

You can:
| Name | Neuromedin-K receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TACR3 |
| Synonym | SP-N receptor Tac3r Nmkr NKR NK3 receptor [ Show all ] |
| Disease | Schizophrenia Schizophrenia; Schizoaffective disorders Psychotic disorders Psychiatric disorder Irritable bowel syndrome [ Show all ] |
| Length | 465 |
| Amino acid sequence | MATLPAAETWIDGGGGVGADAVNLTASLAAGAATGAVETGWLQLLDQAGNLSSSPSALGLPVASPAPSQPWANLTNQFVQPSWRIALWSLAYGVVVAVAVLGNLIVIWIILAHKRMRTVTNYFLVNLAFSDASMAAFNTLVNFIYALHSEWYFGANYCRFQNFFPITAVFASIYSMTAIAVDRYMAIIDPLKPRLSATATKIVIGSIWILAFLLAFPQCLYSKTKVMPGRTLCFVQWPEGPKQHFTYHIIVIILVYCFPLLIMGITYTIVGITLWGGEIPGDTCDKYHEQLKAKRKVVKMMIIVVMTFAICWLPYHIYFILTAIYQQLNRWKYIQQVYLASFWLAMSSTMYNPIIYCCLNKRFRAGFKRAFRWCPFIKVSSYDELELKTTRFHPNRQSSMYTVTRMESMTVVFDPNDADTTRSSRKKRATPRDPSFNGCSRRNSKSASATSSFISSPYTSVDEYS |
| UniProt | P29371 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P29371 |
| 3D structure model | This predicted structure model is from GPCR-EXP P29371. |
| BioLiP | N/A |
| Therapeutic Target Database | T29683 |
| ChEMBL | CHEMBL4429 |
| IUPHAR | 362 |
