

You can:
| Name | Proteinase-activated receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | F2R |
| Synonym | Coagulation factor II receptor PAR-1 coagulation factor II (thrombin) receptor protease-activated receptor 1 Thrombin receptor [ Show all ] |
| Disease | Thrombosis Restenosis Myocardial infarction Cancer Atherosclerosis [ Show all ] |
| Length | 425 |
| Amino acid sequence | MGPRRLLLVAACFSLCGPLLSARTRARRPESKATNATLDPRSFLLRNPNDKYEPFWEDEEKNESGLTEYRLVSINKSSPLQKQLPAFISEDASGYLTSSWLTLFVPSVYTGVFVVSLPLNIMAIVVFILKMKVKKPAVVYMLHLATADVLFVSVLPFKISYYFSGSDWQFGSELCRFVTAAFYCNMYASILLMTVISIDRFLAVVYPMQSLSWRTLGRASFTCLAIWALAIAGVVPLLLKEQTIQVPGLNITTCHDVLNETLLEGYYAYYFSAFSAVFFFVPLIISTVCYVSIIRCLSSSAVANRSKKSRALFLSAAVFCIFIICFGPTNVLLIAHYSFLSHTSTTEAAYFAYLLCVCVSSISCCIDPLIYYYASSECQRYVYSILCCKESSDPSSYNSSGQLMASKMDTCSSNLNNSIYKKLLT |
| UniProt | P25116 |
| Protein Data Bank | 3vw7 |
| GPCR-HGmod model | P25116 |
| 3D structure model | This structure is from PDB ID 3vw7. |
| BioLiP | BL0217099 |
| Therapeutic Target Database | T36483 |
| ChEMBL | CHEMBL3974 |
| IUPHAR | 347 |
| DrugBank | BE0000928 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 535927 | 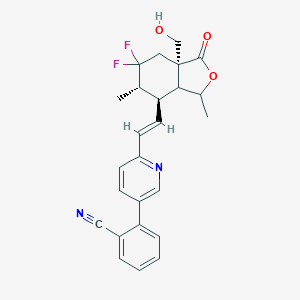 CHEMBL3930467 CHEMBL3930467 | C25H24F2N2O3 | 438.475 | 7 / 1 | 4.5 | Yes |
| 585 | 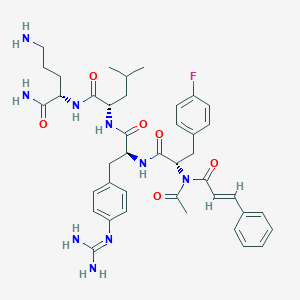 CHEMBL3143254 CHEMBL3143254 | C41H52FN9O6 | 785.922 | 9 / 7 | 2.7 | No |
| 586 | 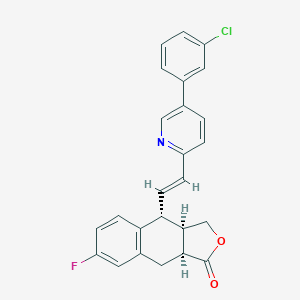 CHEMBL231713 CHEMBL231713 | C25H19ClFNO2 | 419.88 | 4 / 0 | 5.5 | No |
