

You can:
| Name | Histamine H3 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH3 |
| Synonym | HH3R H3R H3 receptor GPCR97 G-protein coupled receptor 97 |
| Disease | Obese insulin-resistant disorders Excessive daytime sleepiness Sleep disorders Schizophrenia Pain [ Show all ] |
| Length | 445 |
| Amino acid sequence | MERAPPDGPLNASGALAGEAAAAGGARGFSAAWTAVLAALMALLIVATVLGNALVMLAFVADSSLRTQNNFFLLNLAISDFLVGAFCIPLYVPYVLTGRWTFGRGLCKLWLVVDYLLCTSSAFNIVLISYDRFLSVTRAVSYRAQQGDTRRAVRKMLLVWVLAFLLYGPAILSWEYLSGGSSIPEGHCYAEFFYNWYFLITASTLEFFTPFLSVTFFNLSIYLNIQRRTRLRLDGAREAAGPEPPPEAQPSPPPPPGCWGCWQKGHGEAMPLHRYGVGEAAVGAEAGEATLGGGGGGGSVASPTSSSGSSSRGTERPRSLKRGSKPSASSASLEKRMKMVSQSFTQRFRLSRDRKVAKSLAVIVSIFGLCWAPYTLLMIIRAACHGHCVPDYWYETSFWLLWANSAVNPVLYPLCHHSFRRAFTKLLCPQKLKIQPHSSLEHCWK |
| UniProt | Q9Y5N1 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | Q9Y5N1 |
| 3D structure model | This predicted structure model is from GPCR-EXP Q9Y5N1. |
| BioLiP | N/A |
| Therapeutic Target Database | T64765 |
| ChEMBL | CHEMBL264 |
| IUPHAR | 264 |
| DrugBank | BE0000968 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 394 | 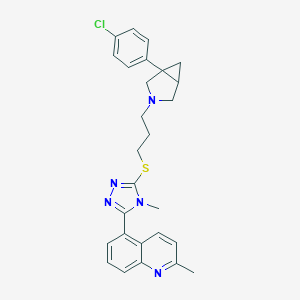 CHEMBL1080117 CHEMBL1080117 | C27H28ClN5S | 490.066 | 5 / 0 | 5.5 | No |
| 421 | 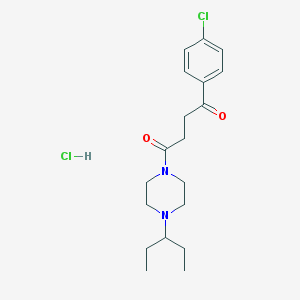 SCHEMBL4376343 SCHEMBL4376343 | C19H28Cl2N2O2 | 387.345 | 3 / 1 | N/A | N/A |
| 616 | 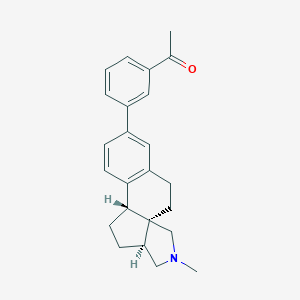 CHEMBL561028 CHEMBL561028 | C24H27NO | 345.486 | 2 / 0 | 4.5 | Yes |
| 619 | 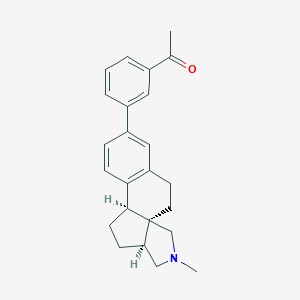 CHEMBL565206 CHEMBL565206 | C24H27NO | 345.486 | 2 / 0 | 4.5 | Yes |
