

You can:
| Name | Muscarinic acetylcholine receptor M3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM3 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 3 cholinergic receptor, muscarinic 3, cardiac Chrm-3 HM4 [ Show all ] |
| Disease | Urinary incontinence Overactive bladder Overactive bladder disorder Postoperative nausea and vomiting Respiratory disease [ Show all ] |
| Length | 590 |
| Amino acid sequence | MTLHNNSTTSPLFPNISSSWIHSPSDAGLPPGTVTHFGSYNVSRAAGNFSSPDGTTDDPLGGHTVWQVVFIAFLTGILALVTIIGNILVIVSFKVNKQLKTVNNYFLLSLACADLIIGVISMNLFTTYIIMNRWALGNLACDLWLAIDYVASNASVMNLLVISFDRYFSITRPLTYRAKRTTKRAGVMIGLAWVISFVLWAPAILFWQYFVGKRTVPPGECFIQFLSEPTITFGTAIAAFYMPVTIMTILYWRIYKETEKRTKELAGLQASGTEAETENFVHPTGSSRSCSSYELQQQSMKRSNRRKYGRCHFWFTTKSWKPSSEQMDQDHSSSDSWNNNDAAASLENSASSDEEDIGSETRAIYSIVLKLPGHSTILNSTKLPSSDNLQVPEEELGMVDLERKADKLQAQKSVDDGGSFPKSFSKLPIQLESAVDTAKTSDVNSSVGKSTATLPLSFKEATLAKRFALKTRSQITKRKRMSLVKEKKAAQTLSAILLAFIITWTPYNIMVLVNTFCDSCIPKTFWNLGYWLCYINSTVNPVCYALCNKTFRTTFKMLLLCQCDKKKRRKQQYQQRQSVIFHKRAPEQAL |
| UniProt | P20309 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | T67684 |
| ChEMBL | CHEMBL245 |
| IUPHAR | 15 |
| DrugBank | BE0000045 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 4 | 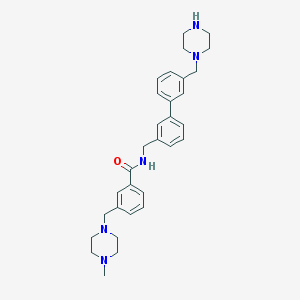 CHEMBL495028 CHEMBL495028 | C31H39N5O | 497.687 | 5 / 2 | 3.1 | Yes |
| 8 | 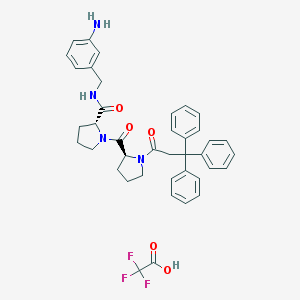 CHEMBL214461 CHEMBL214461 | C40H41F3N4O5 | 714.786 | 9 / 3 | N/A | No |
| 441680 | 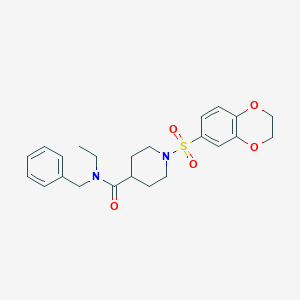 CHEMBL1602658 CHEMBL1602658 | C23H28N2O5S | 444.546 | 6 / 0 | 2.6 | Yes |