| 1903 | 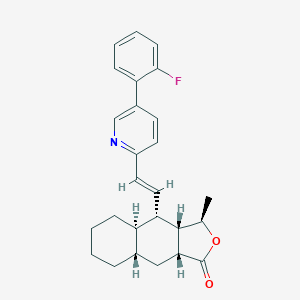 CHEMBL198473 CHEMBL198473 | C26H28FNO2 | 405.513 | 4 / 0 | 6.2 | No |
| 3495 | 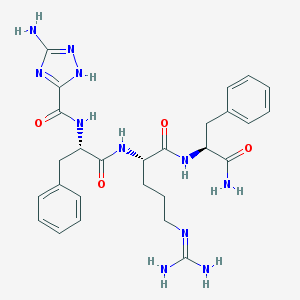 CHEMBL33377 CHEMBL33377 | C27H35N11O4 | 577.65 | 8 / 8 | 0.1 | No |
| 4075 | 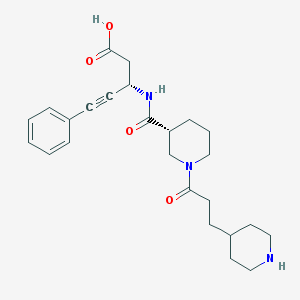 CHEMBL307065 CHEMBL307065 | C25H33N3O4 | 439.556 | 5 / 3 | -0.8 | Yes |
| 4076 | 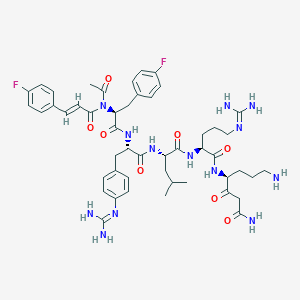 CHEMBL3143278 CHEMBL3143278 | C49H65F2N13O8 | 1002.14 | 13 / 10 | 1.2 | No |
| 5413 | 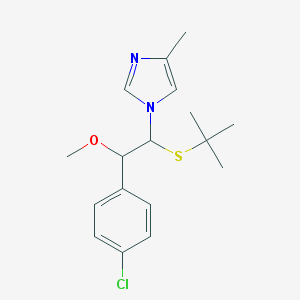 CHEMBL597091 CHEMBL597091 | C17H23ClN2OS | 338.894 | 3 / 0 | 4.1 | Yes |
| 459271 |  CHEMBL3704460 CHEMBL3704460 | C20H25F5N6O3S | 524.511 | 12 / 1 | 5.9 | No |
| 6033 |  CHEMBL338639 CHEMBL338639 | C33H58N12O5 | 702.906 | 8 / 10 | -0.6 | No |
| 6167 |  CHEMBL596706 CHEMBL596706 | C19H25ClN2OS | 364.932 | 3 / 0 | 4.9 | Yes |
| 6826 |  CHEMBL2012502 CHEMBL2012502 | C26H31NO3 | 405.538 | 4 / 1 | 5.4 | No |
| 7116 |  CHEMBL287469 CHEMBL287469 | C24H39N13O4 | 573.663 | 9 / 9 | -0.6 | No |
| 7434 |  CHEMBL232333 CHEMBL232333 | C25H18ClF2NO2 | 437.871 | 5 / 0 | 5.6 | No |
| 7637 |  CHEMBL91987 CHEMBL91987 | C25H31Cl2N3O3 | 492.441 | 3 / 3 | 4.7 | Yes |
| 7704 |  2-bromo-N-[3-(pentanoylamino)phenyl]benzamide 2-bromo-N-[3-(pentanoylamino)phenyl]benzamide | C18H19BrN2O2 | 375.266 | 2 / 2 | 4.0 | Yes |
| 8012 | 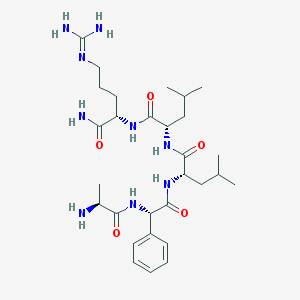 CHEMBL132128 CHEMBL132128 | C29H49N9O5 | 603.769 | 7 / 8 | 0.4 | No |
| 536177 | 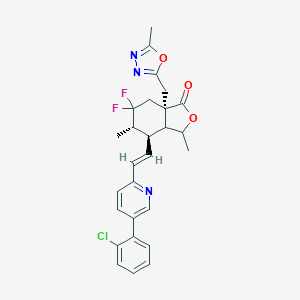 CHEMBL3954641 CHEMBL3954641 | C27H26ClF2N3O3 | 513.97 | 8 / 0 | 5.8 | No |
