) |
 Estimated TM-score=0.55; very hard target. |
>CxC4 like cysteine cluster associated with KDZ transposases (Length=137) PLQQPPALIQLSEKASCPCLDGERAFFDPLQPTITCECKVYTLTEAFSVQIQLQQCPRCPQERHRYIGPEPRDIGLFNLN NSAIFTHELLNEYTSAFSSSETPFEPWVQTIARRYEEMQPSIPFVGGGLFRSVWFAY |
| 3637 |
PF18289 (HU-CCDC81_euk_2) |
 Estimated TM-score=0.55; easy target. |
>CCDC81 eukaryotic HU domain 2 (Length=72) PIVPLNIVIVSLESSLRRDVVQDCLRETLLYLLGLIVEERPITFLFNEIGILRIQNEKVHMTFFKEFLKVMD |
| 3638 |
PF18177 (La_HTH_kDCL) |
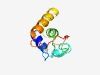 Estimated TM-score=0.55; hard target. |
>La HTH in kinetoplastid DICER domain (Length=89) VMRRLKKMLRFGSPYYEHFVRKGVWAHILCSDGWVPLQLIEEELAADTTTPVAKLPSKEKLKIVESLLCQHDNFQRFEVG YCLLRRSPF |
| 3639 |
PF18174 (HU-CCDC81_bac_1) |
 Estimated TM-score=0.55; easy target. |
>CCDC81-like prokaryotic HU domain 1 (Length=59) MIELERHIEILLLSNDCVIVPGFGGFMAHHVDARYDEQENLFLPPQRTIGFNPQLTLND |
| 3640 |
PF18160 (SLATT_5) |
 Estimated TM-score=0.55; hard target. |
>SMODS and SLOG-associating 2TM effector domain family 5 (Length=203) DKTFLEELNHKIWCTKGTRFNADSRFKKKSKLSNISVSILSAYLIIASLFSVYNINQNSDDNIISYLVTALSILLLVVSM HESNQDYKLRAFNFHSCGLELAEIYNRLRTFKTLEGEKSESEISESEISDFCFEINNKYQSILNKYDNHDDIDYDTFRIK NLDYFEEVFTEKEIKKIERKLNLNIYAWHLSMIIITPIIIIAL |
| 3641 |
PF18142 (SLATT_fungal) |
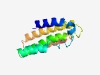 Estimated TM-score=0.55; hard target. |
>SMODS and SLOG-associating 2TM effector domain (Length=123) HAQKARDDFAKRAKLHGYTLNAAIGLQVLLGSLTTGLSAAATSGKSAAVQTTILGGLSTIVASYLARARGSNEPELSNLR VKDLDQFIRDCQALSMDHGYDTTGRFDNDLRVKRERFEELLGN |
| 3642 |
PF17733 (DUF5572) |
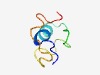 Estimated TM-score=0.55; hard target. |
>Family of unknown function (DUF5572) (Length=48) PYPTSFAHIVELITTGQPIPGIKEIPDTVLEGQGSASTASKRKKPWEK |
| 3643 |
PF17570 (DUF5476) |
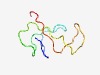 Estimated TM-score=0.55; very hard target. |
>Family of unknown function (DUF5476) (Length=61) MCFSPKVKVPKMDTNQVRAVEPAPLTQEVSGVEFGGSSDETDTEGTEVSGRKDLKVERDDS |
| 3644 |
PF17507 (DUF5439) |
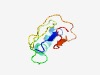 Estimated TM-score=0.55; very hard target. |
>Family of unknown function (DUF5439) (Length=75) MKYDVIVYWYIWKEFYQSRNKRLISHRNTKVNNLIIEDYSVTIVINWYSIIIIDKVLFIHDVMFICWHSSEIDVW |
| 3645 |
PF17376 (DUF5398) |
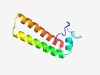 Estimated TM-score=0.55; easy target. |
>Family of unknown function (DUF5398) (Length=80) MFGLEDQKKKKTSEEFVFELEKDLKNPKKNKEIRQQVEERIQKIKEALRSGENQEEFDRFGLLLHGYTSLLKVISRFNPK |