| 441694 | 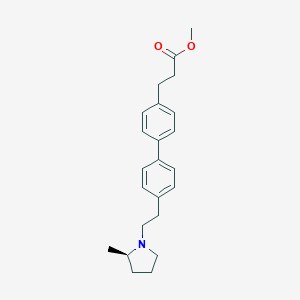 CHEMBL3427232 CHEMBL3427232 | C23H29NO2 | 351.49 | 3 / 0 | 4.9 | Yes |
| 547910 | 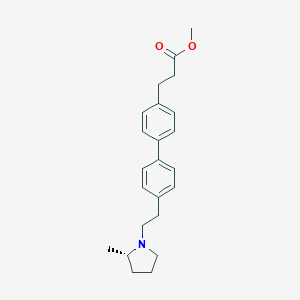 CHEMBL3937066 CHEMBL3937066 | C23H29NO2 | 351.49 | 3 / 0 | 4.9 | Yes |
| 737 | 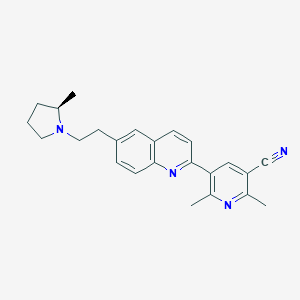 CHEMBL1085560 CHEMBL1085560 | C24H26N4 | 370.5 | 4 / 0 | 4.5 | Yes |
| 739 | 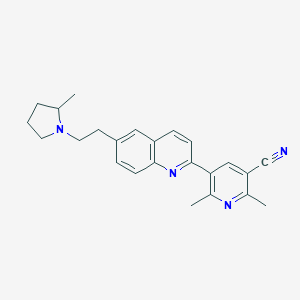 CHEMBL1822863 CHEMBL1822863 | C24H26N4 | 370.5 | 4 / 0 | 4.5 | Yes |
| 463059 | 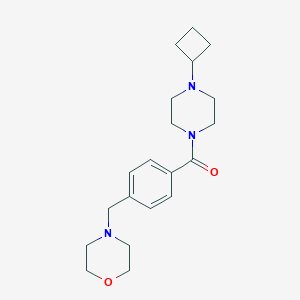 CHEMBL3577956 CHEMBL3577956 | C20H29N3O2 | 343.471 | 4 / 0 | 1.6 | Yes |
| 547911 | 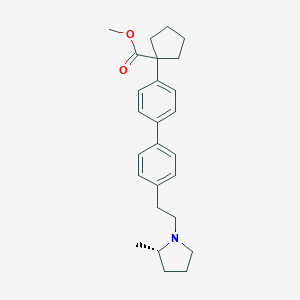 CHEMBL3956769 CHEMBL3956769 | C26H33NO2 | 391.555 | 3 / 0 | 6.0 | No |
| 441706 | 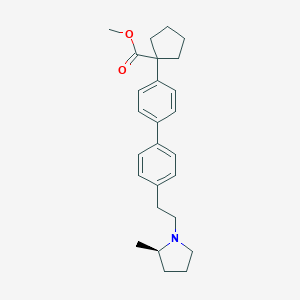 CHEMBL3427239 CHEMBL3427239 | C26H33NO2 | 391.555 | 3 / 0 | 6.0 | No |
| 981 | 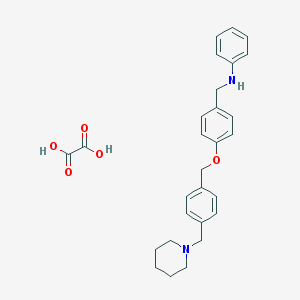 CHEMBL288431 CHEMBL288431 | C28H32N2O5 | 476.573 | 7 / 3 | N/A | N/A |
| 1026 | 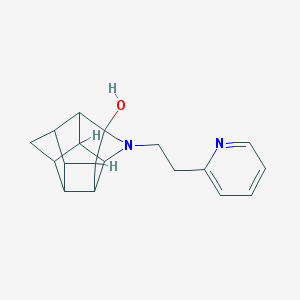 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1135 | 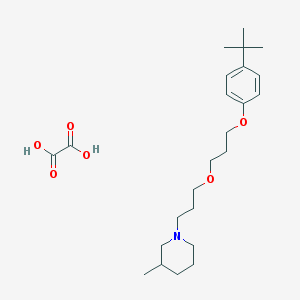 CHEMBL1202076 CHEMBL1202076 | C24H39NO6 | 437.577 | 7 / 2 | N/A | N/A |
| 1166 | 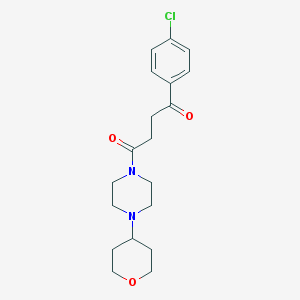 CHEMBL93759 CHEMBL93759 | C19H25ClN2O3 | 364.87 | 4 / 0 | 1.8 | Yes |
| 463131 | 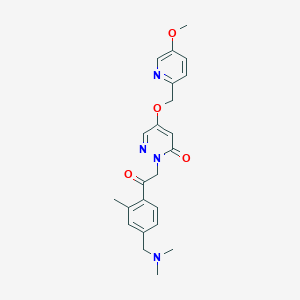 CHEMBL3601377 CHEMBL3601377 | C23H26N4O4 | 422.485 | 7 / 0 | 1.7 | Yes |
| 1480 | 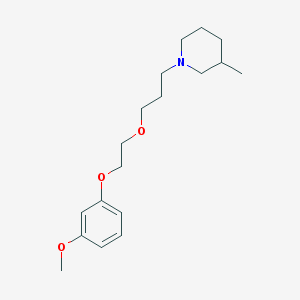 CHEMBL476264 CHEMBL476264 | C18H29NO3 | 307.434 | 4 / 0 | 3.4 | Yes |
