

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM2 |
| Synonym | cholinergic receptor AChR M2 M2 muscarinic acetylcholine receptor M2 receptor Chrm-2 [ Show all ] |
| Disease | Urinary incontinence Heart failure Nausea; Addiction Parkinson's disease Peptic ulcer [ Show all ] |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNSLALTSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKDKKEPVANQDPVSPSLVQGRIVKPNNNNMPSSDDGLEHNKIQNGKAPRDPVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSKDENSKQTCIRIGTKTPKSDSCTPTNTTVEVVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P08172 |
| Protein Data Bank | 5zkc, 4mqs, 4mqt, 5yc8, 5zk3, 5zkb, 5zk8 |
| GPCR-HGmod model | P08172 |
| 3D structure model | This structure is from PDB ID 5zkc. |
| BioLiP | BL0433341, BL0433216, BL0263147, BL0263146, BL0263145, BL0433339, BL0433340, BL0433338 |
| Therapeutic Target Database | T46185 |
| ChEMBL | CHEMBL211 |
| IUPHAR | 14 |
| DrugBank | BE0000560 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 250 | 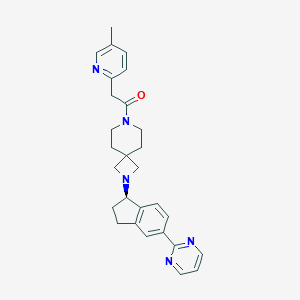 CHEMBL2426667 CHEMBL2426667 | C28H31N5O | 453.59 | 5 / 0 | 2.9 | Yes |
| 441681 | 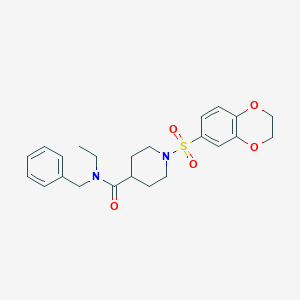 CHEMBL1602658 CHEMBL1602658 | C23H28N2O5S | 444.546 | 6 / 0 | 2.6 | Yes |
| 525 | 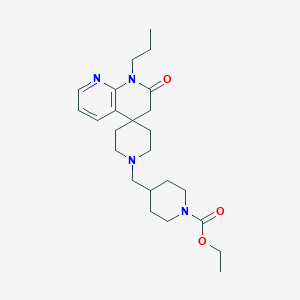 CHEMBL2407175 CHEMBL2407175 | C24H36N4O3 | 428.577 | 5 / 0 | 2.7 | Yes |
| 811 | 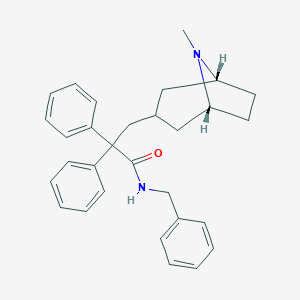 CHEMBL563305 CHEMBL563305 | C30H34N2O | 438.615 | 2 / 1 | 5.9 | No |