| 3646 |
PF17342 (DUF5372) |
 Estimated TM-score=0.55; hard target. |
>Family of unknown function (DUF5372) (Length=78) VTHRFHPLFGRDFEFVAHRQNWGEDRVHLHDENGELFSLPAGWTDIAPVDPFVVVADGRCAFTTDGLLAVADLIDRLR |
| 3647 |
PF17338 (GP88) |
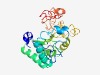 Estimated TM-score=0.55; hard target. |
>Gene product 88 (Length=231) LTQNSEMRQIGVWNWSLPAWAGRLPDGRTYNTCPSAGICAQACYARHGTYTWPVVRAKHQANLTYVLDDLAGWQAAMVAE LSATKFRGAWIRIHDSGDFFSDSYLQAWLDICRARPDTNFYAYTKEVSRFRALVEPDPPPNFLWVYSYGGTQDAALNPAV DRVADVFPDESAIAEAGWASQEASDLLAVLGPRLVGVPANRIPAYLKRLAGRRFSEWQAEVDAERAARRGR |
| 3648 |
PF17249 (DUF5318) |
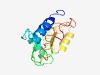 Estimated TM-score=0.55; hard target. |
>Family of unknown function (DUF5318) (Length=130) MSTQRQVVDYALQRRALLAEVYAGRVGTMEVCDASPYLLRAAKFHGRPSDVTCPVCRKEPLTHVSWVYGDELKHAAGSAR TADELARMATLFEEFTVYVVEVCRTCSWNHLVQSYVLGTRGLNTRKPRRR |
| 3649 |
PF17038 (CBP_BcsN) |
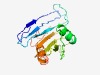 Estimated TM-score=0.55; very hard target. |
>Cellulose biosynthesis protein BcsN (Length=185) ALPPPGGPAVVSIVERRFSNSTQQDIFLFTSASTPGQNVLRVQMFGPVGLQMDGQKALGYSSVRASDIAKEMRRELPGVA LAQSPLYLQNNYGPFSYAYGRGHGNDACLYAWQQIRSPDEARTVFQNRGTIQVRLRLCEDGASEEKLLGTMYGYTIRGAF NAAGWNPYGEPPSVDPTLGRTGNPI |
| 3650 |
PF17032 (zinc_ribbon_15) |
 Estimated TM-score=0.55; hard target. |
>zinc-ribbon family (Length=83) RCINCNSKADLVKYDNVLKLFFVPVWKWPGKKPLLHCNTCNLFFPESLSKTTSSSLPSDNDVLRCHSCARLVDADYRFCP FCG |
| 3651 |
PF16998 (17kDa_Anti_2) |
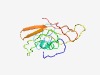 Estimated TM-score=0.55; hard target. |
>17 kDa outer membrane surface antigen (Length=114) GGMDILSSAKVDRSVATGTIPTAPVTTDSVSDETTVRNAVTSADLHKLNGQPVPWANASTGSAGVIDTIVENSASGQVCR QFRTTRHSYQGIANFSGRTCLLGQGEWQLLSFQQ |
| 3652 |
PF16996 (Asp4) |
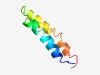 Estimated TM-score=0.55; hard target. |
>Accessory secretory protein Sec Asp4 (Length=55) KKDLFYKEVEGRMESLKRRPAEKEKTTRSEKINVTFNVIIGLVILLGVIFTLFRV |
| 3653 |
PF16995 (tRNA-synt_2_TM) |
 Estimated TM-score=0.55; hard target. |
>Transmembrane region of lysyl-tRNA synthetase (Length=210) RLHQVPHIAGLILGVFSVLVFLWSISPVLRYYVHVPREYVDTYYFDAPDTSLSWALVVALLAAALASRKRIAWWLLTIYL VLFLITNVIVSITDKNVNAMVAAVVQVVLIGILIAARPEFYTRVRRGAGWKALGVLIVGLAIGTLLGWGLVELFPGTLPQ NERFLWALNRVTALAAADNEQFTGRPHGFVNTLLGLFGAIALLAAVITLF |
| 3654 |
PF16977 (ApeC) |
 Estimated TM-score=0.55; hard target. |