| 271 | 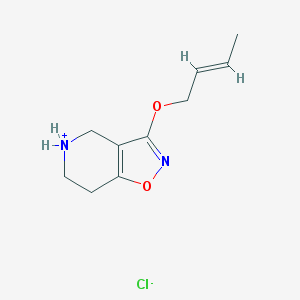 CID 44264495 CID 44264495 | C10H15ClN2O2 | 230.692 | 4 / 1 | N/A | N/A |
| 359 | 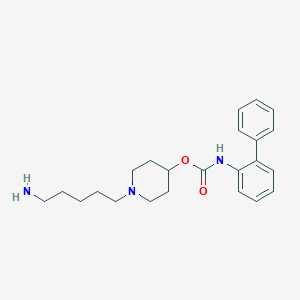 CHEMBL1683923 CHEMBL1683923 | C23H31N3O2 | 381.52 | 4 / 2 | 3.1 | Yes |
| 535921 | 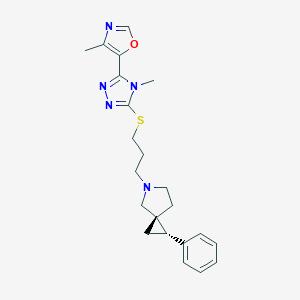 CHEMBL3983937 CHEMBL3983937 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 482 | 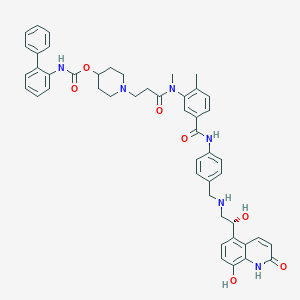 SCHEMBL739957 SCHEMBL739957 | C48H50N6O7 | 822.963 | 9 / 6 | 5.2 | No |
| 524 | 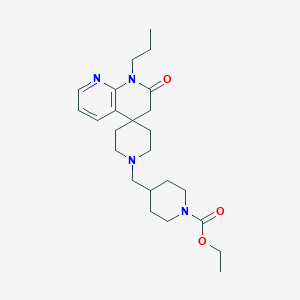 CHEMBL2407175 CHEMBL2407175 | C24H36N4O3 | 428.577 | 5 / 0 | 2.7 | Yes |
| 441691 | 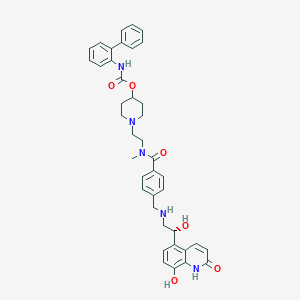 CHEMBL3426702 CHEMBL3426702 | C40H43N5O6 | 689.813 | 8 / 5 | 4.1 | No |
| 810 | 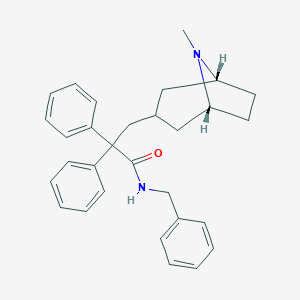 CHEMBL563305 CHEMBL563305 | C30H34N2O | 438.615 | 2 / 1 | 5.9 | No |
| 894 | 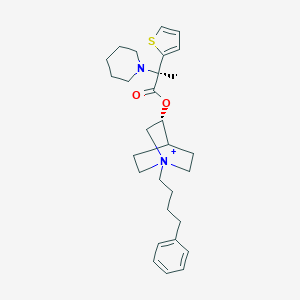 CHEMBL1921928 CHEMBL1921928 | C29H41N2O2S+ | 481.719 | 4 / 0 | 5.9 | No |
| 1017 | 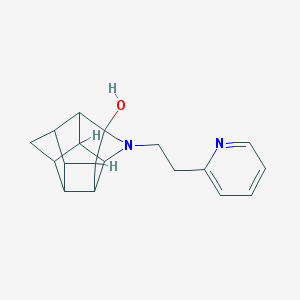 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1055 | 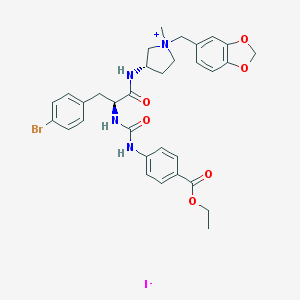 CHEMBL466271 CHEMBL466271 | C32H36BrIN4O6 | 779.47 | 7 / 3 | N/A | No |
| 1279 | 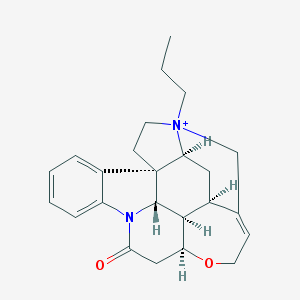 CHEMBL141736 CHEMBL141736 | C24H29N2O2+ | 377.508 | 2 / 0 | 2.8 | Yes |
| 1539 | 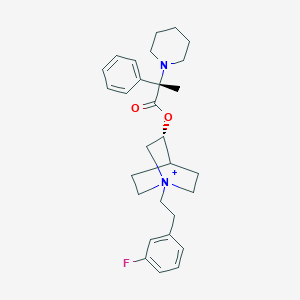 CHEMBL1924050 CHEMBL1924050 | C29H38FN2O2+ | 465.633 | 4 / 0 | 5.6 | No |
| 1939 | 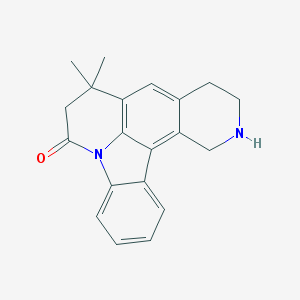 CHEMBL282874 CHEMBL282874 | C20H20N2O | 304.393 | 2 / 1 | 3.5 | Yes |
| 2261 | 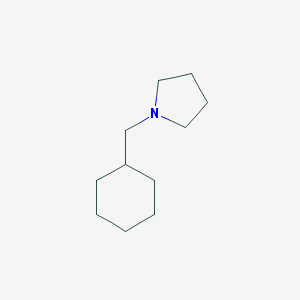 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2907 | 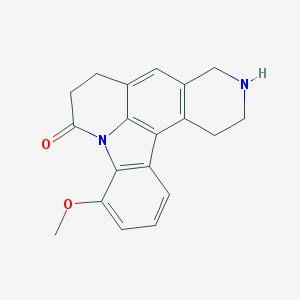 CHEMBL282680 CHEMBL282680 | C19H18N2O2 | 306.365 | 3 / 1 | 2.6 | Yes |