| 1020 |  CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1056 |  CHEMBL466271 CHEMBL466271 | C32H36BrIN4O6 | 779.47 | 7 / 3 | N/A | No |
| 1278 |  CHEMBL141736 CHEMBL141736 | C24H29N2O2+ | 377.508 | 2 / 0 | 2.8 | Yes |
| 1413 |  CHEMBL134294 CHEMBL134294 | C39H65N7O2 | 663.996 | 7 / 3 | 4.9 | No |
| 1941 | 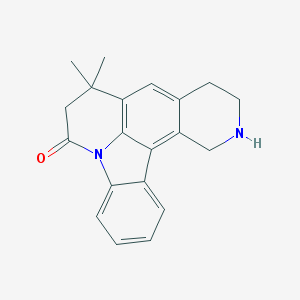 CHEMBL282874 CHEMBL282874 | C20H20N2O | 304.393 | 2 / 1 | 3.5 | Yes |
| 2267 | 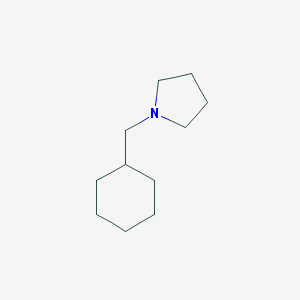 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2322 | 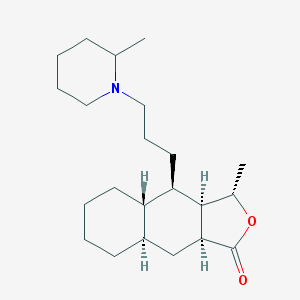 CHEMBL366819 CHEMBL366819 | C22H37NO2 | 347.543 | 3 / 0 | 5.4 | No |
| 2418 | 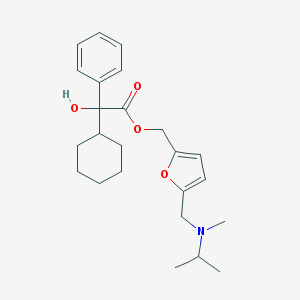 CHEMBL337023 CHEMBL337023 | C24H33NO4 | 399.531 | 5 / 1 | 4.6 | Yes |
| 2906 | 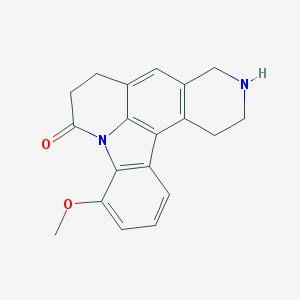 CHEMBL282680 CHEMBL282680 | C19H18N2O2 | 306.365 | 3 / 1 | 2.6 | Yes |
| 3065 | 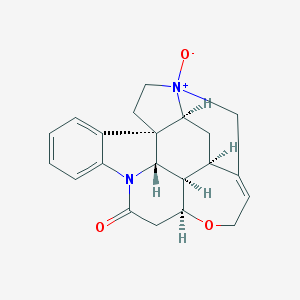 Genostrychnine Genostrychnine | C21H22N2O3 | 350.418 | 3 / 0 | 1.4 | Yes |
| 3148 | 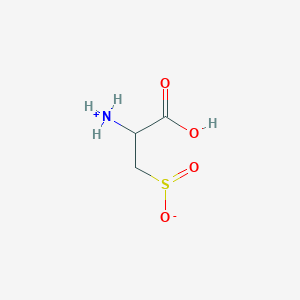 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3640 | 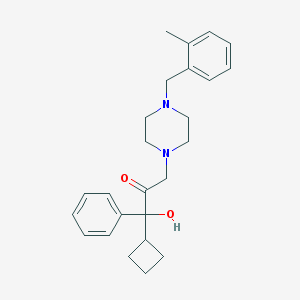 CHEMBL347132 CHEMBL347132 | C25H32N2O2 | 392.543 | 4 / 1 | 3.8 | Yes |
| 3657 | 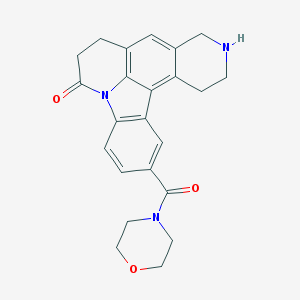 CHEMBL23620 CHEMBL23620 | C23H23N3O3 | 389.455 | 4 / 1 | 1.7 | Yes |
| 3851 | 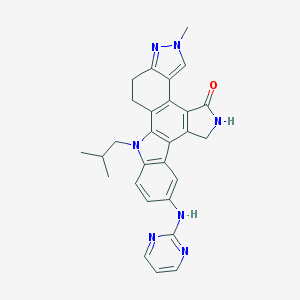 CEP-11981 CEP-11981 | C28H27N7O | 477.572 | 5 / 2 | 3.9 | Yes |
| 4006 | 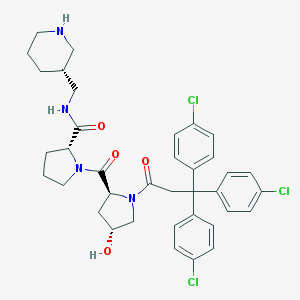 CHEMBL215894 CHEMBL215894 | C37H41Cl3N4O4 | 712.109 | 5 / 3 | 5.9 | No |