>C-terminal domain of apextrin (Length=207) WPAGSYALPMANSGCPNSSGFTWHTGHRTEELENDGNRNAHSTSIHLKAKVGKPDITQHFCVKNSTATDKGKPKWPNGKY CIYKEGAFCPQEFYEGFVRWDDNNEKNGSNRNNKEGKLPAGVYNQDTLIYFCCKTTGSAIKRIALPVIKPFYLMAFGSTT CQEVQGAVYSLEYVVFDTENTDNMDNRVYPFPYGADRKEPTIHYCYY |
| 3655 |
PF16945 (Phage_r1t_holin) |
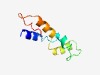 Estimated TM-score=0.55; hard target. |
>Putative lactococcus lactis phage r1t holin (Length=70) GNFLIDLADRVGRTFVGALVGAIPATGTLADIDWQAALSIAATTAIVSAITSILSINLGTAKGLPSLAPV |
| 3656 |
PF16722 (SAPIS-gp6) |
 Estimated TM-score=0.55; trivial target. |
>Pathogenicity island protein gp6 in Staphylococcus (Length=72) METKYELNNTKKVANAFGLNEEDTNLLINAVDLDIKNNMQEISSELQQSEQSKQKQYGTTLQNLAKQNRIIK |
| 3657 |
PF16665 (NCOA_u2) |
 Estimated TM-score=0.55; very hard target. |
>Unstructured region on nuclear receptor coactivator protein (Length=113) YGLNMNSPPHGSPGLGSNQQNIMISPRNRGSPKMPSHQFSPVAGMHSPMGSASNTGGNSFSSSSLSALQAISEGVGTSLL STLSSPGPKLDNSPNMNINQQSKIGSQDSKSPA |
| 3658 |
PF16447 (DUF5044) |
 Estimated TM-score=0.55; very hard target. |
>Domain of unknown function (DUF5044) (Length=179) PFLTARGALAATQERYAFGPGEVIARFSPQQEGMPPLRYYIVRDGDWFAWCSVEGDGLFWRPGALTAACPDGEAPLAILA SPTFGGTSPELVVVSSDPDIAAVELDYLVKSAIIDTVVYVHVEMARQEPLEDSCFYFSLEDAYSRLFYETPSTVFRVRYY DADGKLLYESPYPARWLEY |
| 3659 |
PF16434 (DUF5031) |
 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF5031) (Length=362) MKTIQYISRISILLIVGSIASSCVKEEVVTPIPALAPEGMPVAINFKDPKPSLADLKYSLYLFSHAKGSSDTYIMEELKN PLQNDSRLKFRNSELREKDYRFLFVATPGTTPEIQVYKNTLNNPPATGDNWEDIRIGILSPKLSLHNYYQVKDISGDELL ANDSVRGELSRMVGQMEFRFFKIGNDITNPIPIDLTGIYSIFDRIYKIDITYTGYTTSLSFGEGNILQPETSNNEEFKQE INVALNDRFGIEIPNGQLDTLAGGATAGGKFKGYCFLPANNNVRAVLKIYYYDTTPTCDNLIVPHTANCYEQRQLELRIP AEATAGLPIEANTYTVNSAGIRCNRIIDIHVDHNTNIFTQWD |
| 3660 |
PF16424 (DUF5021) |
 Estimated TM-score=0.55; very hard target. |
>Domain of unknown function (DUF5021) (Length=178) VTSANSTAASIKKQIDNFLTDADTAGYGMKQSSAAKANITFKINADGEWEASVVTGTYTGGAAGGALTDAFKTGGSIQWG ASAADITKDTPKSSAGDATALLTIDLASVFPDVKSSYIYAYCEGGKTLYVAYTADGNTKPTSMPGEADFKKGTYTWDGNT AGITSDGITLGTAPALSL |
| 3661 |
PF16334 (DUF4964) |
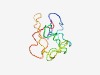 Estimated TM-score=0.55; very hard target. |
>Domain of unknown function (DUF4964) (Length=81) VFCSCETQVEQHEKNELRAPAYPLVTIDPYTSAWSTTDNLYDSPVKHWTGKDFPLLGVAKVDGQTYRFMGTEELELRPLV K |
| 3662 |
PF16266 (DUF4919) |