| DrugBank | BE0002371 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 14 | 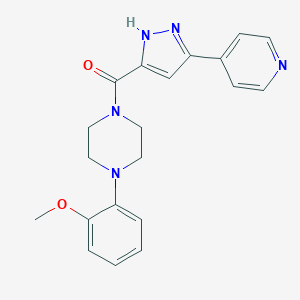 CHEMBL1760216 CHEMBL1760216 | C20H21N5O2 | 363.421 | 5 / 1 | 2.2 | Yes |
| 397 | 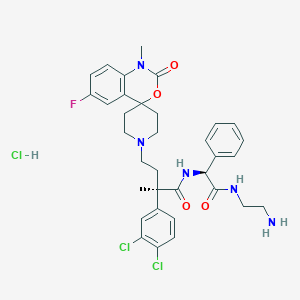 CHEMBL3288163 CHEMBL3288163 | C34H39Cl3FN5O4 | 707.065 | 7 / 4 | N/A | No |
| 1068 | 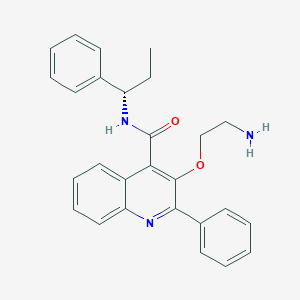 CHEMBL2113679 CHEMBL2113679 | C27H27N3O2 | 425.532 | 4 / 2 | 4.6 | Yes |
| 2490 | 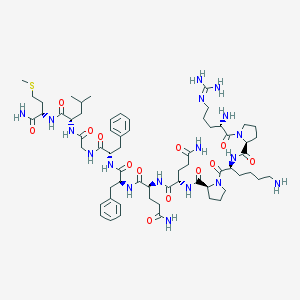 Substance P Substance P | C63H98N18O13S | 1347.65 | 17 / 15 | -2.3 | No |
| 2801 | 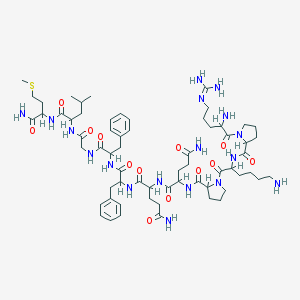 sub-stance p sub-stance p | C63H98N18O13S | 1347.65 | 17 / 15 | -2.3 | No |
| 3201 | 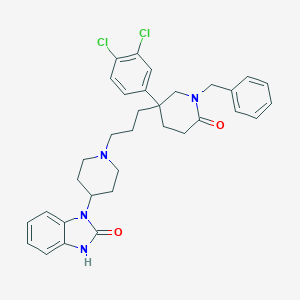 CHEMBL26790 CHEMBL26790 | C33H36Cl2N4O2 | 591.577 | 3 / 1 | 6.1 | No |
| 3449 | 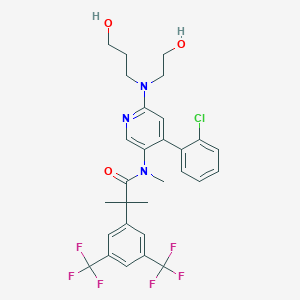 CHEMBL1084165 CHEMBL1084165 | C29H30ClF6N3O3 | 618.017 | 11 / 2 | 6.2 | No |
| 536027 | 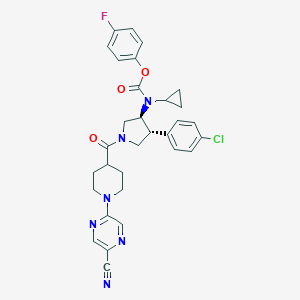 SCHEMBL1993496 SCHEMBL1993496 | C31H30ClFN6O3 | 589.068 | 8 / 0 | 4.6 | No |
| 5214 | 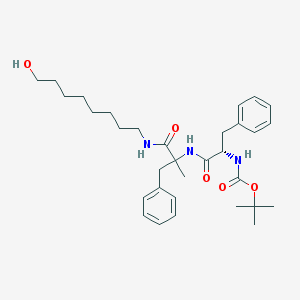 CHEMBL298272 CHEMBL298272 | C32H47N3O5 | 553.744 | 5 / 4 | 5.6 | No |