| 8544 | 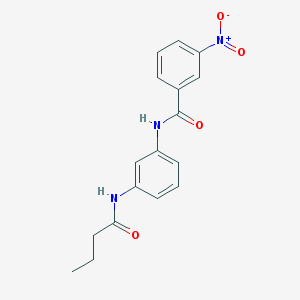 CHEMBL2048428 CHEMBL2048428 | C17H17N3O4 | 327.34 | 4 / 2 | 2.6 | Yes |
| 8913 | 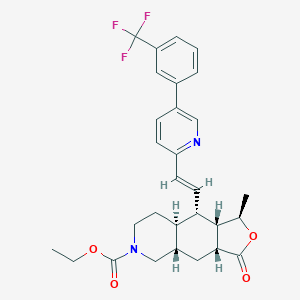 CHEMBL410157 CHEMBL410157 | C29H31F3N2O4 | 528.572 | 8 / 0 | 5.5 | No |
| 9015 |  CHEMBL411798 CHEMBL411798 | C61H64FN15O11 | 1202.28 | 17 / 11 | 5.2 | No |
| 9057 |  CHEMBL340389 CHEMBL340389 | C31H53N9O6 | 647.822 | 8 / 8 | -0.2 | No |
| 459299 |  SCHEMBL1204282 SCHEMBL1204282 | C22H27ClN6O3 | 458.947 | 7 / 1 | 3.2 | Yes |
| 9688 |  CHEMBL2012501 CHEMBL2012501 | C26H31NO3 | 405.538 | 4 / 1 | 5.4 | No |
| 10842 |  CHEMBL371751 CHEMBL371751 | C26H28ClNO2 | 421.965 | 3 / 0 | 6.7 | No |
| 11309 |  CHEMBL337852 CHEMBL337852 | C30H52N10O5 | 632.811 | 8 / 9 | -1.0 | No |
| 11760 |  CHEMBL71411 CHEMBL71411 | C39H41Cl2F2N7O3S | 796.76 | 8 / 5 | 5.8 | No |
| 536331 |  CHEMBL3945945 CHEMBL3945945 | C25H22F2N6O2 | 476.488 | 9 / 0 | 4.2 | Yes |
| 12990 | 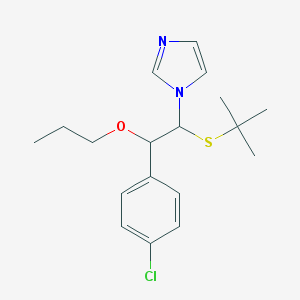 CHEMBL590626 CHEMBL590626 | C18H25ClN2OS | 352.921 | 3 / 0 | 4.6 | Yes |
| 13295 | 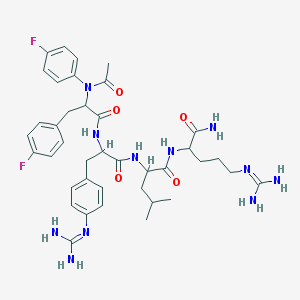 CHEMBL147872 CHEMBL147872 | C39H51F2N11O5 | 791.906 | 9 / 8 | 1.9 | No |
| 13296 | 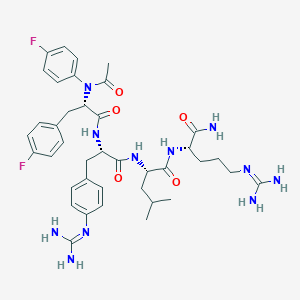 CHEMBL3143306 CHEMBL3143306 | C39H51F2N11O5 | 791.906 | 9 / 8 | 1.9 | No |
| 14385 | 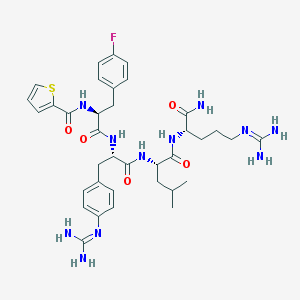 CHEMBL3143277 CHEMBL3143277 | C36H48FN11O5S | 765.91 | 9 / 9 | 1.7 | No |
| 14386 |  CHEMBL144504 CHEMBL144504 | C36H48FN11O5S | 765.91 | 9 / 9 | 1.7 | No |