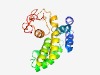 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4919) (Length=183) IESSIRDFNSEMYYPRLMRRYINNDTTLTLNEFRHLYLGYSFQESYNPYRISPHIEKLANLYARYEHSTTECDSIIKYAT LAVDDFPFDLRQINMLIYAYKQKKMKDIAAIWEYKLKNIVNAILSTGNGEKPETAWYVIYPTHEYDVVNRMGFTGAKYTF IEPTFDYLEVEKNPLNVKGYYFN |
| 3663 |
PF16110 (DUF4828) |
 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4828) (Length=78) FYAGTWQFVDPHNQRHHRLEISPNLDVHIDGRSLEVRVQELNEHQLTFQDKFGYHLVIHANEQQPISFFDEADDCTYT |
| 3664 |
PF16097 (FXR_C3) |
 Estimated TM-score=0.55; very hard target. |
>Fragile X-related 1 protein C-terminal region 3 (Length=69) PKDNLKKNKKEMGQVDLIEEHSTLEKMINGPSNGSGDELGKFQCTPTERTQKVTYQGDGNNQETVLNGV |
| 3665 |
PF15940 (YjcB) |
 Estimated TM-score=0.55; hard target. |
>Family of unknown function (Length=92) MGTLTASLVLMRWELVSAVMMFIASTFNVKCRQASHKGIAFVFTGVGIGMSCWFVTGLLGITLSMDNLHNFWHITKDVFI DVMSHTPTDYPM |
| 3666 |
PF15884 (QIL1) |
 Estimated TM-score=0.55; hard target. |
>MICOS complex subunit MIC13, QIL1 (Length=76) TIDKGIWKDSATTSAFYEELESGVSPYVGELKKQIPYELPPLPSNDRLSYLMKYYWNCGVKTTFNFLVDLPTHTSN |
| 3667 |
PF15805 (SCNM1_acidic) |
 Estimated TM-score=0.55; hard target. |
>Acidic C-terminal region of sodium channel modifier 1 SCNM1 (Length=46) PEKRRTLEHYLKLKSSGWIQDGSGKWVKDENVEFDSDEEEPPMLPP |
| 3668 |
PF15674 (CCDC23) |
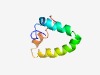 Estimated TM-score=0.55; easy target. |
>Coiled-coil domain-containing protein 23 (Length=63) PARKEKSKVKEPAFRVEKAKQKSAQQELKQRQRAEIYALNRVMTELEQQQFDEFCKQMQPPGE |
| 3669 |
PF15657 (Tox-HNH-EHHH) |
 Estimated TM-score=0.55; very hard target. |
>HNH/Endo VII superfamily nuclease toxins (Length=69) VLGENGRPVMTREYTYVRPDGSKVIIQDHSAGHRFGEGGIGDRGPHFNVRPPENTRSGHVPGTDDHYPF |
| 3670 |
PF15652 (Tox-SHH) |
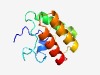 Estimated TM-score=0.55; hard target. |
>HNH/Endo VII superfamily toxin with a SHH signature (Length=102) NSPLENHHGVMDVWAKHNVPNYVSRGSNTPTVALTKEQHNATKKVYREWLFEKTGKKVGGKVNWKEVSPREIQELTEKMF DAANVPKEARQQYYNAFNQYNF |
| 3671 |
PF15647 (Tox-REase-3) |
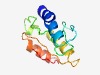 Estimated TM-score=0.55; hard target. |
>Restriction endonuclease fold toxin 3 (Length=106) QNAKYREALDIHYEDLIRRKTDGSSKFINGREIDAVTNDALIQAKRTISAIDKPKNFLNQKNRKQIKATIEAANQQGKRA EFWFKYGVHSQVKSYIESKGGIVKTG |
| 3672 |
PF15640 (Tox-MPTase4) |
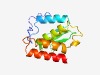 Estimated TM-score=0.55; hard target. |
>Metallopeptidase toxin 4 (Length=132) EEMPVSGQRRLTSTGHRIMDISEMKSFKKEMGAIDIKVLIDKKDKILPENAAAGFNPATGEIVLRKEPTNLSAIHESYHA KQWKELGKDNYLQQSTLEREEFVYNEIMKNKVLFNDGEILFSQKYIYKLRNG |
| 3673 |