| 3064 | 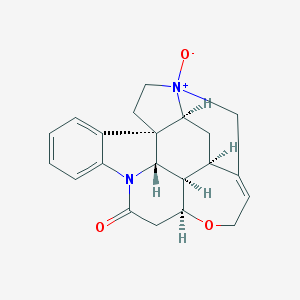 Genostrychnine Genostrychnine | C21H22N2O3 | 350.418 | 3 / 0 | 1.4 | Yes |
| 3160 | 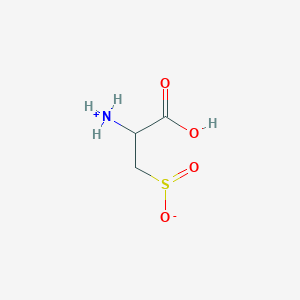 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3658 | 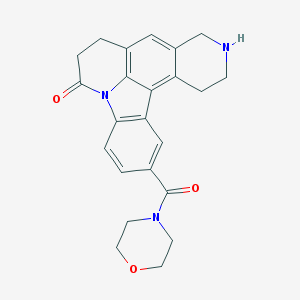 CHEMBL23620 CHEMBL23620 | C23H23N3O3 | 389.455 | 4 / 1 | 1.7 | Yes |
| 4007 | 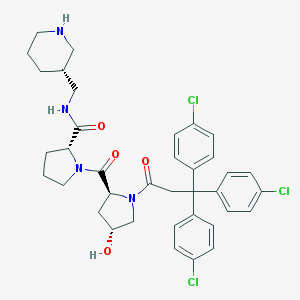 CHEMBL215894 CHEMBL215894 | C37H41Cl3N4O4 | 712.109 | 5 / 3 | 5.9 | No |
| 4080 | 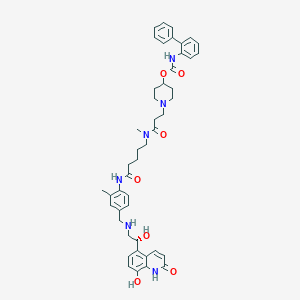 SCHEMBL738884 SCHEMBL738884 | C46H54N6O7 | 802.973 | 9 / 6 | 4.3 | No |
| 4110 | 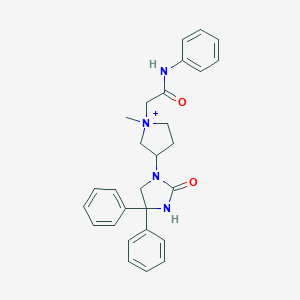 CHEMBL222634 CHEMBL222634 | C28H31N4O2+ | 455.582 | 2 / 2 | 3.5 | Yes |
| 4326 |  sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 4474 |  CHEMBL1773264 CHEMBL1773264 | C42H57N3O6S | 731.993 | 9 / 5 | 7.2 | No |
| 4988 |  CHEMBL255692 CHEMBL255692 | C36H46Cl2N2O2 | 609.676 | 4 / 4 | N/A | No |
| 5351 |  171722-92-2 171722-92-2 | C18H20N2O2 | 296.37 | 3 / 2 | 2.5 | Yes |
| 5741 | 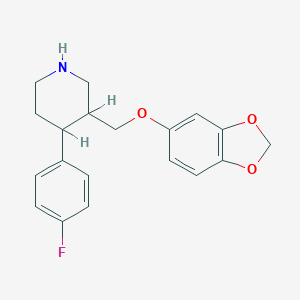 AHOUBRCZNHFOSL-UHFFFAOYSA-N AHOUBRCZNHFOSL-UHFFFAOYSA-N | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5760 | 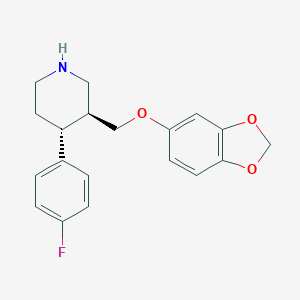 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 6120 | 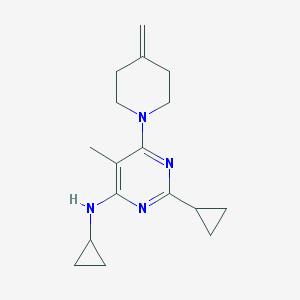 CHEMBL206480 CHEMBL206480 | C17H24N4 | 284.407 | 4 / 1 | 3.2 | Yes |
| 6130 | 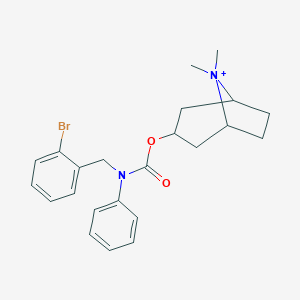 CHEMBL237313 CHEMBL237313 | C23H28BrN2O2+ | 444.393 | 2 / 0 | 5.1 | No |