| 5218 | 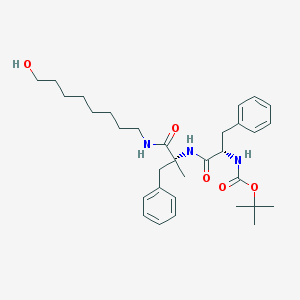 CHEMBL296014 CHEMBL296014 | C32H47N3O5 | 553.744 | 5 / 4 | 5.6 | No |
| 5220 | 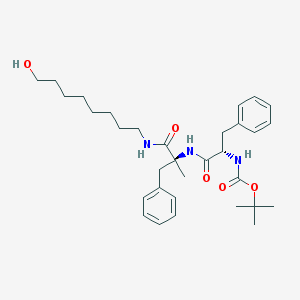 CHEMBL430576 CHEMBL430576 | C32H47N3O5 | 553.744 | 5 / 4 | 5.6 | No |
| 5596 | 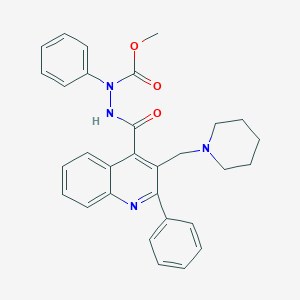 CHEMBL221077 CHEMBL221077 | C30H30N4O3 | 494.595 | 5 / 1 | 5.5 | No |
| 5861 | 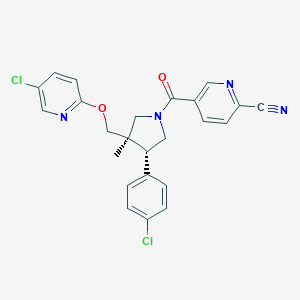 CHEMBL3648215 CHEMBL3648215 | C24H20Cl2N4O2 | 467.35 | 5 / 0 | 4.7 | Yes |
| 7227 | 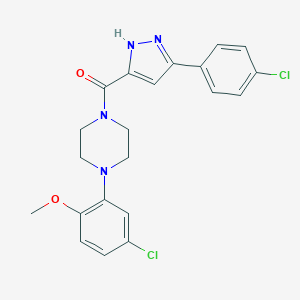 CHEMBL1760331 CHEMBL1760331 | C21H20Cl2N4O2 | 431.317 | 4 / 1 | 4.5 | Yes |
| 7718 | 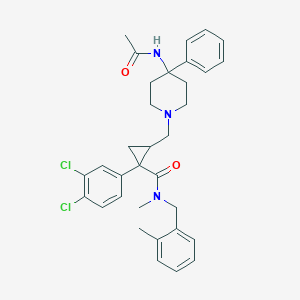 CHEMBL1682628 CHEMBL1682628 | C33H37Cl2N3O2 | 578.578 | 3 / 1 | 5.9 | No |
| 7874 | 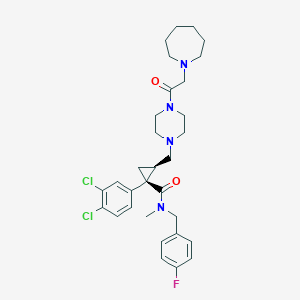 CHEMBL1682682 CHEMBL1682682 | C31H39Cl2FN4O2 | 589.577 | 5 / 0 | 5.0 | No |
| 442075 | 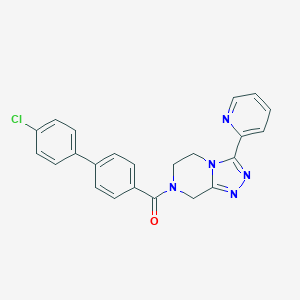 CHEMBL3421991 CHEMBL3421991 | C23H18ClN5O | 415.881 | 4 / 0 | 3.0 | Yes |
| 9348 | 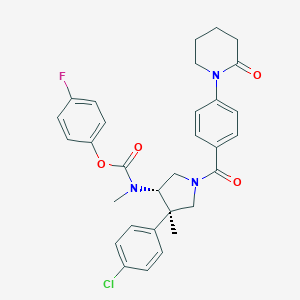 CHEMBL3680211 CHEMBL3680211 | C31H31ClFN3O4 | 564.054 | 5 / 0 | 5.5 | No |