| 4111 | 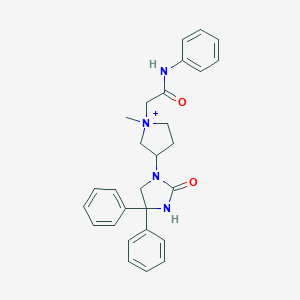 CHEMBL222634 CHEMBL222634 | C28H31N4O2+ | 455.582 | 2 / 2 | 3.5 | Yes |
| 4917 | 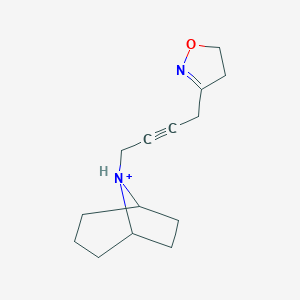 CHEMBL239657 CHEMBL239657 | C14H21N2O+ | 233.335 | 2 / 1 | 1.8 | Yes |
| 4987 | 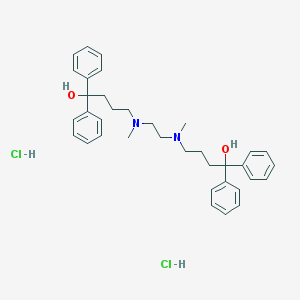 CHEMBL255692 CHEMBL255692 | C36H46Cl2N2O2 | 609.676 | 4 / 4 | N/A | No |
| 5355 | 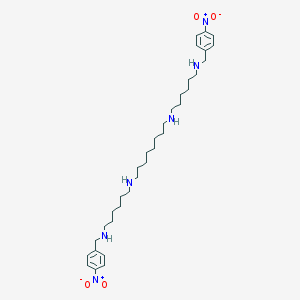 CHEMBL166803 CHEMBL166803 | C34H56N6O4 | 612.86 | 8 / 4 | 6.5 | No |
| 5669 | 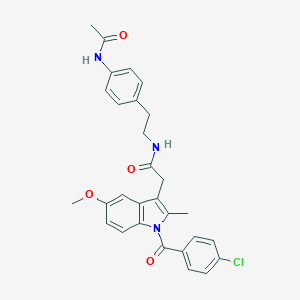 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 5752 | 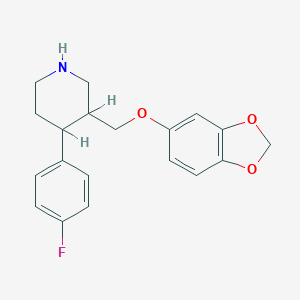 AHOUBRCZNHFOSL-UHFFFAOYSA-N AHOUBRCZNHFOSL-UHFFFAOYSA-N | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5769 | 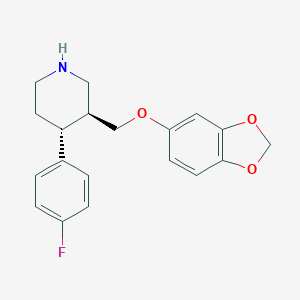 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5894 | 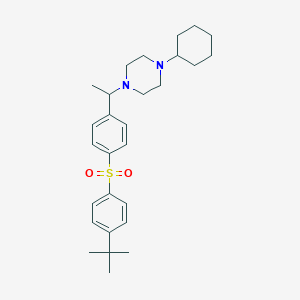 CHEMBL73661 CHEMBL73661 | C28H40N2O2S | 468.7 | 4 / 0 | 6.2 | No |
| 6132 | 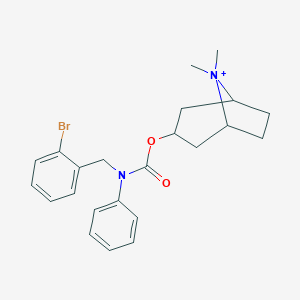 CHEMBL237313 CHEMBL237313 | C23H28BrN2O2+ | 444.393 | 2 / 0 | 5.1 | No |
| 6211 | 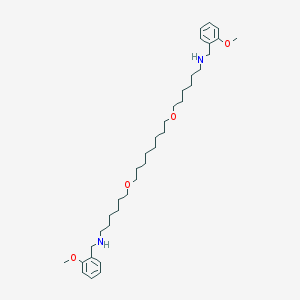 CHEMBL294865 CHEMBL294865 | C36H60N2O4 | 584.886 | 6 / 2 | 7.4 | No |
| 553274 | 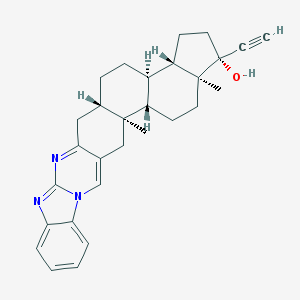 WIN-51708 WIN-51708 | C29H33N3O | 439.603 | 3 / 1 | 6.5 | No |
| 6321 | 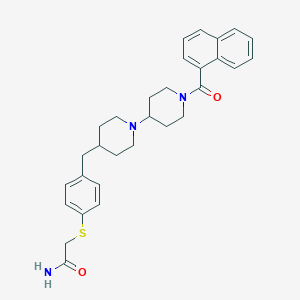 CHEMBL12085 CHEMBL12085 | C30H35N3O2S | 501.689 | 4 / 1 | 5.4 | No |
| 6332 | 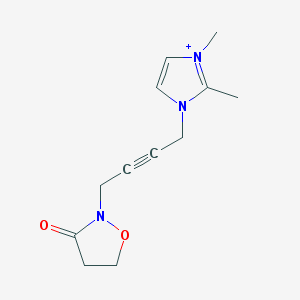 BDBM86295 BDBM86295 | C12H16N3O2+ | 234.279 | 2 / 0 | 0.0 | Yes |