| 553275 | 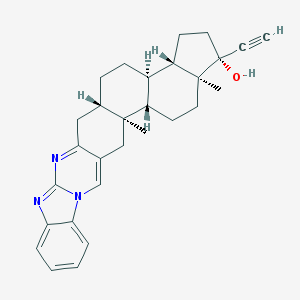 WIN-51708 WIN-51708 | C29H33N3O | 439.603 | 3 / 1 | 6.5 | No |
| 6330 | 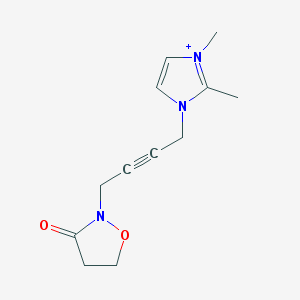 BDBM86295 BDBM86295 | C12H16N3O2+ | 234.279 | 2 / 0 | 0.0 | Yes |
| 6354 | 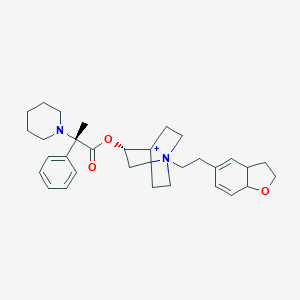 CHEMBL1921911 CHEMBL1921911 | C31H43N2O3+ | 491.696 | 4 / 0 | 4.7 | Yes |
| 6613 | 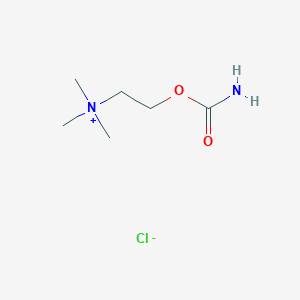 carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 6684 | 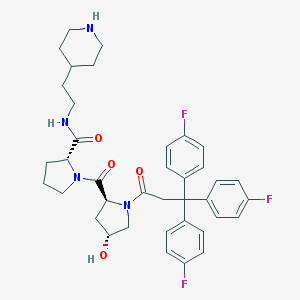 CHEMBL214029 CHEMBL214029 | C38H43F3N4O4 | 676.781 | 8 / 3 | 4.7 | No |
| 6742 | 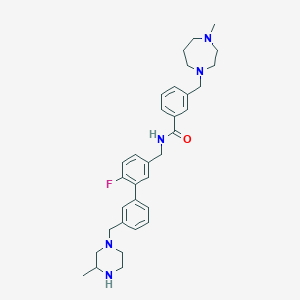 CHEMBL523044 CHEMBL523044 | C33H42FN5O | 543.731 | 6 / 2 | 4.0 | No |
| 6743 | 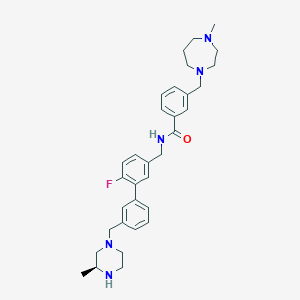 CHEMBL523044 CHEMBL523044 | C33H42FN5O | 543.731 | 6 / 2 | 4.0 | No |
| 6752 | 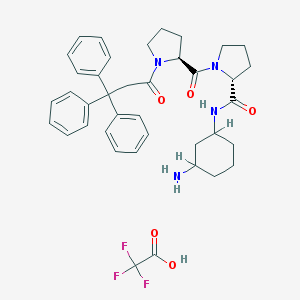 CHEMBL212530 CHEMBL212530 | C39H45F3N4O5 | 706.807 | 9 / 3 | N/A | No |
| 441971 | 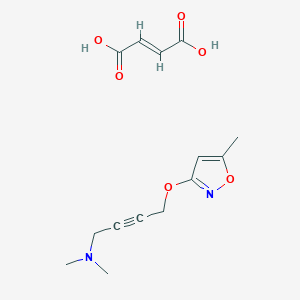 CID 49797317 CID 49797317 | C14H18N2O6 | 310.306 | 8 / 2 | N/A | N/A |
| 6882 | 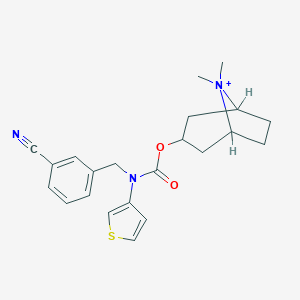 CHEMBL400038 CHEMBL400038 | C22H26N3O2S+ | 396.529 | 4 / 0 | 3.8 | Yes |
| 6943 | 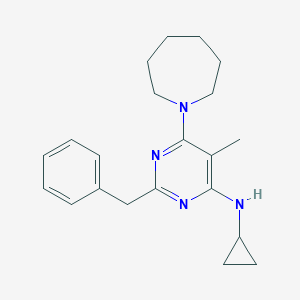 CHEMBL208300 CHEMBL208300 | C21H28N4 | 336.483 | 4 / 1 | 5.1 | No |
| 463739 | 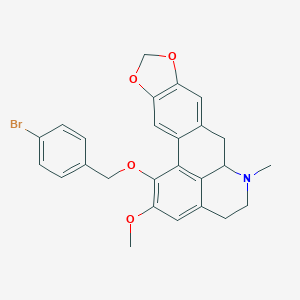 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 533936 | 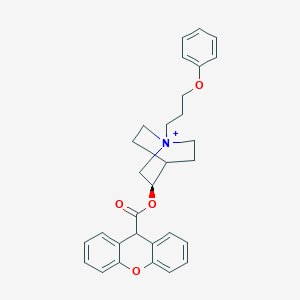 CHEMBL3932738 CHEMBL3932738 | C30H32NO4+ | 470.589 | 4 / 0 | 5.7 | No |
| 7187 | 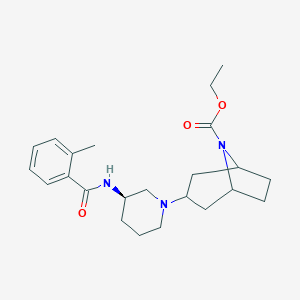 CHEMBL2024331 CHEMBL2024331 | C23H33N3O3 | 399.535 | 4 / 1 | 3.6 | Yes |