PF15606 (Ntox34) |
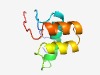 Estimated TM-score=0.55; hard target. |
>Bacterial toxin 34 (Length=75) PGLALDITMIASRGNVADTGITDRVNDIINDRFWSDGKKPDRCDVLQELIDCGDISAKDAKSTQKAWNCRHSRQS |
| 3674 |
PF15574 (Imm48) |
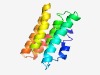 Estimated TM-score=0.55; hard target. |
>Immunity protein 48 (Length=123) YVSESLRIANDIIQLVKIDLKDEMNRQILASYIFGVLNAKAIQESISPIDVQVTMIRVGIEALGYSPEAATQMTQFVIDA TDKNFHPTVYAIIHRGIEAFYLYSNEKYEQLKEDFDNIMASIK |
| 3675 |
PF15536 (Ntox3) |
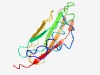 Estimated TM-score=0.55; hard target. |
>Bacterial toxin 3 (Length=132) RAGIAVPGAEARYERQEPTATAQQRARIEASPDVRVAVPGTRITYTLVRDAELHAQGAHYQYQWYFLNDPETSQTLGHPA RVEASEGPRVDARARFVGDHKIICKEVYHPADGDPQAPVFYEVPLRVVSEGD |
| 3676 |
PF15529 (Ntox24) |
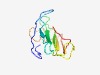 Estimated TM-score=0.55; hard target. |
>Bacterial toxin 24 (Length=94) KGTGGRAGNNLPVENGPKDGTLYKTDPQTGKVTNYTTYDSEGRAVKRVDLEGRPHGGVDTPHVVEYERNTNPKTGQVFVR PSNEVRAAFPWEIP |
| 3677 |
PF15506 (OCC1) |
 Estimated TM-score=0.55; very hard target. |
>OCC1 family (Length=60) MGCGNSTSASTAAGPSESAIDVQDDSSMDDEKRRNYGGVYVGLPADLSNVATGQTPSTQK |
| 3678 |
PF15451 (DUF4632) |
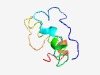 Estimated TM-score=0.55; very hard target. |
>Domain of unknown function (DUF4632) (Length=71) AGNPAKESSDADGEAEEEGESEKGAGPRSAGWRALRRLWDRVLAPARRWRRPLPSNVLYCPEIKDIAHLTR |
| 3679 |
PF15386 (Tantalus) |
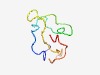 Estimated TM-score=0.55; very hard target. |
>Drosophila Tantalus-like (Length=58) LTPMGLPRPKRLKKKEFSLEEIYTNKNYKSPPANRCLETIFEEPKERNGTLISVSQQK |
| 3680 |
PF15165 (REC114-like) |
 Estimated TM-score=0.55; easy target. |
>Meiotic recombination protein REC114-like (Length=241) WPLQRYARCIPSNTRDPPGPCLEAGTAPCPTWKVFDSNEESGYLVLTIVISGHFFIFQGQTLLEGFSLIGSKDWLKIVRR VDCLLFGTTIKDKSRLFRVQFSGESKEQALEHCCSCVQKLAQYITVQVPDGNIQELQLIPGPPRATESQGKDSAKSVPRQ PGSHQHSEQQQVCVTAGTGAPDGRTSLTQLAQTLLASEELPHVYEQSAWGAEELGPFLRLCLMDQNFPAFVEEVEKELKK L |
| 3681 |
PF15159 (PIG-Y) |
 Estimated TM-score=0.55; hard target. |
>Phosphatidylinositol N-acetylglucosaminyltransferase subunit Y (Length=69) GWFLLFAAYFIFVFSMYAIVVSKFVPETGNKTLDWIKKDEYYCLLVPLTLPVTIYAVFFNWLGMKFFRH |
| 3682 |
PF15022 (DUF4522) |
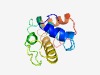 Estimated TM-score=0.55; very hard target. |
>Protein of unknown function (DUF4522) (Length=117) MAYGLPRKNTVKTILRGSCYNVQEPWDLALLTETWYTNLANIKLPFLGEISFGSPLHLKKTKTIKNALLPSAESMKLERE YEMKRLNRLKCQENVAEEIQLSLRERQVGLRRPLPPK |