| 9409 | 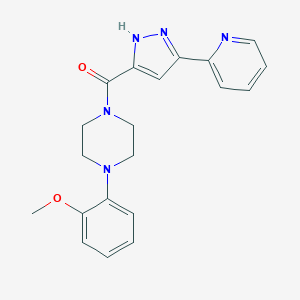 CHEMBL1760214 CHEMBL1760214 | C20H21N5O2 | 363.421 | 5 / 1 | 2.2 | Yes |
| 536242 | 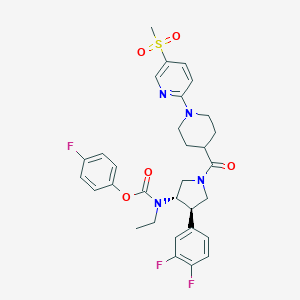 SCHEMBL1990036 SCHEMBL1990036 | C31H33F3N4O5S | 630.683 | 10 / 0 | 4.2 | No |
| 521744 | 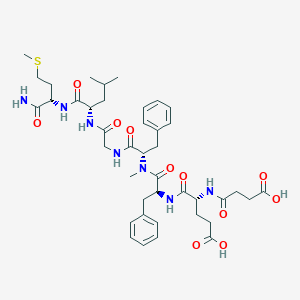 CHEMBL3735396 CHEMBL3735396 | C41H57N7O11S | 856.005 | 12 / 8 | 1.5 | No |
| 521748 | 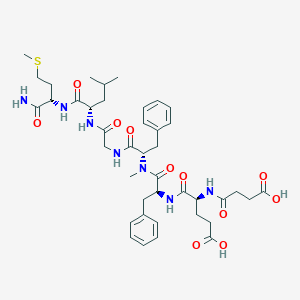 CHEMBL3735210 CHEMBL3735210 | C41H57N7O11S | 856.005 | 12 / 8 | 1.5 | No |
| 536282 | 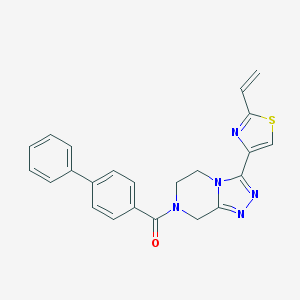 CHEMBL3940252 CHEMBL3940252 | C23H19N5OS | 413.499 | 5 / 0 | 3.3 | Yes |
| 536311 | 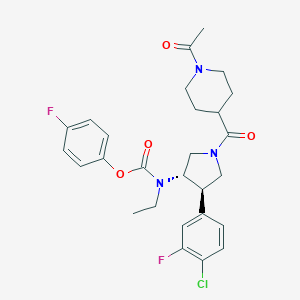 SCHEMBL1984740 SCHEMBL1984740 | C27H30ClF2N3O4 | 534.001 | 6 / 0 | 3.9 | No |
| 521827 | 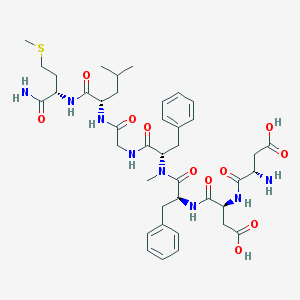 CHEMBL3736313 CHEMBL3736313 | C40H56N8O11S | 856.993 | 13 / 9 | -2.0 | No |
| 13001 | 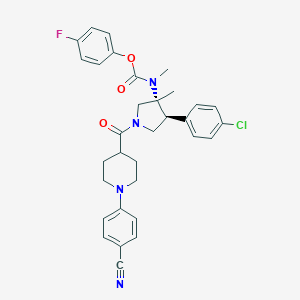 CHEMBL3680187 CHEMBL3680187 | C32H32ClFN4O3 | 575.081 | 6 / 0 | 5.7 | No |
| 521850 | 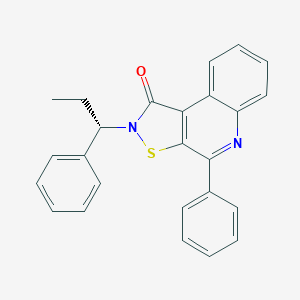 CHEMBL3814985 CHEMBL3814985 | C25H20N2OS | 396.508 | 3 / 0 | 5.8 | No |
| 13772 | 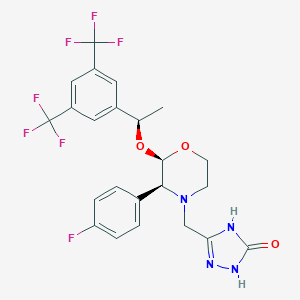 Aprepitant Aprepitant | C23H21F7N4O3 | 534.435 | 12 / 2 | 4.2 | No |