| 6608 | 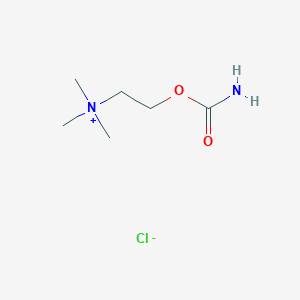 carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 6683 | 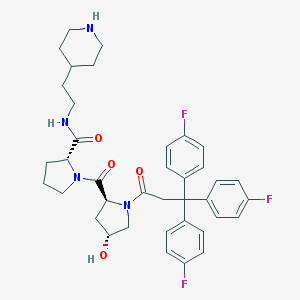 CHEMBL214029 CHEMBL214029 | C38H43F3N4O4 | 676.781 | 8 / 3 | 4.7 | No |
| 6835 | 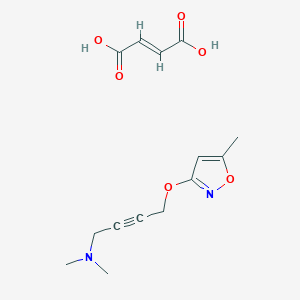 CID 49797317 CID 49797317 | C14H18N2O6 | 310.306 | 8 / 2 | N/A | N/A |
| 6850 | 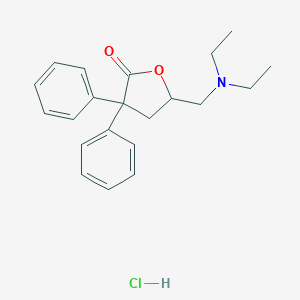 29712-11-6 29712-11-6 | C21H26ClNO2 | 359.894 | 3 / 1 | N/A | N/A |
| 6884 |  CHEMBL400038 CHEMBL400038 | C22H26N3O2S+ | 396.529 | 4 / 0 | 3.8 | Yes |
| 463733 |  CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7186 |  CHEMBL2024331 CHEMBL2024331 | C23H33N3O3 | 399.535 | 4 / 1 | 3.6 | Yes |
| 7294 |  CHEMBL1779132 CHEMBL1779132 | C26H31BrN2O2S2 | 547.57 | 5 / 0 | N/A | No |
| 7488 |  CHEMBL445134 CHEMBL445134 | C21H35NO3 | 349.515 | 4 / 1 | 4.0 | Yes |
| 7653 |  CHEMBL84113 CHEMBL84113 | C14H17NO2 | 231.295 | 3 / 1 | 2.5 | Yes |
| 7777 |  CHEMBL376295 CHEMBL376295 | C19H25N5O5S2 | 467.559 | 12 / 0 | 2.8 | No |
| 7922 |  CHEMBL432170 CHEMBL432170 | C29H40F3N5O | 531.668 | 8 / 0 | 5.1 | No |
| 8022 | 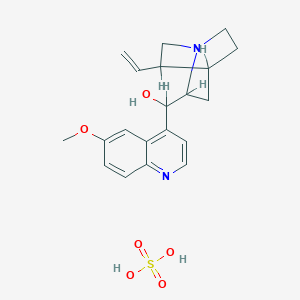 QUININE ACID SULFATE QUININE ACID SULFATE | C20H26N2O6S | 422.496 | 8 / 3 | N/A | N/A |
| 8673 | 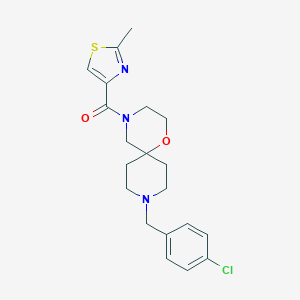 CHEMBL1289545 CHEMBL1289545 | C20H24ClN3O2S | 405.941 | 5 / 0 | 3.3 | Yes |
| 8925 | 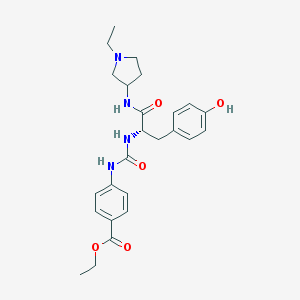 CHEMBL513228 CHEMBL513228 | C25H32N4O5 | 468.554 | 6 / 4 | 2.7 | Yes |