| 7295 | 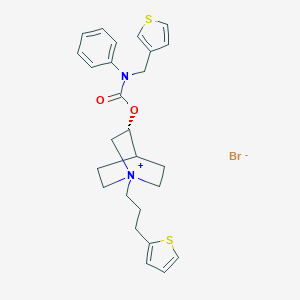 CHEMBL1779132 CHEMBL1779132 | C26H31BrN2O2S2 | 547.57 | 5 / 0 | N/A | No |
| 7494 | 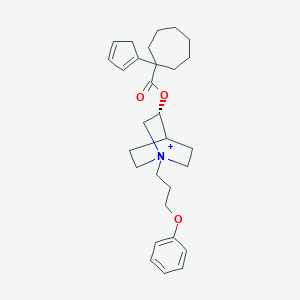 CHEMBL3088086 CHEMBL3088086 | C29H40NO3+ | 450.643 | 3 / 0 | 6.4 | No |
| 7656 | 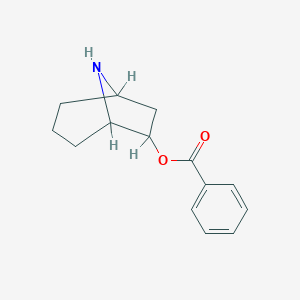 CHEMBL84113 CHEMBL84113 | C14H17NO2 | 231.295 | 3 / 1 | 2.5 | Yes |
| 7780 | 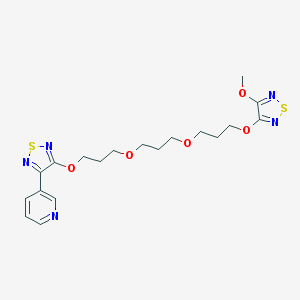 CHEMBL376295 CHEMBL376295 | C19H25N5O5S2 | 467.559 | 12 / 0 | 2.8 | No |
| 7925 | 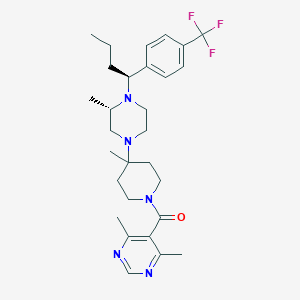 CHEMBL432170 CHEMBL432170 | C29H40F3N5O | 531.668 | 8 / 0 | 5.1 | No |
| 8002 | 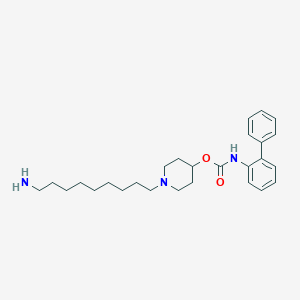 CHEMBL1683927 CHEMBL1683927 | C27H39N3O2 | 437.628 | 4 / 2 | 5.8 | No |
| 8036 | 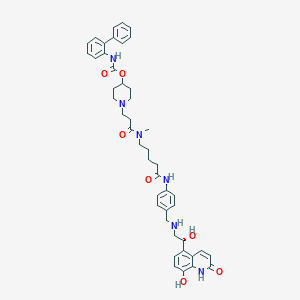 US8551978, I-14 US8551978, I-14 | C45H52N6O7 | 788.946 | 9 / 6 | 3.9 | No |
| 8672 | 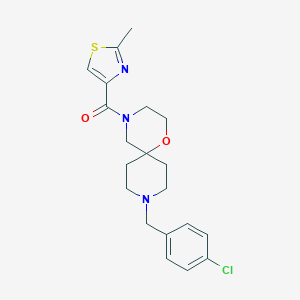 CHEMBL1289545 CHEMBL1289545 | C20H24ClN3O2S | 405.941 | 5 / 0 | 3.3 | Yes |
| 8750 | 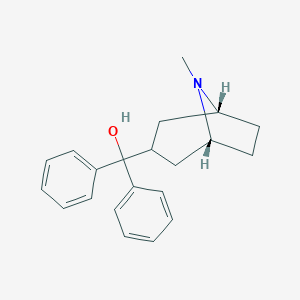 CHEMBL1782087 CHEMBL1782087 | C21H25NO | 307.437 | 2 / 1 | 4.0 | Yes |
| 8841 | 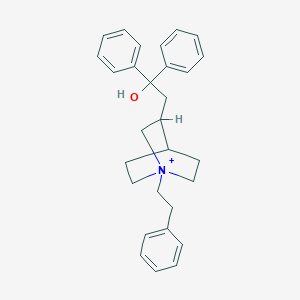 CHEMBL478069 CHEMBL478069 | C29H34NO+ | 412.597 | 1 / 1 | 5.7 | No |
| 8924 | 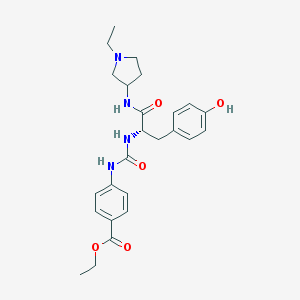 CHEMBL513228 CHEMBL513228 | C25H32N4O5 | 468.554 | 6 / 4 | 2.7 | Yes |
| 9106 | 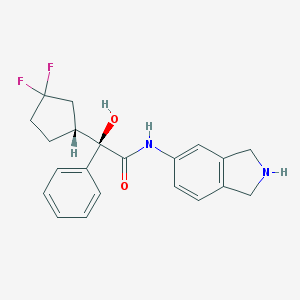 CHEMBL307554 CHEMBL307554 | C21H22F2N2O2 | 372.416 | 5 / 3 | 2.7 | Yes |
| 9203 | 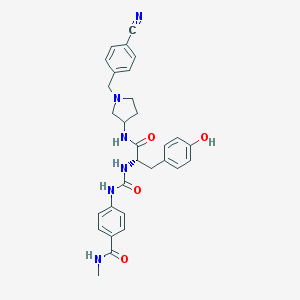 CHEMBL519893 CHEMBL519893 | C30H32N6O4 | 540.624 | 6 / 5 | 2.6 | No |