| 15166 |  CHEMBL132254 CHEMBL132254 | C31H53N9O5 | 631.823 | 7 / 7 | 0.8 | No |
| 15224 |  CHEMBL98231 CHEMBL98231 | C30H42N4O2 | 490.692 | 4 / 2 | 6.3 | No |
| 15615 |  CHEMBL415138 CHEMBL415138 | C49H77N15O15 | 1116.24 | 16 / 15 | -4.8 | No |
| 15750 | 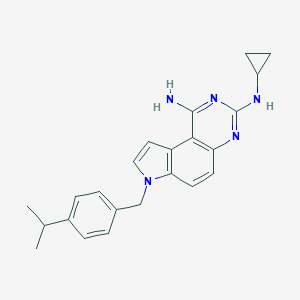 CHEMBL63426 CHEMBL63426 | C23H25N5 | 371.488 | 4 / 2 | 4.9 | Yes |
| 15902 | 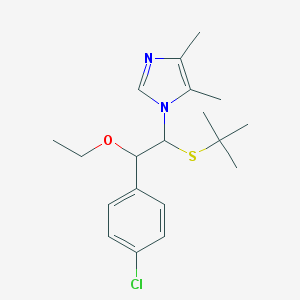 CHEMBL596686 CHEMBL596686 | C19H27ClN2OS | 366.948 | 3 / 0 | 4.9 | Yes |
| 16112 | 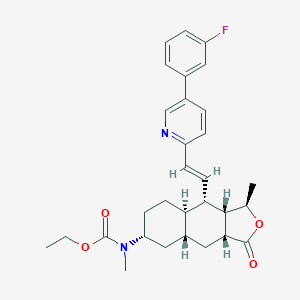 CHEMBL500551 CHEMBL500551 | C30H35FN2O4 | 506.618 | 6 / 0 | 5.5 | No |
| 16705 | 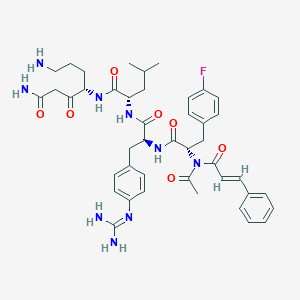 CHEMBL3143253 CHEMBL3143253 | C43H54FN9O7 | 827.959 | 10 / 7 | 2.5 | No |
| 17521 | 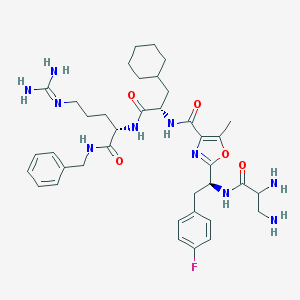 CHEMBL42224 CHEMBL42224 | C38H53FN10O5 | 748.905 | 10 / 8 | 2.3 | No |
| 17841 | 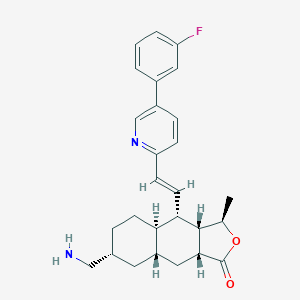 CHEMBL3109580 CHEMBL3109580 | C27H31FN2O2 | 434.555 | 5 / 1 | 4.6 | Yes |
| 18575 | 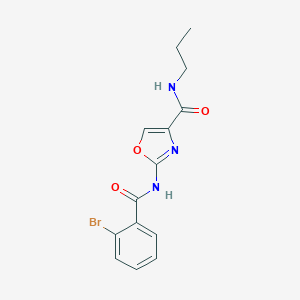 CHEMBL2047280 CHEMBL2047280 | C14H14BrN3O3 | 352.188 | 4 / 2 | 2.7 | Yes |
| 536481 | 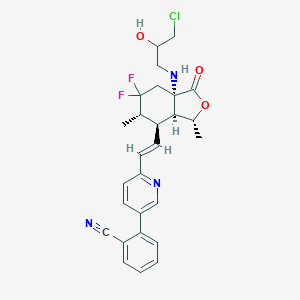 CHEMBL3952681 CHEMBL3952681 | C27H28ClF2N3O3 | 515.986 | 8 / 2 | 4.3 | No |
| 20491 | 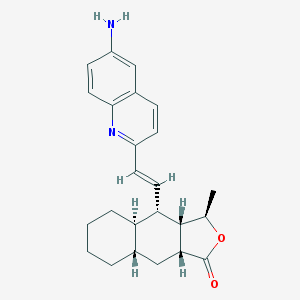 CHEMBL204955 CHEMBL204955 | C24H28N2O2 | 376.5 | 4 / 1 | 5.1 | No |
| 20630 | 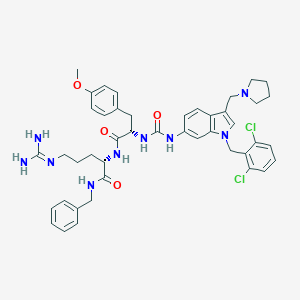 CHEMBL7610 CHEMBL7610 | C44H51Cl2N9O4 | 840.851 | 6 / 6 | 5.5 | No |