| 8949 | 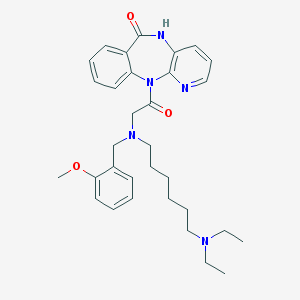 CHEMBL112208 CHEMBL112208 | C32H41N5O3 | 543.712 | 6 / 1 | 4.0 | No |
| 9108 | 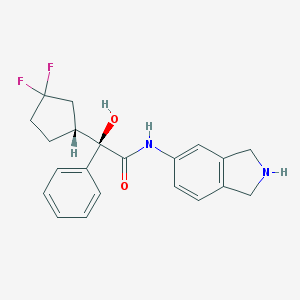 CHEMBL307554 CHEMBL307554 | C21H22F2N2O2 | 372.416 | 5 / 3 | 2.7 | Yes |
| 9201 | 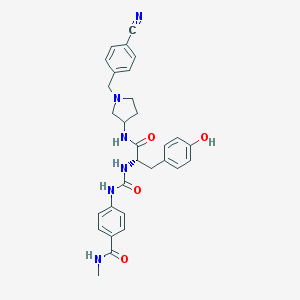 CHEMBL519893 CHEMBL519893 | C30H32N6O4 | 540.624 | 6 / 5 | 2.6 | No |
| 9329 | 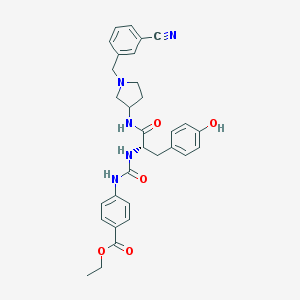 CHEMBL521244 CHEMBL521244 | C31H33N5O5 | 555.635 | 7 / 4 | 3.6 | No |
| 9419 | 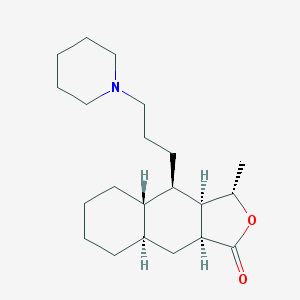 CHEMBL170205 CHEMBL170205 | C21H35NO2 | 333.516 | 3 / 0 | 5.0 | Yes |
| 547992 | 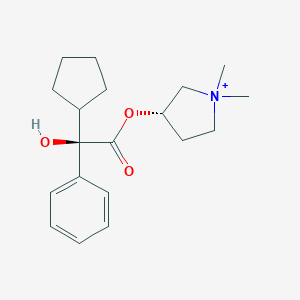 Glycopyrrolate cation Glycopyrrolate cation | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9531 | 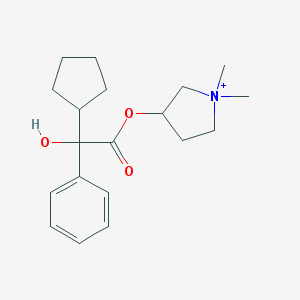 Glycopyrronium Glycopyrronium | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9862 | 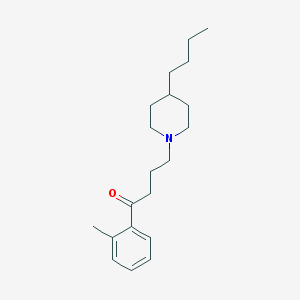 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9898 | 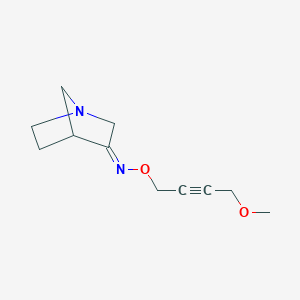 CHEMBL168991 CHEMBL168991 | C11H16N2O2 | 208.261 | 4 / 0 | 0.5 | Yes |
| 10002 | 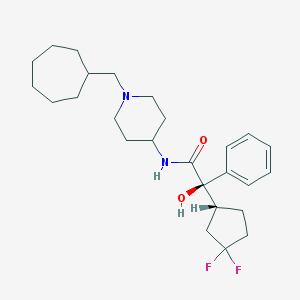 CHEMBL358289 CHEMBL358289 | C26H38F2N2O2 | 448.599 | 5 / 2 | 5.7 | No |
| 10075 | 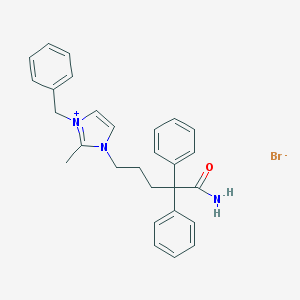 CHEMBL102749 CHEMBL102749 | C28H30BrN3O | 504.472 | 2 / 1 | N/A | No |
| 10133 | 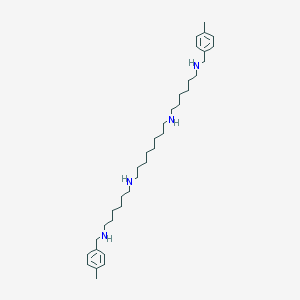 CHEMBL168304 CHEMBL168304 | C36H62N4 | 550.92 | 4 / 4 | 7.6 | No |
| 10184 | 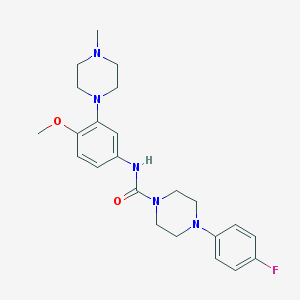 CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10389 | 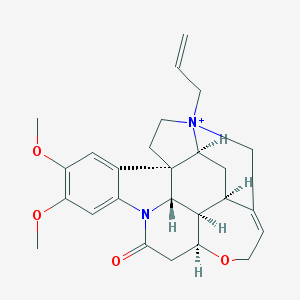 N-Allyl Brucine Iodide N-Allyl Brucine Iodide | C26H31N2O4+ | 435.544 | 4 / 0 | 1.6 | Yes |