| 557681 | 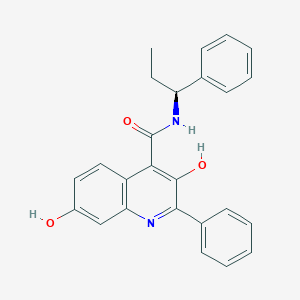 CHEMBL3813735 CHEMBL3813735 | C25H22N2O3 | 398.462 | 4 / 3 | 5.4 | No |
| 521894 | 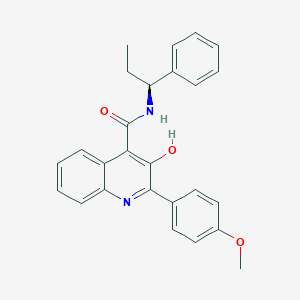 CHEMBL3814793 CHEMBL3814793 | C26H24N2O3 | 412.489 | 4 / 2 | 5.7 | No |
| 536375 | 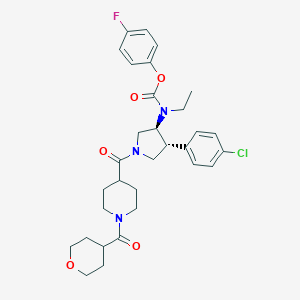 SCHEMBL1989445 SCHEMBL1989445 | C31H37ClFN3O5 | 586.101 | 6 / 0 | 4.1 | No |
| 548063 | 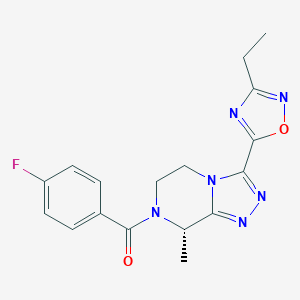 CHEMBL3984151 CHEMBL3984151 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 557731 | 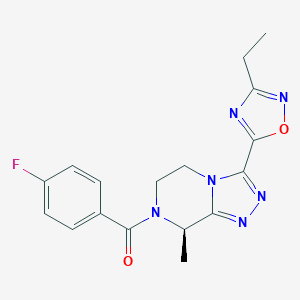 SCHEMBL16143091 SCHEMBL16143091 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 536394 | 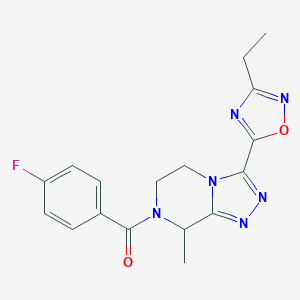 CHEMBL3942295 CHEMBL3942295 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 16171 | 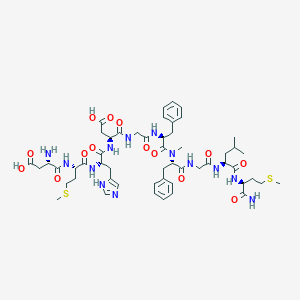 CHEMBL2347498 CHEMBL2347498 | C53H75N13O14S2 | 1182.38 | 18 / 13 | -2.7 | No |
| 16539 | 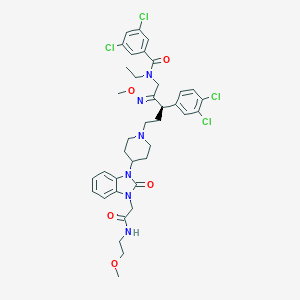 CHEMBL75478 CHEMBL75478 | C38H44Cl4N6O5 | 806.607 | 7 / 1 | 7.1 | No |
| 16666 | 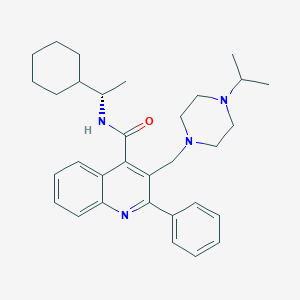 CHEMBL47775 CHEMBL47775 | C32H42N4O | 498.715 | 4 / 1 | 6.4 | No |