| 20690 | 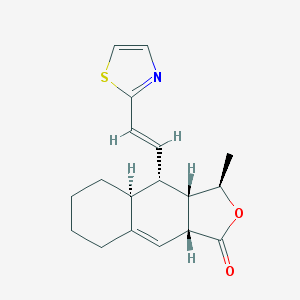 CHEMBL232120 CHEMBL232120 | C18H21NO2S | 315.431 | 4 / 0 | 3.9 | Yes |
| 20720 | 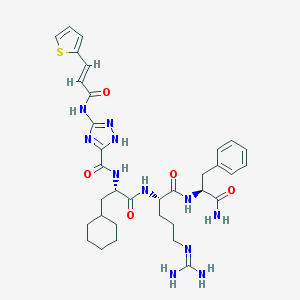 CHEMBL278216 CHEMBL278216 | C34H45N11O5S | 719.866 | 9 / 8 | 2.9 | No |
| 20747 | 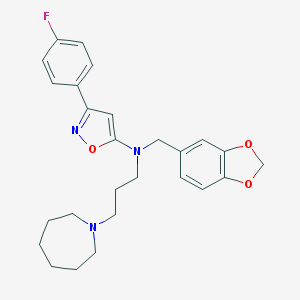 CHEMBL323054 CHEMBL323054 | C26H30FN3O3 | 451.542 | 7 / 0 | 5.5 | No |
| 20774 | 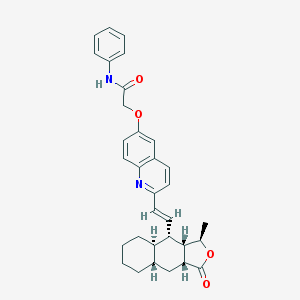 CHEMBL382830 CHEMBL382830 | C32H34N2O4 | 510.634 | 5 / 1 | 6.7 | No |
| 21520 | 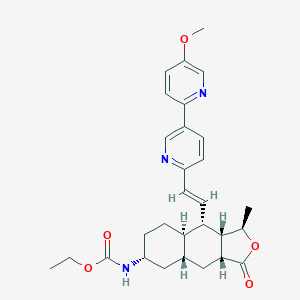 CHEMBL1271353 CHEMBL1271353 | C29H35N3O5 | 505.615 | 7 / 1 | 4.1 | No |
| 21532 | 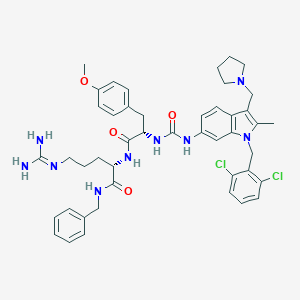 CHEMBL504651 CHEMBL504651 | C45H53Cl2N9O4 | 854.878 | 6 / 6 | 5.9 | No |
| 21991 | 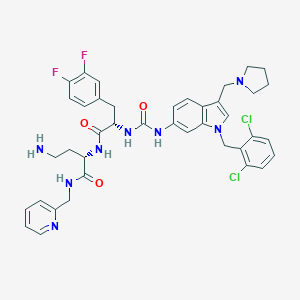 CHEMBL308052 CHEMBL308052 | C40H42Cl2F2N8O3 | 791.726 | 8 / 5 | 5.0 | No |
| 21995 | 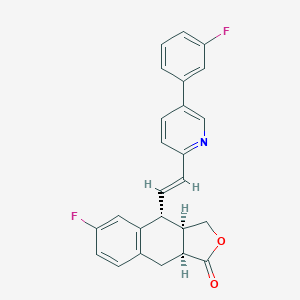 CHEMBL269006 CHEMBL269006 | C25H19F2NO2 | 403.429 | 5 / 0 | 5.0 | Yes |
| 22154 | 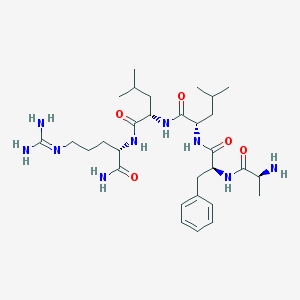 CHEMBL337875 CHEMBL337875 | C30H51N9O5 | 617.796 | 7 / 8 | -0.8 | No |
| 22155 | 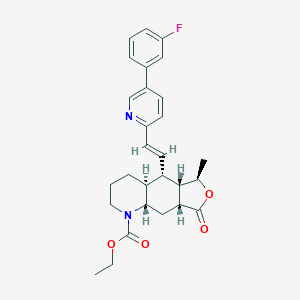 CHEMBL244484 CHEMBL244484 | C28H31FN2O4 | 478.564 | 6 / 0 | 4.8 | Yes |
| 536556 |  CHEMBL3909563 CHEMBL3909563 | C25H23ClF2N2O2 | 456.918 | 6 / 0 | 5.3 | No |
| 22816 |  CHEMBL326456 CHEMBL326456 | C26H30ClN3O3 | 467.994 | 6 / 0 | 6.0 | No |
| 23669 |  CHEMBL198175 CHEMBL198175 | C27H31NO3 | 417.549 | 4 / 0 | 6.1 | No |
| 24412 |  CHEMBL275946 CHEMBL275946 | C43H50FN9O3 | 759.931 | 6 / 6 | 4.4 | No |