| 521480 | 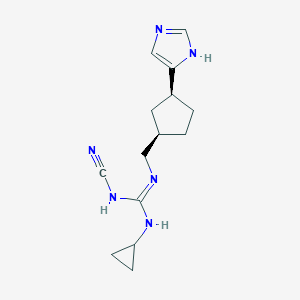 CHEMBL3806146 CHEMBL3806146 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 521487 | 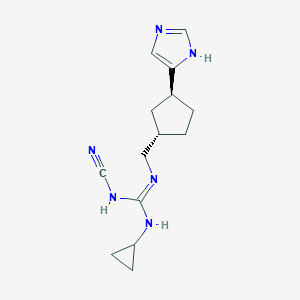 CHEMBL3804837 CHEMBL3804837 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 1786 | 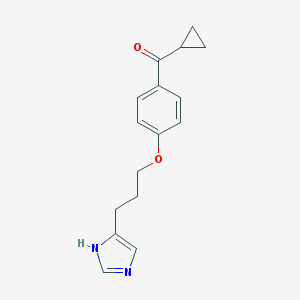 ciproxifan ciproxifan | C16H18N2O2 | 270.332 | 3 / 1 | 2.5 | Yes |
| 1854 |  CHEMBL245716 CHEMBL245716 | C25H35ClN2O2 | 431.017 | 4 / 0 | 5.7 | No |
| 2031 |  CHEMBL1222596 CHEMBL1222596 | C28H32N8O | 496.619 | 8 / 0 | 4.0 | Yes |
| 2138 |  CHEMBL87173 CHEMBL87173 | C22H23N3O2 | 361.445 | 5 / 0 | 4.3 | Yes |
| 2161 |  donepezil donepezil | C24H29NO3 | 379.5 | 4 / 0 | 4.3 | Yes |
| 2203 |  CHEMBL458722 CHEMBL458722 | C26H38N2 | 378.604 | 2 / 0 | 6.3 | No |
| 2274 |  1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2311 |  CHEMBL522894 CHEMBL522894 | C25H25FN4O2 | 432.499 | 6 / 0 | 3.8 | Yes |
| 2401 |  CHEMBL1783908 CHEMBL1783908 | C16H27N3O3 | 309.41 | 6 / 0 | 1.4 | Yes |
| 3020 | 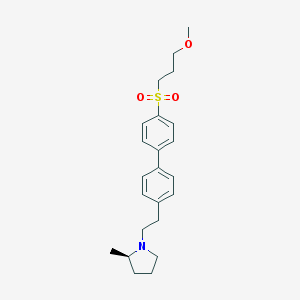 UNII-10521U3EQA UNII-10521U3EQA | C23H31NO3S | 401.565 | 4 / 0 | 4.2 | Yes |
| 441833 | 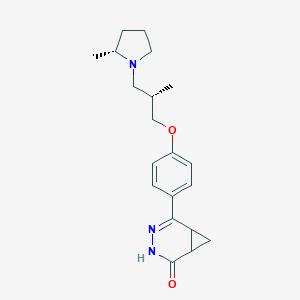 CHEMBL3417505 CHEMBL3417505 | C20H27N3O2 | 341.455 | 4 / 1 | 2.8 | Yes |
| 3166 | 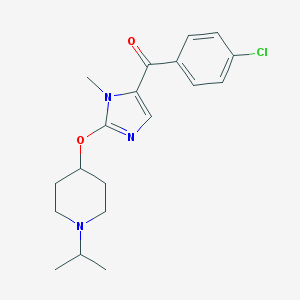 CHEMBL520100 CHEMBL520100 | C19H24ClN3O2 | 361.87 | 4 / 0 | 4.0 | Yes |
| 3257 | 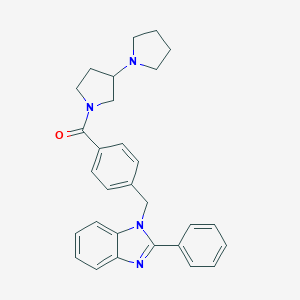 CHEMBL605141 CHEMBL605141 | C29H30N4O | 450.586 | 3 / 0 | 4.9 | Yes |