| 10689 | 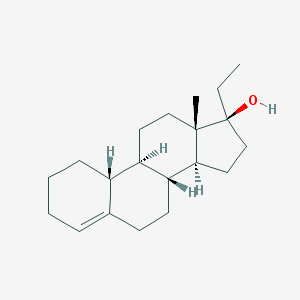 ethylestrenol ethylestrenol | C20H32O | 288.475 | 1 / 1 | 5.1 | No |
| 10709 | 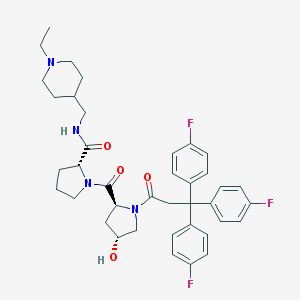 CHEMBL213709 CHEMBL213709 | C39H45F3N4O4 | 690.808 | 8 / 2 | 5.1 | No |
| 10748 | 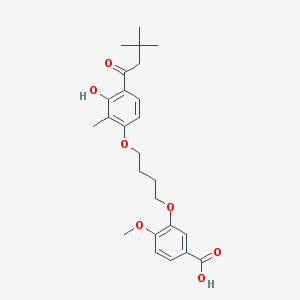 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10801 | 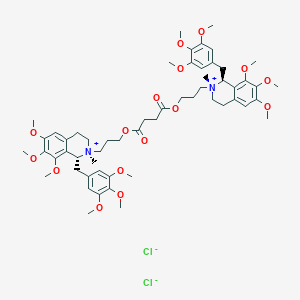 DOXACURIUM CHLORIDE DOXACURIUM CHLORIDE | C56H78Cl2N2O16 | 1106.14 | 18 / 0 | N/A | No |
| 10912 | 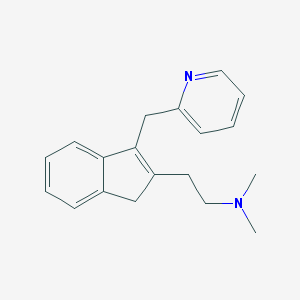 CHEMBL169494 CHEMBL169494 | C19H22N2 | 278.399 | 2 / 0 | 2.4 | Yes |
| 11009 | 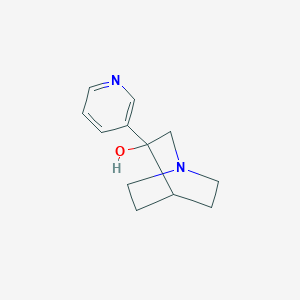 CHEMBL423746 CHEMBL423746 | C12H16N2O | 204.273 | 3 / 1 | 0.4 | Yes |
| 11383 | 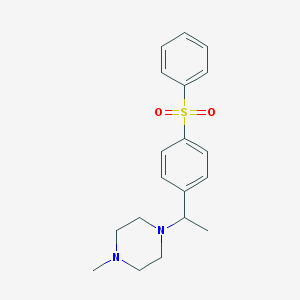 CHEMBL73410 CHEMBL73410 | C19H24N2O2S | 344.473 | 4 / 0 | 2.8 | Yes |
| 521782 | 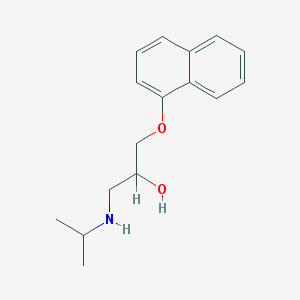 propranolol propranolol | C16H21NO2 | 259.349 | 3 / 2 | 3.0 | Yes |
| 11764 | 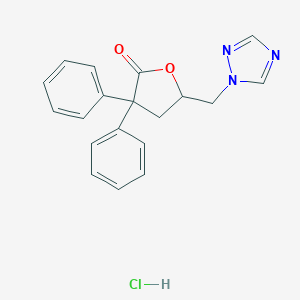 CHEMBL556590 CHEMBL556590 | C19H18ClN3O2 | 355.822 | 4 / 1 | N/A | N/A |
| 11917 | 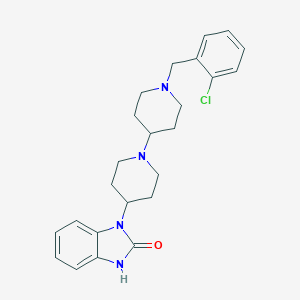 CHEMBL477821 CHEMBL477821 | C24H29ClN4O | 424.973 | 3 / 1 | 4.1 | Yes |
| 12096 | 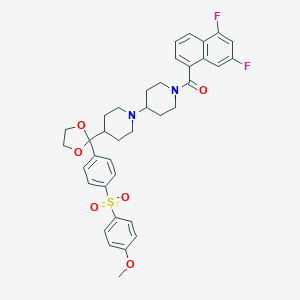 CHEMBL75108 CHEMBL75108 | C37H38F2N2O6S | 676.776 | 9 / 0 | 5.9 | No |
| 12117 | 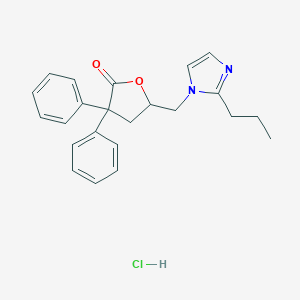 CHEMBL555173 CHEMBL555173 | C23H25ClN2O2 | 396.915 | 3 / 1 | N/A | N/A |