| 24517 | 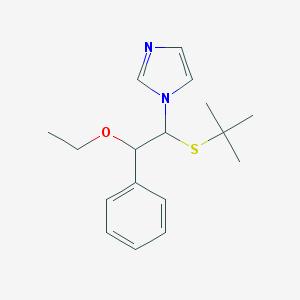 CHEMBL599363 CHEMBL599363 | C17H24N2OS | 304.452 | 3 / 0 | 3.5 | Yes |
| 24550 | 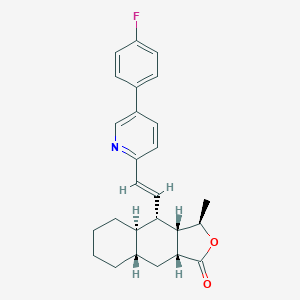 CHEMBL199101 CHEMBL199101 | C26H28FNO2 | 405.513 | 4 / 0 | 6.2 | No |
| 24600 | 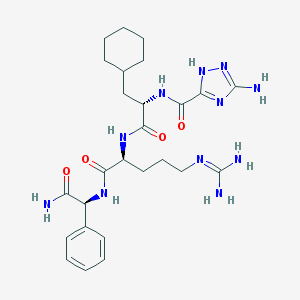 CHEMBL33940 CHEMBL33940 | C26H39N11O4 | 569.671 | 8 / 8 | 0.9 | No |
| 24637 | 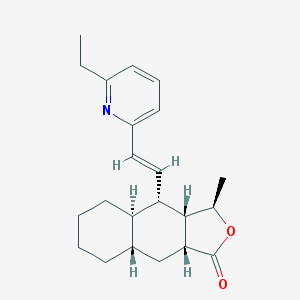 CHEMBL197791 CHEMBL197791 | C22H29NO2 | 339.479 | 3 / 0 | 5.3 | No |
| 24638 |  CHEMBL195909 CHEMBL195909 | C22H29NO2 | 339.479 | 3 / 0 | 5.3 | No |
| 25333 |  CHEMBL1081087 CHEMBL1081087 | C18H14F2N2O | 312.32 | 5 / 0 | 3.5 | Yes |
| 459453 |  CHEMBL342337 CHEMBL342337 | C40H53FN12O6 | 816.94 | 10 / 9 | 1.5 | No |
| 27034 |  CHEMBL3143263 CHEMBL3143263 | C40H53FN12O6 | 816.94 | 10 / 9 | 1.5 | No |
| 27273 | 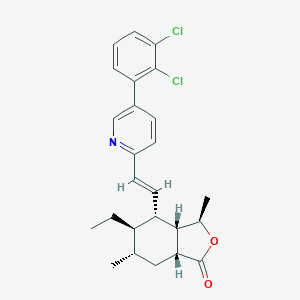 CHEMBL2012497 CHEMBL2012497 | C25H27Cl2NO2 | 444.396 | 3 / 0 | 7.0 | No |
| 28274 | 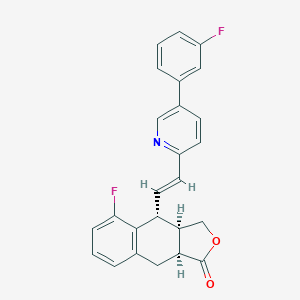 CHEMBL233163 CHEMBL233163 | C25H19F2NO2 | 403.429 | 5 / 0 | 5.0 | Yes |
| 28418 | 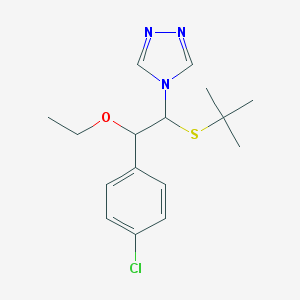 CHEMBL596676 CHEMBL596676 | C16H22ClN3OS | 339.882 | 4 / 0 | 3.5 | Yes |
| 536735 | 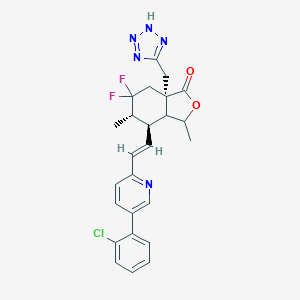 CHEMBL3964337 CHEMBL3964337 | C25H24ClF2N5O2 | 499.947 | 8 / 1 | 5.1 | No |
| 30189 |  2-methyl-N-[2-methyl-3-(propionylamino)phenyl]benzamide 2-methyl-N-[2-methyl-3-(propionylamino)phenyl]benzamide | C18H20N2O2 | 296.37 | 2 / 2 | 2.9 | Yes |
| 30268 |  CHEMBL436130 CHEMBL436130 | C27H31NO2 | 401.55 | 3 / 0 | 6.5 | No |