| 3683 |
PF14998 (Ripply) |
 Estimated TM-score=0.55; hard target. |
>Transcription Regulator (Length=84) FWRPWIATPKDAERERGQNHSALCSLPEITENSAKLTQYTHPVRLFWPKSKCFDYLYQEAEALLRNFPVQATISFYEDSE SEEE |
| 3684 |
PF14980 (TIP39) |
 Estimated TM-score=0.55; hard target. |
>TIP39 peptide (Length=50) DWPSRVGHQQKRNIVVADDAAFREKSKLLTAMERQKWLNSYMQKLLVVNS |
| 3685 |
PF14974 (P_C10) |
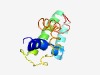 Estimated TM-score=0.55; hard target. |
>Protein C10 (Length=103) FTLETAKAILTDILTALNTPENLQKLAEAKENSGNEMLKMMQFVFPLVTQIQMDVIKNYGFPEGREGTVQFAQLIRTLER EDAEVAQLHSQVRSYFLPPVRIN |
| 3686 |
PF14908 (HU-CCDC81_euk_1) |
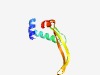 Estimated TM-score=0.55; easy target. |
>CCDC81 eukaryotic HU domain 1 (Length=85) IEENCKLFYPTLKNLHVDEITNIWDKVSEFVHHNLCMNKGVQIPGLGIFSFARIKLDLSGNKFLVTQKPVFLLSEKLCQA YRLKQ |
| 3687 |
PF14797 (SEEEED) |
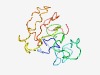 Estimated TM-score=0.55; very hard target. |
>Serine-rich region of AP3B1, clathrin-adaptor complex (Length=130) AKKFYSESEEEEDSSDSSSDSESESGSESGEQGESGEEGDSNEDSSEDSSSEQDSESGRESGLENKRTAKRNSKAKGKSD SEDGEKENEKSKTSDSSNAESSSIEDSSSDSESESESESESESRKVTQEK |
| 3688 |
PF14736 (N_Asn_amidohyd) |
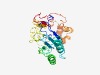 Estimated TM-score=0.55; hard target. |
>Protein N-terminal asparagine amidohydrolase (Length=265) VEVDPKRLLYVQQREFAATTPADDCVSVIGSDDATTCHLVVLRHTGSGAVCLAHCDGSSTWSEIQLITKAVTSLSNVRDD GRFELHLVGGFNDDAKTSQKLSLSILAAFQKLEEEIHLETCCITEMNDAVVNGIHRPTVYGIGVNVKTGEVFPASFPHKG PAEELRSARTFTGGEMAGIYDSSRGLVEIGPCHWTSDMDIAFWLSQSDDTILKYLSTSPEAEPPHFVQHMKSTIQFLLSH PDSDSLFPGGQPQLYRRSESGAWER |
| 3689 |
PF14545 (DBB) |
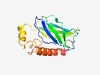 Estimated TM-score=0.55; hard target. |
>Dof, BCAP, and BANK (DBB) motif, (Length=139) VVQPDRIRCGAETTVYVIVRCKLDEKVSTEVEFSPEDSPSVRMEGTLENEYTVSVKAPDLSSGNVSLKVYSGDLVVCETT ISYYTDMEEIGNLLSSAANPVEFMCQAFKIVPYNTETLDKLLTESLKNNIPASGLHLFG |
| 3690 |
PF14342 (DUF4396) |
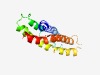 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4396) (Length=143) WQSVGVGASHCGSGCTLGDIIAEWFVFFVPIVIFGSHVVGTWVLDYIVAFVFGIAFQYFTIKPMRELSVGEGLKAAVKAD TLSLTAWQIGMYGWMAISFFLLFSPETLRPNNPVFWFMMQIGMVCGFLTSYPVNWWLLSKKIK |
| 3691 |
PF14250 (AbrB-like) |
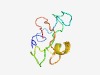 Estimated TM-score=0.55; easy target. |
>AbrB-like transcriptional regulator (Length=69) KFYNALIEAEGVELDSNTSAQGRGGRAATYRISVQSNGNLLIGSAYTKKMNLKPGDEFEISLGRKHIHL |