| 3327 | 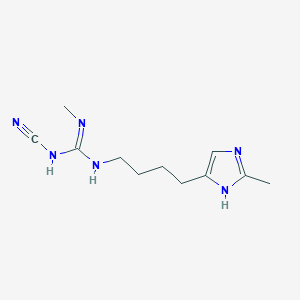 CHEMBL3220637 CHEMBL3220637 | C11H18N6 | 234.307 | 3 / 3 | 0.8 | Yes |
| 3413 | 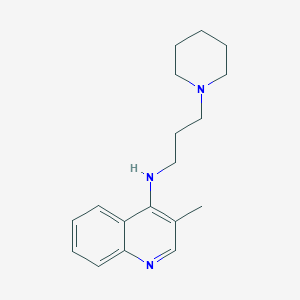 CHEMBL275874 CHEMBL275874 | C18H25N3 | 283.419 | 3 / 1 | 3.8 | Yes |
| 3464 | 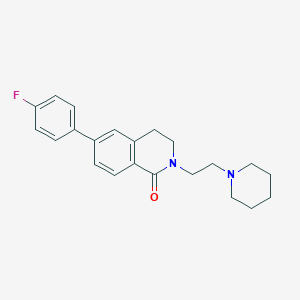 CHEMBL2031757 CHEMBL2031757 | C22H25FN2O | 352.453 | 3 / 0 | 4.0 | Yes |
| 3700 | 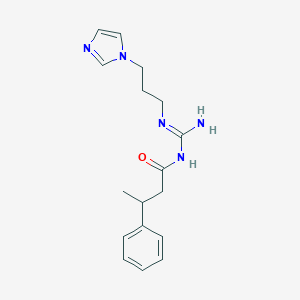 CHEMBL3220892 CHEMBL3220892 | C17H23N5O | 313.405 | 3 / 2 | 1.7 | Yes |
| 3930 |  CHEMBL1079480 CHEMBL1079480 | C23H25F3N6S | 474.55 | 9 / 0 | 3.4 | Yes |
| 4091 |  CHEMBL1829483 CHEMBL1829483 | C19H22N4O2 | 338.411 | 5 / 1 | 2.3 | Yes |
| 4134 |  SCHEMBL4089524 SCHEMBL4089524 | C28H36N2O | 416.609 | 3 / 0 | 5.8 | No |
| 4217 |  CHEMBL202638 CHEMBL202638 | C12H23N3 | 209.337 | 2 / 2 | 2.0 | Yes |
| 4268 | 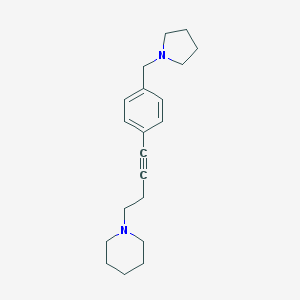 CHEMBL574712 CHEMBL574712 | C20H28N2 | 296.458 | 2 / 0 | 3.8 | Yes |
| 4292 | 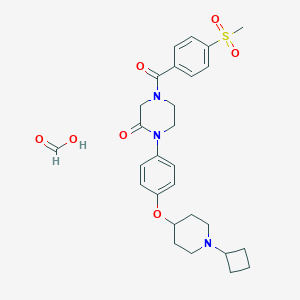 CHEMBL398996 CHEMBL398996 | C28H35N3O7S | 557.662 | 8 / 1 | N/A | No |
| 4379 | 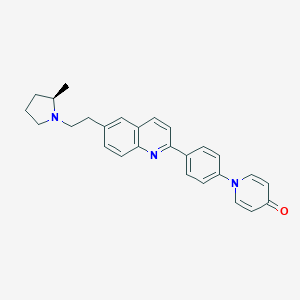 CHEMBL1086514 CHEMBL1086514 | C27H27N3O | 409.533 | 4 / 0 | 5.0 | Yes |
| 536043 | 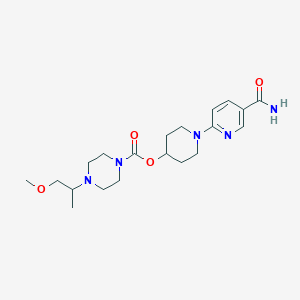 CHEMBL3935833 CHEMBL3935833 | C20H31N5O4 | 405.499 | 7 / 1 | 0.7 | Yes |
| 4637 | 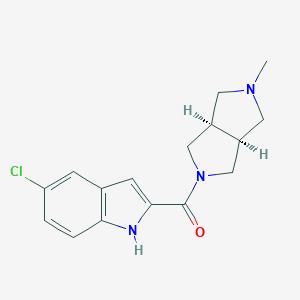 CHEMBL1915345 CHEMBL1915345 | C16H18ClN3O | 303.79 | 2 / 1 | 2.6 | Yes |
| 4798 | 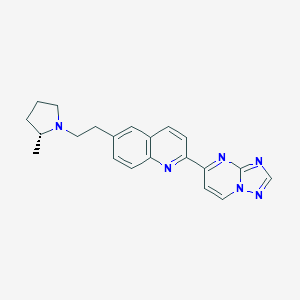 CHEMBL1084616 CHEMBL1084616 | C21H22N6 | 358.449 | 5 / 0 | 3.3 | Yes |