| 31298 |  CHEMBL3091980 CHEMBL3091980 | C24H29FN2O2S | 428.566 | 5 / 0 | 5.2 | No |
| 31322 |  CHEMBL198375 CHEMBL198375 | C26H28BrNO2 | 466.419 | 3 / 0 | 6.8 | No |
| 536820 | 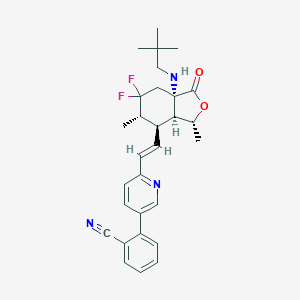 CHEMBL3924976 CHEMBL3924976 | C29H33F2N3O2 | 493.599 | 7 / 1 | 6.1 | No |
| 32083 | 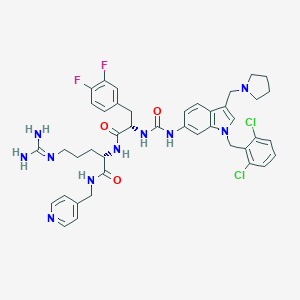 CHEMBL267059 CHEMBL267059 | C42H46Cl2F2N10O3 | 847.794 | 8 / 6 | 4.7 | No |
| 32913 | 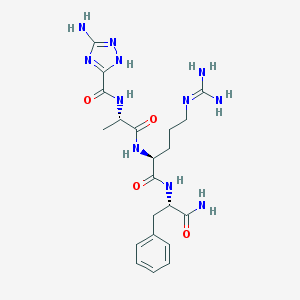 CHEMBL287156 CHEMBL287156 | C21H31N11O4 | 501.552 | 8 / 8 | -1.5 | No |
| 33244 | 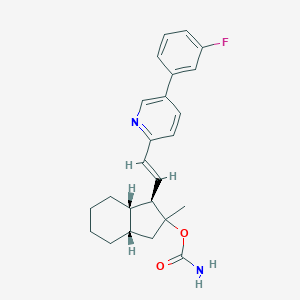 CHEMBL3091993 CHEMBL3091993 | C24H27FN2O2 | 394.49 | 4 / 1 | 5.5 | No |
| 33319 | 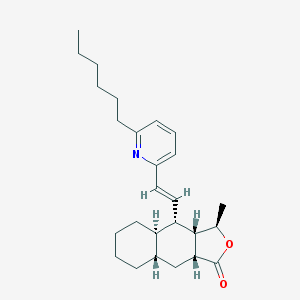 CHEMBL382733 CHEMBL382733 | C26H37NO2 | 395.587 | 3 / 0 | 7.3 | No |
| 33322 | 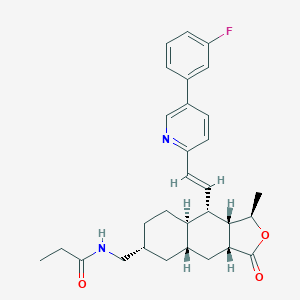 CHEMBL3109584 CHEMBL3109584 | C30H35FN2O3 | 490.619 | 5 / 1 | 5.4 | No |
| 33574 | 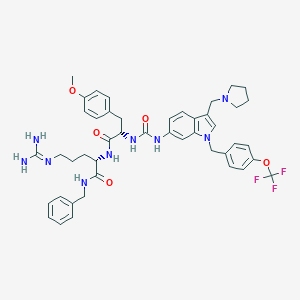 CHEMBL268328 CHEMBL268328 | C45H52F3N9O5 | 855.964 | 10 / 6 | 5.5 | No |
| 33647 | 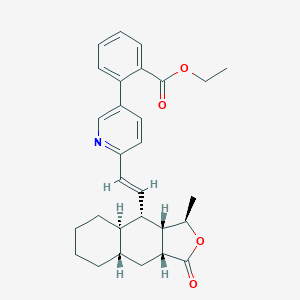 CHEMBL383606 CHEMBL383606 | C29H33NO4 | 459.586 | 5 / 0 | 6.3 | No |
| 34568 |  CHEMBL3143281 CHEMBL3143281 | C48H65F2N15O7 | 1002.14 | 12 / 11 | 0.7 | No |
| 35157 |  CHEMBL219282 CHEMBL219282 | C27H31NO3 | 417.549 | 4 / 1 | 4.8 | Yes |
| 35877 |  CHEMBL2049112 CHEMBL2049112 | C18H19BrN2O2 | 375.266 | 2 / 2 | 3.8 | Yes |
| 36360 |  CHEMBL108604 CHEMBL108604 | C29H33N3O | 439.603 | 4 / 0 | 6.8 | No |