| 3692 |
PF14184 (YrvL) |
 Estimated TM-score=0.55; hard target. |
>Regulatory protein YrvL (Length=126) EKMKTFIICGMIFMGMLSIIALVCGAIMRFLGFQYESAGSIVLFFTIATVISYPLNLVAGALPKALYELKRISKLCALVL YLILDTTATSLGLTAVDNCMSSVSATTISIVIISFLLALPGKDDFK |
| 3693 |
PF14178 (YppF) |
 Estimated TM-score=0.55; easy target. |
>YppF-like protein (Length=59) MNMAELKNKFIAVKDYEPTDMNELLCFARQMYLRGEITIAQYRDLVRELELAGAYKPDN |
| 3694 |
PF14164 (YqzH) |
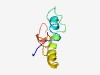 Estimated TM-score=0.55; hard target. |
>YqzH-like protein (Length=64) MDERWLQKQVRNCLRHYGYDDDMMPLGEDDWRELCTRIAAAKERQPNADIYELVHDIVYRFITS |
| 3695 |
PF14133 (DUF4300) |
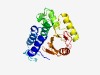 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4300) (Length=252) TYSNLVDSLSQDEVKKAMESAGITSENIDLFFQSVNDFNTTIEKKSLVKDGFITVDNLKPEYDEVAIQEIWNAKNPKFIG YNCRITSFALMKDLISIGNPNTKNSNNLFMDEDALENSPKKLFNPVEQKYFESLFSFVPTENTKDISIHLNKVKEDWKSK DITFSNKKKASLISVFFHSHEDSYLFIGHAGVLIPTKDGKLLFIEKLSFNEPYQVIKFDNRIELNDYLMNKYDVSWNQPN AKPFIMENDQLL |
| 3696 |
PF14123 (DUF4290) |
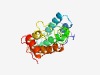 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4290) (Length=173) MDYNTNEKRLVLPEYGRNIQNMVDYCLTIQDREERKKCANTIINIMGNMFPHLRDVNDFKHILWDHLAIMADFKLDIDYP YEIIEKENLHTRPPRIPYNSSRIRYRHYGRTLEMMIQKAVELEDGEERDALIRLLANHMKKSFMAWNKESVDDRKILSDL AELSEGKITLNEE |
| 3697 |
PF14030 (DUF4245) |
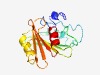 Estimated TM-score=0.55; hard target. |
>Protein of unknown function (DUF4245) (Length=165) RGTQTVRDMFLSMAVIGLVVAGIYMFIPHDDKADPVKAVDYRVELATARRAAPYPVVAPAGLADTWKPTSVSYKRQSADA WHLGYLAPDGQYVAVEQSTAPPAKYVSDVTQRAEDTGRTQQVAGETWQRWKGPKYDALVRADGDATTVVTGTASYERLAE MAAAL |
| 3698 |
PF14029 (DUF4244) |
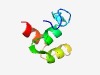 Estimated TM-score=0.55; hard target. |
>Protein of unknown function (DUF4244) (Length=50) RRMRSDAGMTTSEYAVGTIAACAFAAVLYKVVTSGTVQGALQSLIERALS |
| 3699 |
PF13952 (DUF4216) |
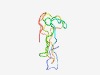 Estimated TM-score=0.55; hard target. |
>Domain of unknown function (DUF4216) (Length=77) LEIQYMNNKSVMLFQCDWFEVPPQGRSQSRGYKKDEYGFVCVDITRLHYTSDPFILGSQAQSVYYVKHGQSEKWHSV |
| 3700 |
PF13906 (AA_permease_C) |
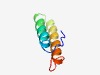 Estimated TM-score=0.55; easy target. |
>C-terminus of AA_permease (Length=51) FKVPLVPLLPLISVFFNLYLMFQLDTGTWIRFSVWIAVGYFIYFSYGIRHS |
[an error occurred while processing this directive]
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218