| 9330 | 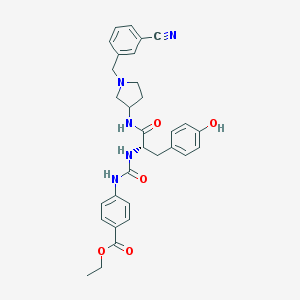 CHEMBL521244 CHEMBL521244 | C31H33N5O5 | 555.635 | 7 / 4 | 3.6 | No |
| 547993 |  Glycopyrrolate cation Glycopyrrolate cation | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9532 |  Glycopyrronium Glycopyrronium | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9855 |  AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9995 |  CHEMBL516121 CHEMBL516121 | C21H26NO+ | 308.445 | 1 / 1 | 3.3 | Yes |
| 10001 | 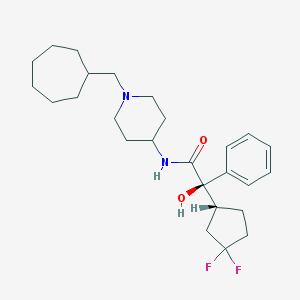 CHEMBL358289 CHEMBL358289 | C26H38F2N2O2 | 448.599 | 5 / 2 | 5.7 | No |
| 10181 | 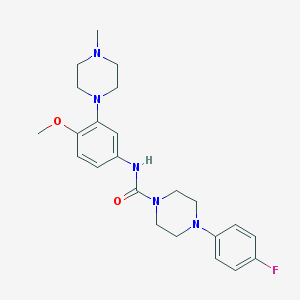 CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10387 | 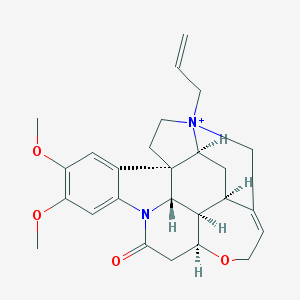 N-Allyl Brucine Iodide N-Allyl Brucine Iodide | C26H31N2O4+ | 435.544 | 4 / 0 | 1.6 | Yes |
| 10706 | 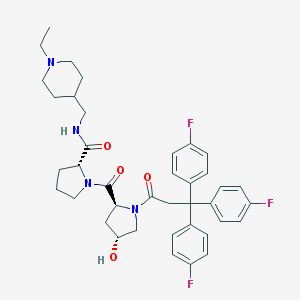 CHEMBL213709 CHEMBL213709 | C39H45F3N4O4 | 690.808 | 8 / 2 | 5.1 | No |
| 10722 | 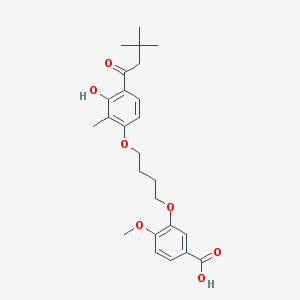 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10796 | 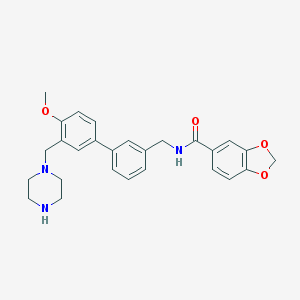 CHEMBL492756 CHEMBL492756 | C27H29N3O4 | 459.546 | 6 / 2 | 3.3 | Yes |
| 10911 | 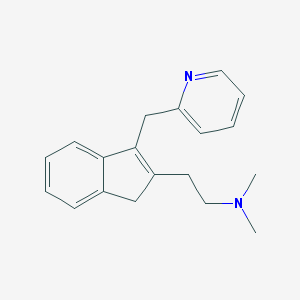 CHEMBL169494 CHEMBL169494 | C19H22N2 | 278.399 | 2 / 0 | 2.4 | Yes |
| 10992 | 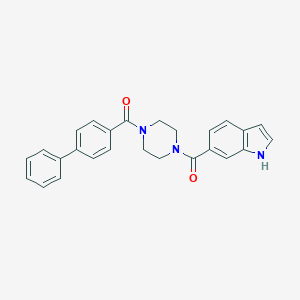 CHEMBL1076586 CHEMBL1076586 | C26H23N3O2 | 409.489 | 2 / 1 | 4.1 | Yes |
| 11573 | 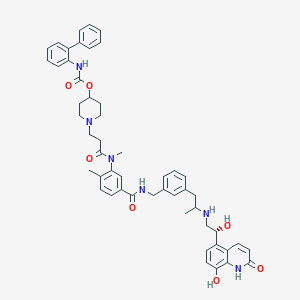 SCHEMBL741284 SCHEMBL741284 | C51H56N6O7 | 865.044 | 9 / 6 | 6.0 | No |
| 521778 | 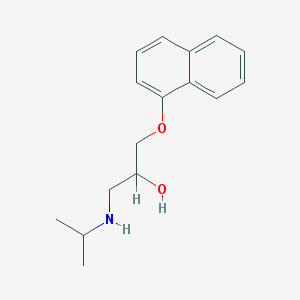 propranolol propranolol | C16H21NO2 | 259.349 | 3 / 2 | 3.0 | Yes |