| 36434 | 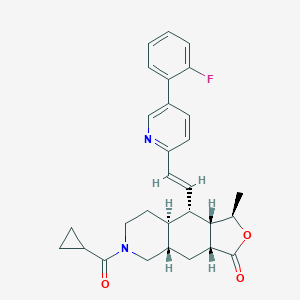 CHEMBL244299 CHEMBL244299 | C29H31FN2O3 | 474.576 | 5 / 0 | 4.4 | Yes |
| 37285 | 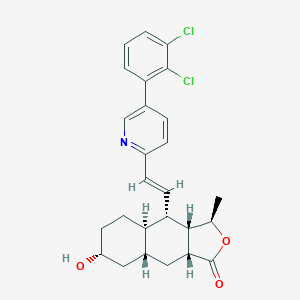 CHEMBL374503 CHEMBL374503 | C26H27Cl2NO3 | 472.406 | 4 / 1 | 5.7 | No |
| 37540 | 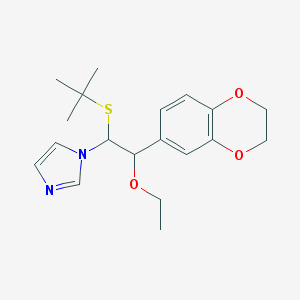 CHEMBL598962 CHEMBL598962 | C19H26N2O3S | 362.488 | 5 / 0 | 3.2 | Yes |
| 37898 | 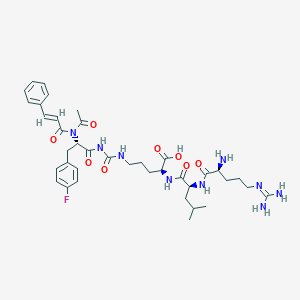 CHEMBL3143312 CHEMBL3143312 | C38H52FN9O8 | 781.887 | 11 / 8 | -0.1 | No |
| 37943 | 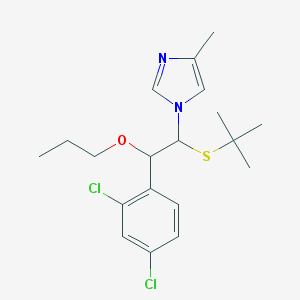 CHEMBL604175 CHEMBL604175 | C19H26Cl2N2OS | 401.39 | 3 / 0 | 5.6 | No |
| 39130 | 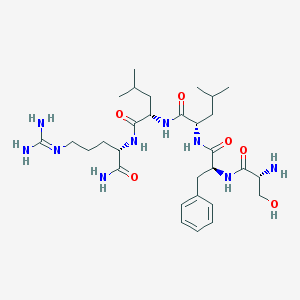 CHEMBL130226 CHEMBL130226 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39131 | 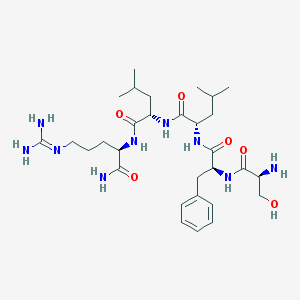 CHEMBL336277 CHEMBL336277 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39132 | 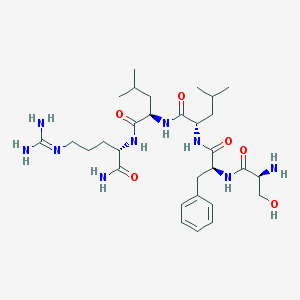 CHEMBL134820 CHEMBL134820 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39134 | 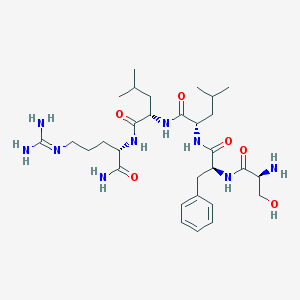 H-SER-PHE-LEU-LEU-ARG-NH2 H-SER-PHE-LEU-LEU-ARG-NH2 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39135 | 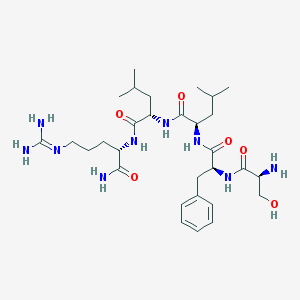 CHEMBL133618 CHEMBL133618 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39136 | 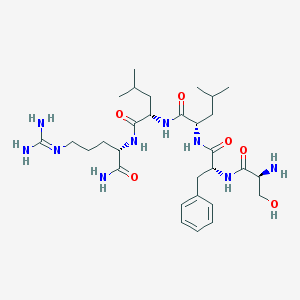 CHEMBL335036 CHEMBL335036 | C30H51N9O6 | 633.795 | 8 / 9 | -0.5 | No |
| 39685 | 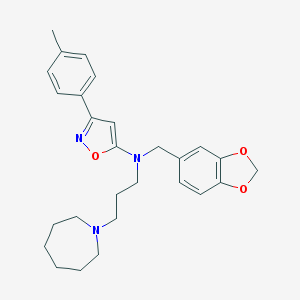 CHEMBL111258 CHEMBL111258 | C27H33N3O3 | 447.579 | 6 / 0 | 5.8 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417