| 4831 | 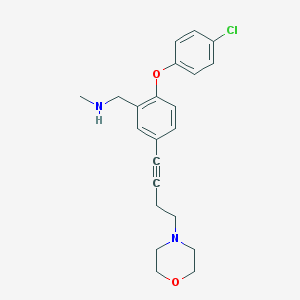 CHEMBL236475 CHEMBL236475 | C22H25ClN2O2 | 384.904 | 4 / 1 | 3.8 | Yes |
| 5083 | 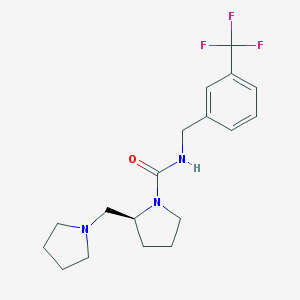 CHEMBL218930 CHEMBL218930 | C18H24F3N3O | 355.405 | 5 / 1 | 3.0 | Yes |
| 5557 | 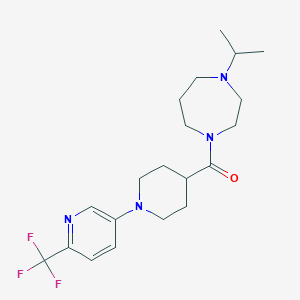 GSK334429 GSK334429 | C20H29F3N4O | 398.474 | 7 / 0 | 2.8 | Yes |
| 536080 | 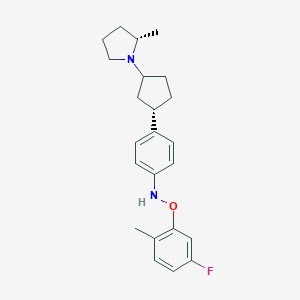 CHEMBL3978813 CHEMBL3978813 | C23H29FN2O | 368.496 | 4 / 1 | 6.2 | No |
| 557439 | 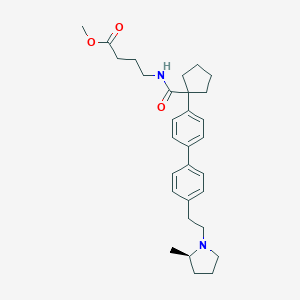 SCHEMBL15875079 SCHEMBL15875079 | C30H40N2O3 | 476.661 | 4 / 1 | 5.6 | No |
| 547960 | 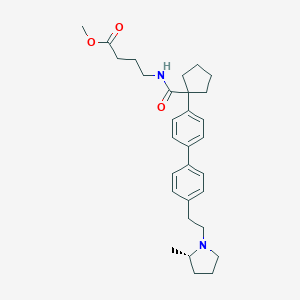 CHEMBL3958533 CHEMBL3958533 | C30H40N2O3 | 476.661 | 4 / 1 | 5.6 | No |
| 5732 |  CHEMBL233054 CHEMBL233054 | C25H33FN2O2 | 412.549 | 5 / 0 | 4.5 | Yes |
| 5733 |  CHEMBL235824 CHEMBL235824 | C25H33FN2O2 | 412.549 | 5 / 0 | 4.5 | Yes |
| 5779 |  CHEMBL161202 CHEMBL161202 | C29H36FN5O5 | 553.635 | 9 / 1 | 4.4 | No |
| 5865 |  CHEMBL1923030 CHEMBL1923030 | C18H31BrN4S | 415.438 | 3 / 3 | N/A | N/A |
| 5937 | 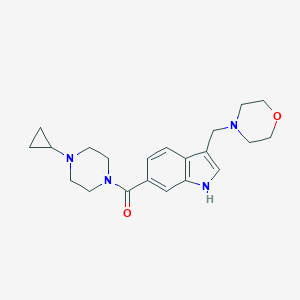 CHEMBL1271276 CHEMBL1271276 | C21H28N4O2 | 368.481 | 4 / 1 | 1.3 | Yes |
| 5984 | 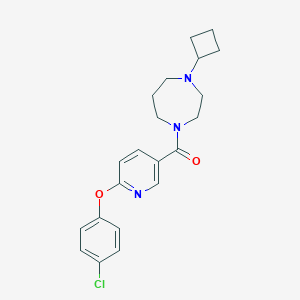 CHEMBL1171886 CHEMBL1171886 | C21H24ClN3O2 | 385.892 | 4 / 0 | 3.9 | Yes |
| 6188 | 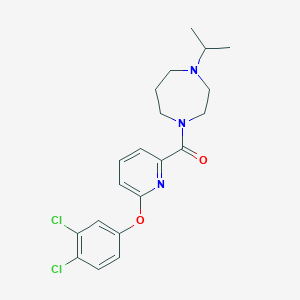 CHEMBL1172272 CHEMBL1172272 | C20H23Cl2N3O2 | 408.323 | 4 / 0 | 4.7 | Yes |
| 6212 | 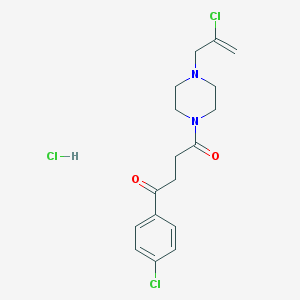 SCHEMBL4379186 SCHEMBL4379186 | C17H21Cl3N2O2 | 391.717 | 3 / 1 | N/A | N/A |