| 11774 | 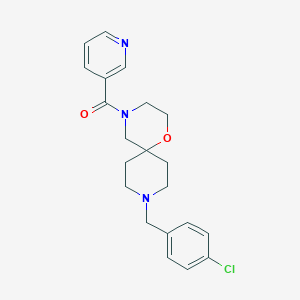 CHEMBL1289433 CHEMBL1289433 | C21H24ClN3O2 | 385.892 | 4 / 0 | 2.5 | Yes |
| 11919 | 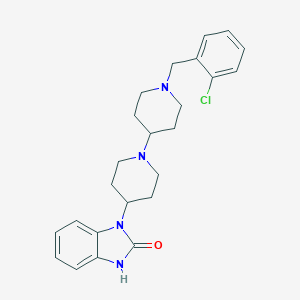 CHEMBL477821 CHEMBL477821 | C24H29ClN4O | 424.973 | 3 / 1 | 4.1 | Yes |
| 12184 | 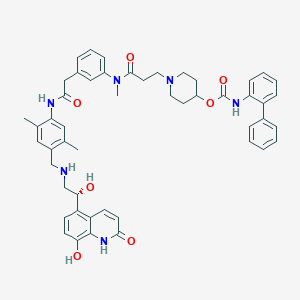 SCHEMBL738913 SCHEMBL738913 | C50H54N6O7 | 851.017 | 9 / 6 | 5.5 | No |
| 12217 | 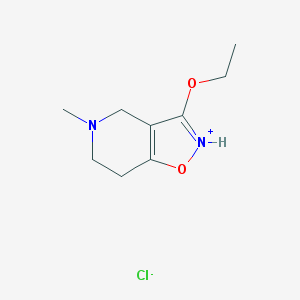 SCHEMBL10653962 SCHEMBL10653962 | C9H15ClN2O2 | 218.681 | 4 / 1 | N/A | N/A |
| 12305 | 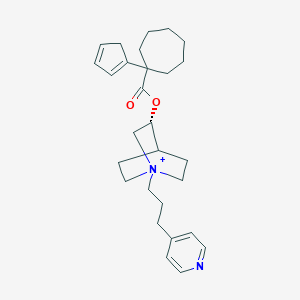 CHEMBL3087229 CHEMBL3087229 | C28H39N2O2+ | 435.632 | 3 / 0 | 5.6 | No |
| 12319 | 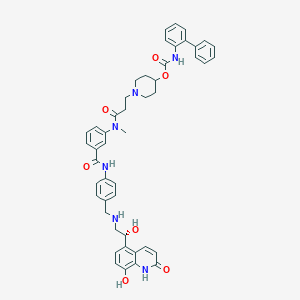 US8551978, I-25 US8551978, I-25 | C47H48N6O7 | 808.936 | 9 / 6 | 4.8 | No |
| 12541 | 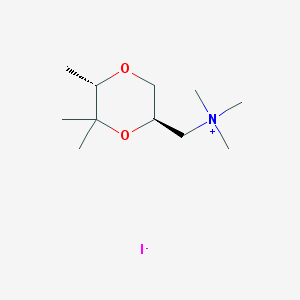 CHEMBL593620 CHEMBL593620 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12546 | 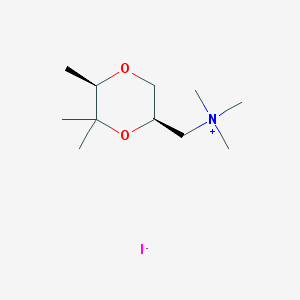 CHEMBL593685 CHEMBL593685 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12598 | 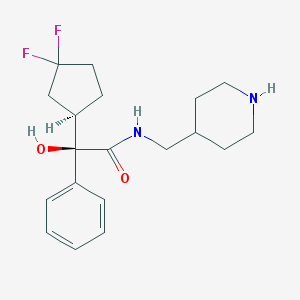 CHEMBL306903 CHEMBL306903 | C19H26F2N2O2 | 352.426 | 5 / 3 | 2.5 | Yes |
| 12702 | 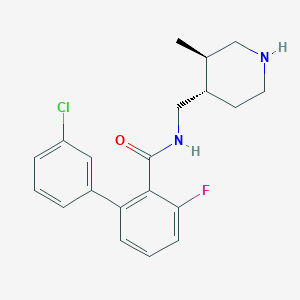 CHEMBL1083925 CHEMBL1083925 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 12706 | 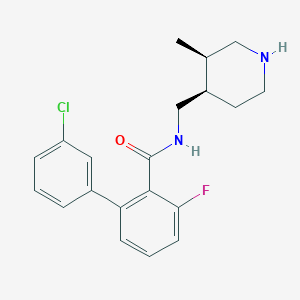 CHEMBL1083926 CHEMBL1083926 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 442176 | 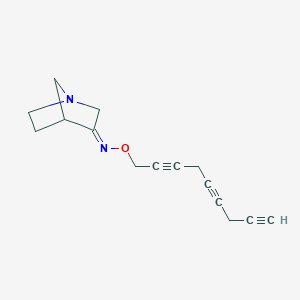 CHEMBL83074 CHEMBL83074 | C15H16N2O | 240.306 | 3 / 0 | 2.1 | Yes |
| 12820 | 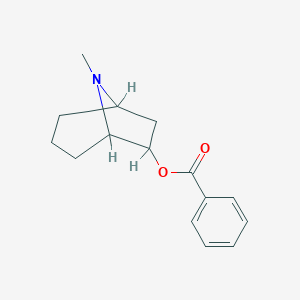 CHEMBL85190 CHEMBL85190 | C15H19NO2 | 245.322 | 3 / 0 | 3.0 | Yes |