| 16900 | 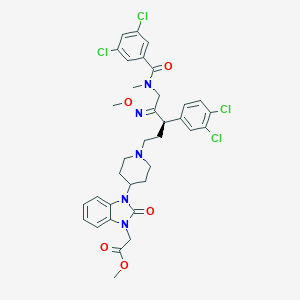 CHEMBL78851 CHEMBL78851 | C35H37Cl4N5O5 | 749.511 | 7 / 0 | 7.4 | No |
| 17232 | 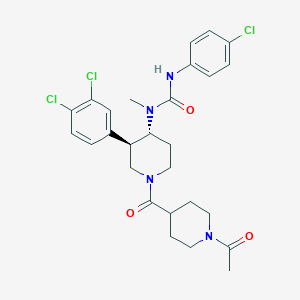 SCHEMBL1973042 SCHEMBL1973042 | C27H31Cl3N4O3 | 565.92 | 3 / 1 | 4.4 | No |
| 17256 | 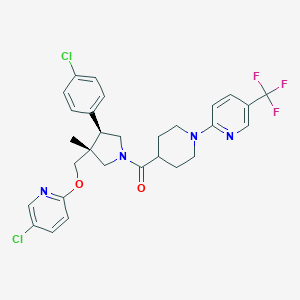 SCHEMBL6837152 SCHEMBL6837152 | C29H29Cl2F3N4O2 | 593.472 | 8 / 0 | 6.5 | No |
| 17486 | 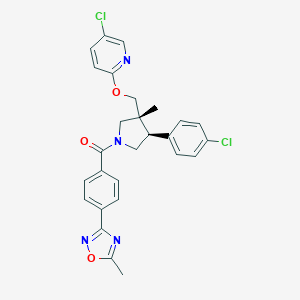 SCHEMBL6837264 SCHEMBL6837264 | C27H24Cl2N4O3 | 523.414 | 6 / 0 | 6.0 | No |
| 17564 | 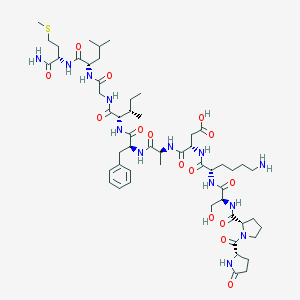 ELEDOISIN ELEDOISIN | C54H85N13O15S | 1188.41 | 17 / 14 | -3.6 | No |
| 536444 | 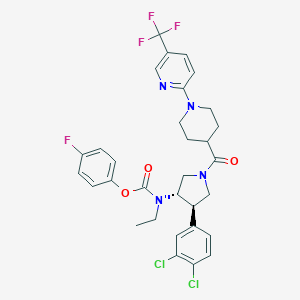 SCHEMBL1988590 SCHEMBL1988590 | C31H30Cl2F4N4O3 | 653.5 | 9 / 0 | 6.9 | No |
| 17811 | 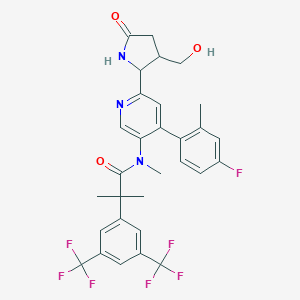 CHEMBL1911964 CHEMBL1911964 | C30H28F7N3O3 | 611.561 | 11 / 2 | 5.1 | No |
| 17861 | 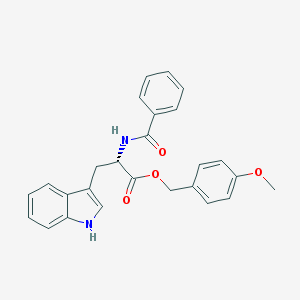 CHEMBL282728 CHEMBL282728 | C26H24N2O4 | 428.488 | 4 / 2 | 4.5 | Yes |
| 19039 | 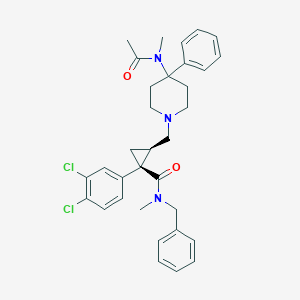 CHEMBL1682666 CHEMBL1682666 | C33H37Cl2N3O2 | 578.578 | 3 / 0 | 5.7 | No |