| 6290 | 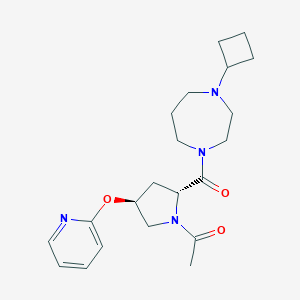 CHEMBL1097065 CHEMBL1097065 | C21H30N4O3 | 386.496 | 5 / 0 | 1.5 | Yes |
| 6441 | 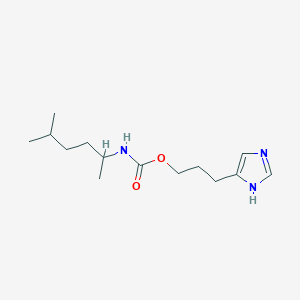 CHEMBL1081720 CHEMBL1081720 | C14H25N3O2 | 267.373 | 3 / 2 | 2.9 | Yes |
| 6457 | 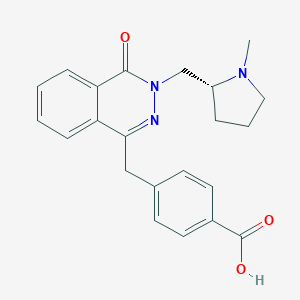 CHEMBL1767144 CHEMBL1767144 | C22H23N3O3 | 377.444 | 5 / 1 | 0.6 | Yes |
| 6478 | 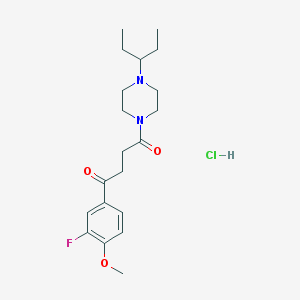 CHEMBL1202893 CHEMBL1202893 | C20H30ClFN2O3 | 400.919 | 5 / 1 | N/A | N/A |
| 6628 | 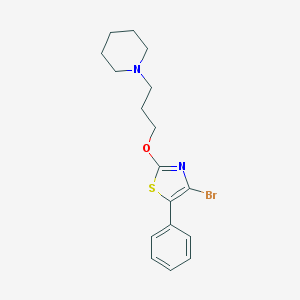 CHEMBL1257582 CHEMBL1257582 | C17H21BrN2OS | 381.332 | 4 / 0 | 5.2 | No |
| 6651 | 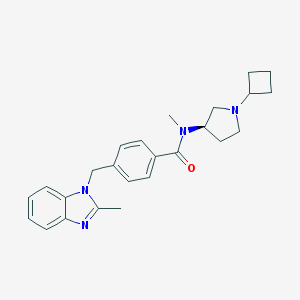 CHEMBL1835993 CHEMBL1835993 | C25H30N4O | 402.542 | 3 / 0 | 4.0 | Yes |
| 6842 | 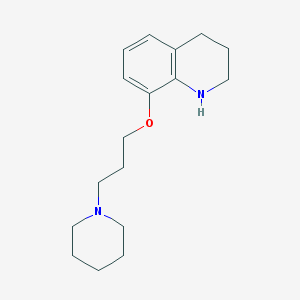 CHEMBL212278 CHEMBL212278 | C17H26N2O | 274.408 | 3 / 1 | 3.5 | Yes |
| 463713 | 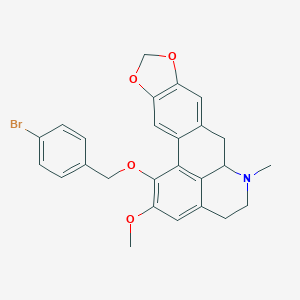 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 6954 | 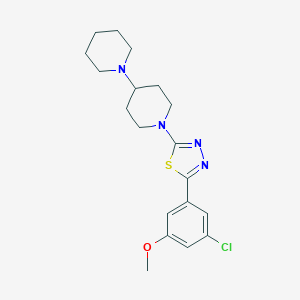 CHEMBL1641831 CHEMBL1641831 | C19H25ClN4OS | 392.946 | 6 / 0 | 4.5 | Yes |
| 7101 | 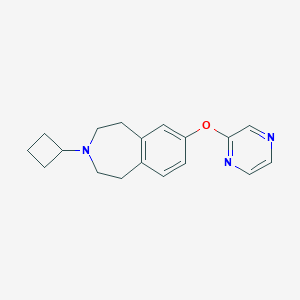 SCHEMBL3090788 SCHEMBL3090788 | C18H21N3O | 295.386 | 4 / 0 | 2.9 | Yes |
| 7153 | 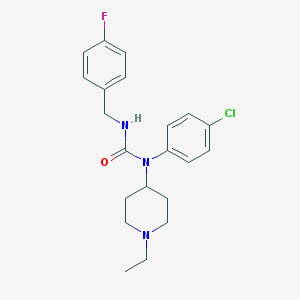 CHEMBL1092105 CHEMBL1092105 | C21H25ClFN3O | 389.899 | 3 / 1 | 4.3 | Yes |
| 7230 | 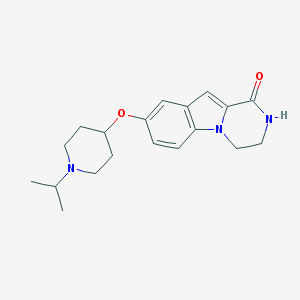 CHEMBL1258754 CHEMBL1258754 | C19H25N3O2 | 327.428 | 3 / 1 | 2.7 | Yes |
| 7324 | 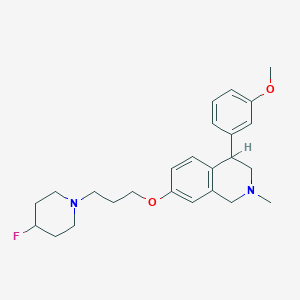 CHEMBL397299 CHEMBL397299 | C25H33FN2O2 | 412.549 | 5 / 0 | 4.5 | Yes |