| 12119 | 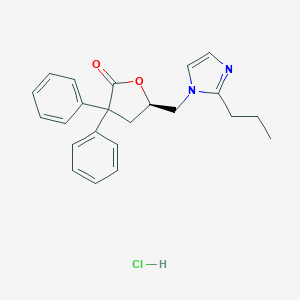 CHEMBL2093910 CHEMBL2093910 | C23H25ClN2O2 | 396.915 | 3 / 1 | N/A | N/A |
| 12542 | 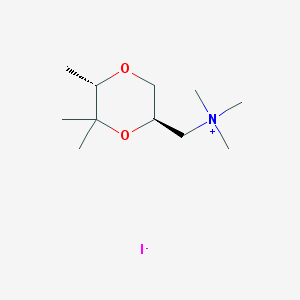 CHEMBL593620 CHEMBL593620 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12547 | 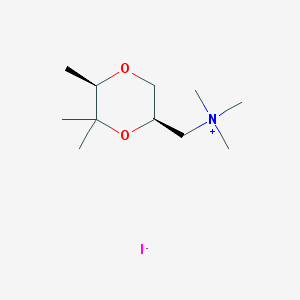 CHEMBL593685 CHEMBL593685 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12599 | 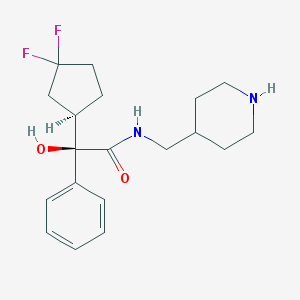 CHEMBL306903 CHEMBL306903 | C19H26F2N2O2 | 352.426 | 5 / 3 | 2.5 | Yes |
| 12673 | 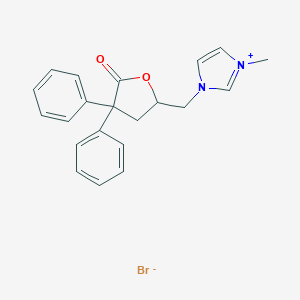 CHEMBL343112 CHEMBL343112 | C21H21BrN2O2 | 413.315 | 3 / 0 | N/A | N/A |
| 12700 | 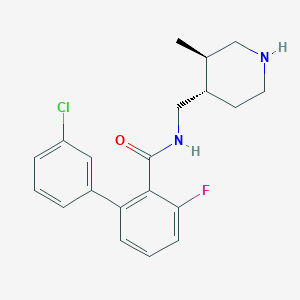 CHEMBL1083925 CHEMBL1083925 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 12752 | 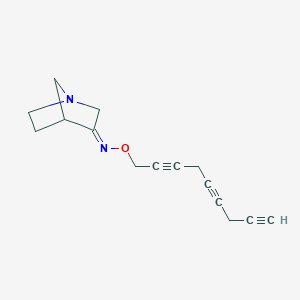 CHEMBL83074 CHEMBL83074 | C15H16N2O | 240.306 | 3 / 0 | 2.1 | Yes |
| 12817 | 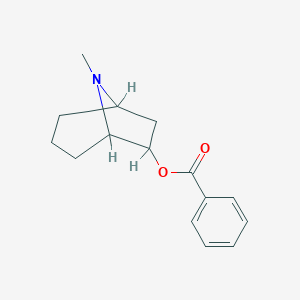 CHEMBL85190 CHEMBL85190 | C15H19NO2 | 245.322 | 3 / 0 | 3.0 | Yes |
| 12869 | 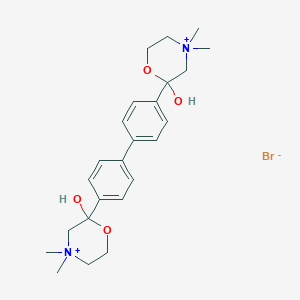 SMR000058619 SMR000058619 | C24H34BrN2O4+ | 494.45 | 5 / 2 | N/A | N/A |
| 13060 | 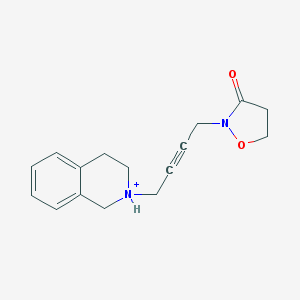 CHEMBL397538 CHEMBL397538 | C16H19N2O2+ | 271.34 | 2 / 1 | 1.1 | Yes |
| 13175 | 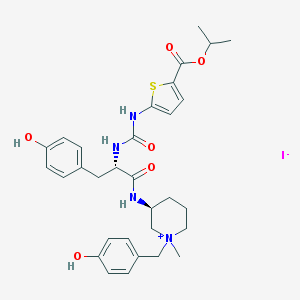 CHEMBL540148 CHEMBL540148 | C31H39IN4O6S | 722.639 | 8 / 5 | N/A | No |
| 13307 |  Aclidinium Aclidinium | C26H30NO4S2+ | 484.649 | 6 / 1 | 4.7 | Yes |
| 13782 |  CHEMBL557808 CHEMBL557808 | C19H30INO2S | 463.418 | 4 / 0 | N/A | N/A |