| 13111 | 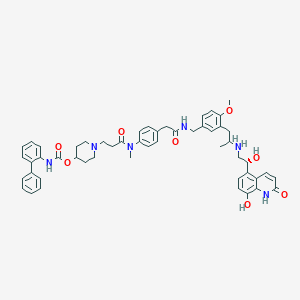 SCHEMBL741232 SCHEMBL741232 | C52H58N6O8 | 895.07 | 10 / 6 | 5.6 | No |
| 13174 | 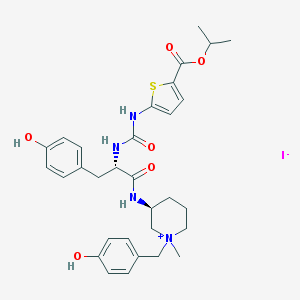 CHEMBL540148 CHEMBL540148 | C31H39IN4O6S | 722.639 | 8 / 5 | N/A | No |
| 13257 | 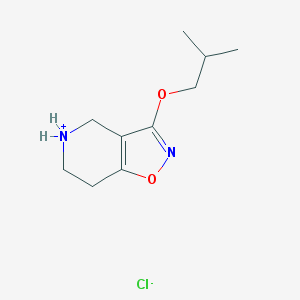 CID 44264496 CID 44264496 | C10H17ClN2O2 | 232.708 | 4 / 1 | N/A | N/A |
| 13305 | 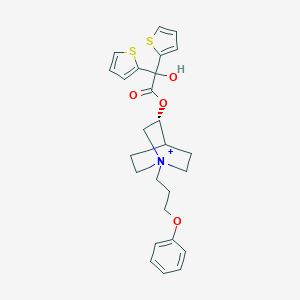 Aclidinium Aclidinium | C26H30NO4S2+ | 484.649 | 6 / 1 | 4.7 | Yes |
| 13784 | 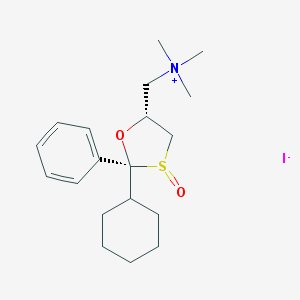 CHEMBL557808 CHEMBL557808 | C19H30INO2S | 463.418 | 4 / 0 | N/A | N/A |
| 15157 | 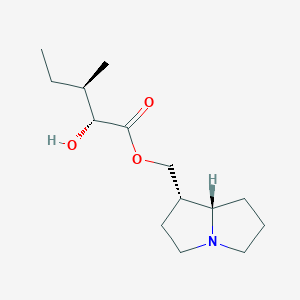 cremastrine cremastrine | C14H25NO3 | 255.358 | 4 / 1 | 2.2 | Yes |
| 15427 | 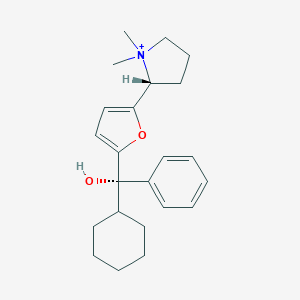 CHEMBL568775 CHEMBL568775 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15435 | 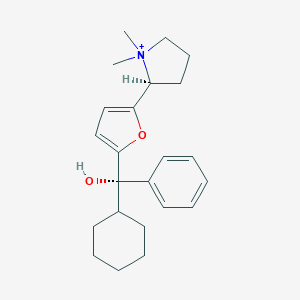 CHEMBL569089 CHEMBL569089 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15437 | 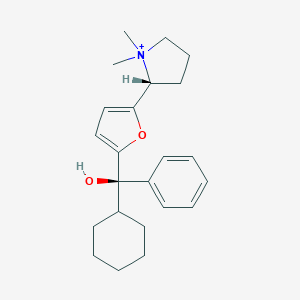 CHEMBL569307 CHEMBL569307 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15446 | 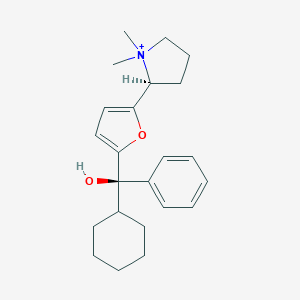 CHEMBL568870 CHEMBL568870 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15448 | 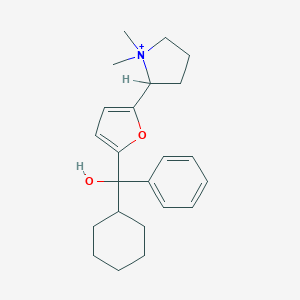 CHEMBL569760 CHEMBL569760 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15462 | 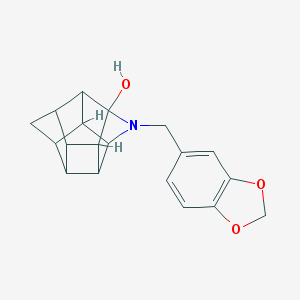 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15785 | 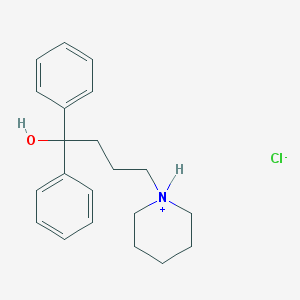 CID 44448670 CID 44448670 | C21H28ClNO | 345.911 | 2 / 2 | N/A | N/A |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417