| 7339 | 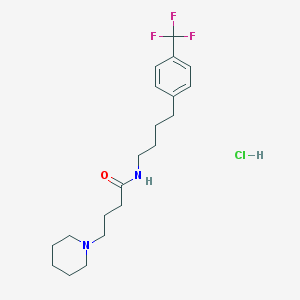 CHEMBL2441948 CHEMBL2441948 | C20H30ClF3N2O | 406.918 | 5 / 2 | N/A | N/A |
| 7352 | 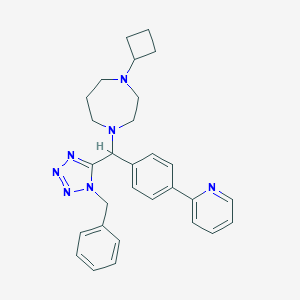 CHEMBL1222594 CHEMBL1222594 | C29H33N7 | 479.632 | 6 / 0 | 4.4 | Yes |
| 7371 | 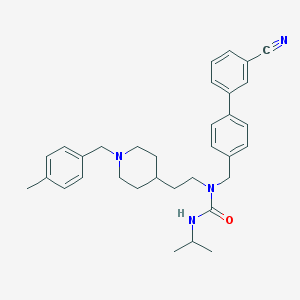 CHEMBL240116 CHEMBL240116 | C33H40N4O | 508.71 | 3 / 1 | 6.2 | No |
| 459285 |  CHEMBL3700332 CHEMBL3700332 | C18H27N5O3 | 361.446 | 5 / 1 | 0.5 | Yes |
| 7475 |  CHEMBL1783896 CHEMBL1783896 | C18H29N3O2 | 319.449 | 5 / 0 | 1.6 | Yes |
| 7497 |  CHEMBL158056 CHEMBL158056 | C27H34FN5O5 | 527.597 | 9 / 1 | 3.9 | No |
| 7711 |  CHEMBL453654 CHEMBL453654 | C25H29N3O3 | 419.525 | 5 / 0 | 4.0 | Yes |
| 7843 | 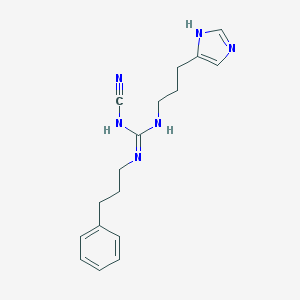 CHEMBL1078210 CHEMBL1078210 | C17H22N6 | 310.405 | 3 / 3 | 2.3 | Yes |
| 8018 | 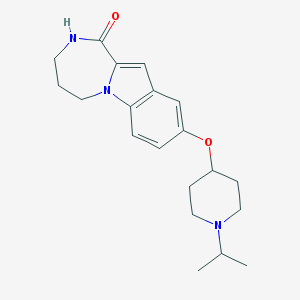 CHEMBL1258755 CHEMBL1258755 | C20H27N3O2 | 341.455 | 3 / 1 | 3.0 | Yes |
| 8104 | 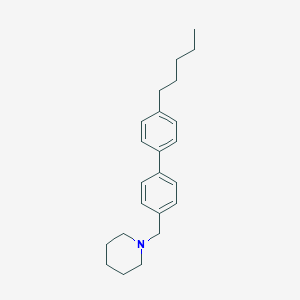 CHEMBL1934066 CHEMBL1934066 | C23H31N | 321.508 | 1 / 0 | 6.7 | No |
| 8181 | 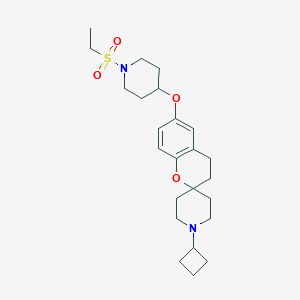 CHEMBL2013044 CHEMBL2013044 | C24H36N2O4S | 448.622 | 6 / 0 | 3.8 | Yes |
| 8279 | 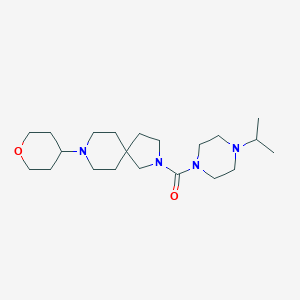 CHEMBL3127683 CHEMBL3127683 | C21H38N4O2 | 378.561 | 4 / 0 | 1.5 | Yes |
| 8415 | 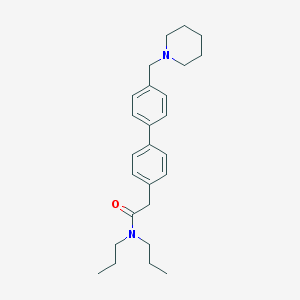 CHEMBL458507 CHEMBL458507 | C26H36N2O | 392.587 | 2 / 0 | 5.4 | No |
| 8419 | 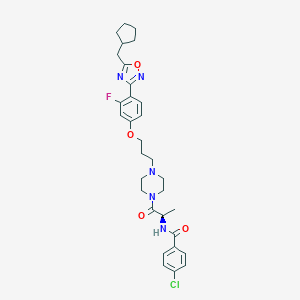 CHEMBL346711 CHEMBL346711 | C31H37ClFN5O4 | 598.116 | 8 / 1 | 6.1 | No |