| 14206 |  CHEMBL165035 CHEMBL165035 | C33H41N3O3S | 559.769 | 5 / 0 | 5.6 | No |
| 14211 |  CHEMBL122499 CHEMBL122499 | C35H50N4O4+2 | 590.809 | 4 / 0 | 5.0 | No |
| 14249 | 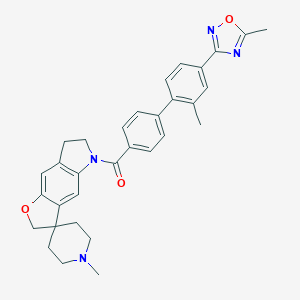 180083-23-2 180083-23-2 | C32H32N4O3 | 520.633 | 6 / 0 | 5.6 | No |
| 14649 | 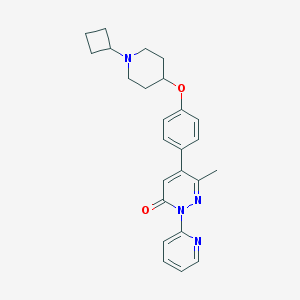 CHEMBL1945848 CHEMBL1945848 | C25H28N4O2 | 416.525 | 5 / 0 | 3.7 | Yes |
| 15156 | 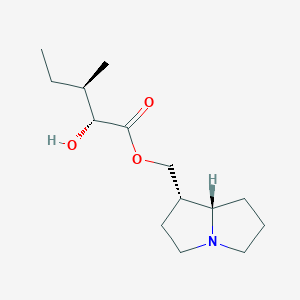 cremastrine cremastrine | C14H25NO3 | 255.358 | 4 / 1 | 2.2 | Yes |
| 15429 | 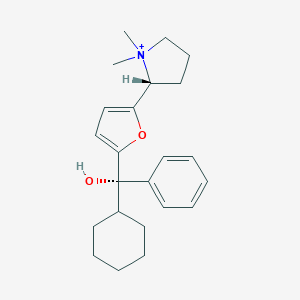 CHEMBL568775 CHEMBL568775 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15433 |  CHEMBL569089 CHEMBL569089 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15441 |  CHEMBL569307 CHEMBL569307 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15442 |  CHEMBL568870 CHEMBL568870 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15450 |  CHEMBL569760 CHEMBL569760 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15461 | 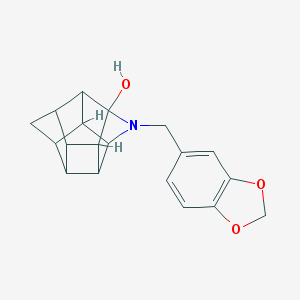 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15553 | 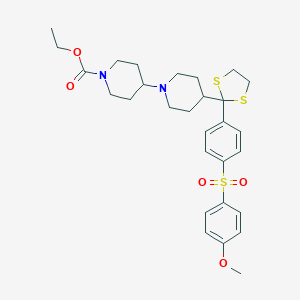 CHEMBL94247 CHEMBL94247 | C29H38N2O5S3 | 590.812 | 8 / 0 | 5.1 | No |
| 15784 | 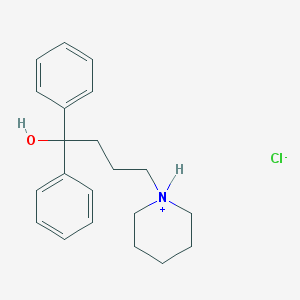 CID 44448670 CID 44448670 | C21H28ClNO | 345.911 | 2 / 2 | N/A | N/A |
| 15830 | 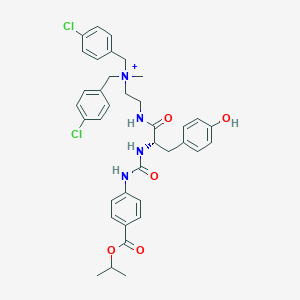 CHEMBL2206602 CHEMBL2206602 | C37H41Cl2N4O5+ | 692.658 | 5 / 4 | 6.9 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417