| 8501 | 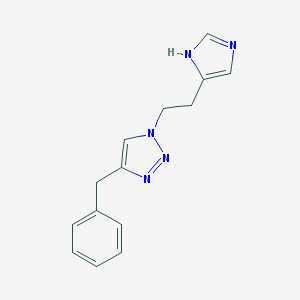 CHEMBL1688939 CHEMBL1688939 | C14H15N5 | 253.309 | 3 / 1 | 1.7 | Yes |
| 8526 | 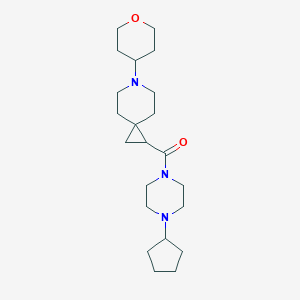 SCHEMBL2796894 SCHEMBL2796894 | C22H37N3O2 | 375.557 | 4 / 0 | 2.0 | Yes |
| 521698 | 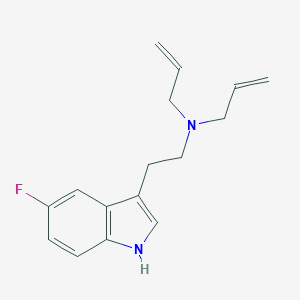 CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8605 | 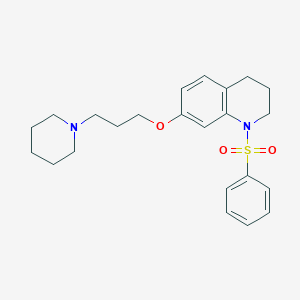 CHEMBL209139 CHEMBL209139 | C23H30N2O3S | 414.564 | 5 / 0 | 4.2 | Yes |
| 8614 | 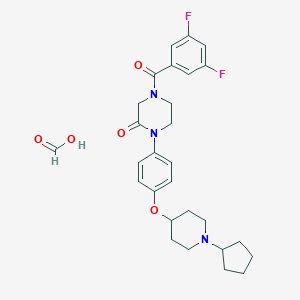 CHEMBL240567 CHEMBL240567 | C28H33F2N3O5 | 529.585 | 8 / 1 | N/A | No |
| 8690 | 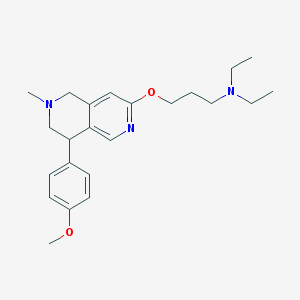 CHEMBL247267 CHEMBL247267 | C23H33N3O2 | 383.536 | 5 / 0 | 3.6 | Yes |
| 8988 | 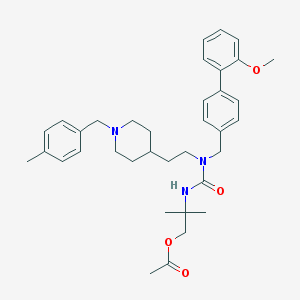 CHEMBL392013 CHEMBL392013 | C36H47N3O4 | 585.789 | 5 / 1 | 6.1 | No |
| 9013 | 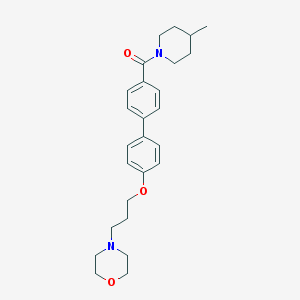 CHEMBL27545 CHEMBL27545 | C26H34N2O3 | 422.569 | 4 / 0 | 4.3 | Yes |
| 9061 | 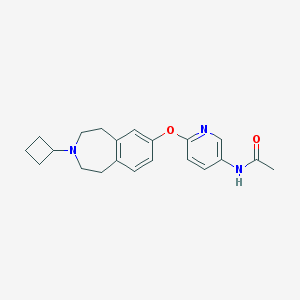 CHEMBL3092827 CHEMBL3092827 | C21H25N3O2 | 351.45 | 4 / 1 | 3.1 | Yes |
| 9453 | 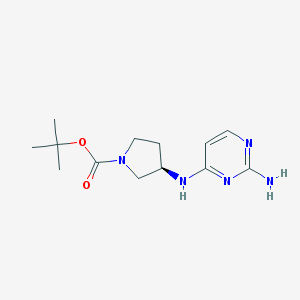 SCHEMBL3778815 SCHEMBL3778815 | C13H21N5O2 | 279.344 | 6 / 2 | 1.3 | Yes |
| 9477 | 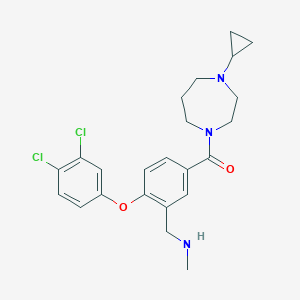 CHEMBL257738 CHEMBL257738 | C23H27Cl2N3O2 | 448.388 | 4 / 1 | 4.3 | Yes |
| 9813 | 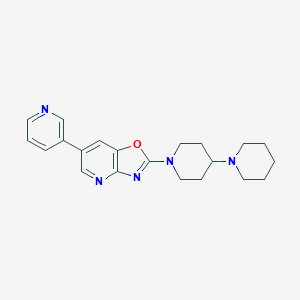 CHEMBL1958156 CHEMBL1958156 | C21H25N5O | 363.465 | 6 / 0 | 3.5 | Yes |
| 9885 | 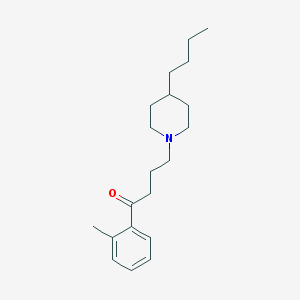 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417