| 19622 | 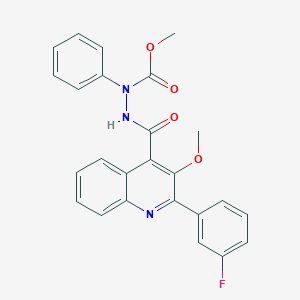 CHEMBL375640 CHEMBL375640 | C25H20FN3O4 | 445.45 | 6 / 1 | 4.9 | Yes |
| 19868 | 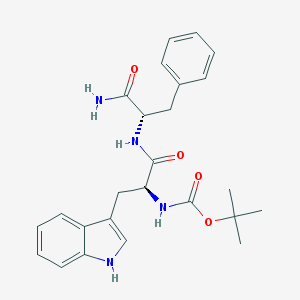 CHEMBL32301 CHEMBL32301 | C25H30N4O4 | 450.539 | 4 / 4 | 3.0 | Yes |
| 442447 | 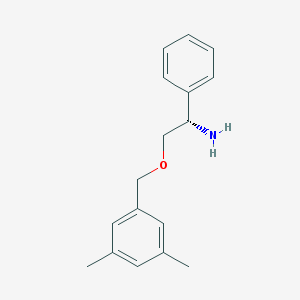 CHEMBL71498 CHEMBL71498 | C17H21NO | 255.361 | 2 / 1 | 2.9 | Yes |
| 442472 | 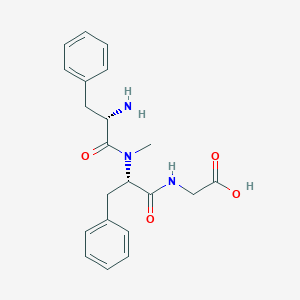 CHEMBL3361403 CHEMBL3361403 | C21H25N3O4 | 383.448 | 5 / 3 | -0.5 | Yes |
| 20917 | 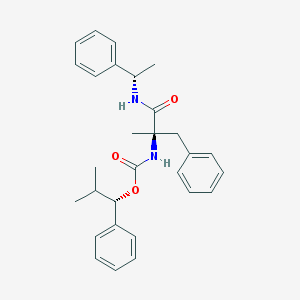 CHEMBL45278 CHEMBL45278 | C29H34N2O3 | 458.602 | 3 / 2 | 6.2 | No |
| 20918 | 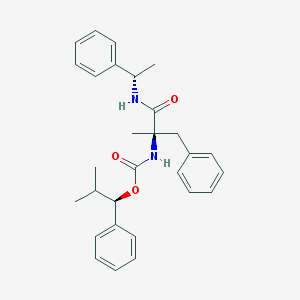 CHEMBL44686 CHEMBL44686 | C29H34N2O3 | 458.602 | 3 / 2 | 6.2 | No |
| 21189 | 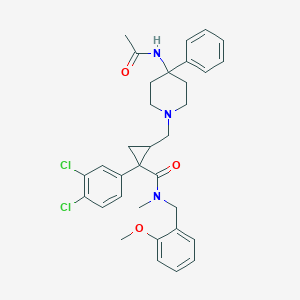 CHEMBL1682629 CHEMBL1682629 | C33H37Cl2N3O3 | 594.577 | 4 / 1 | 5.5 | No |
| 21530 | 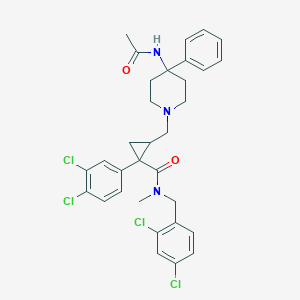 CHEMBL1682640 CHEMBL1682640 | C32H33Cl4N3O2 | 633.435 | 3 / 1 | 6.8 | No |
| 536527 | 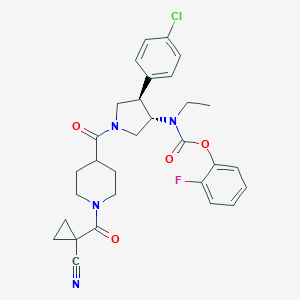 SCHEMBL1990943 SCHEMBL1990943 | C30H32ClFN4O4 | 567.058 | 6 / 0 | 4.1 | No |
| 21725 | 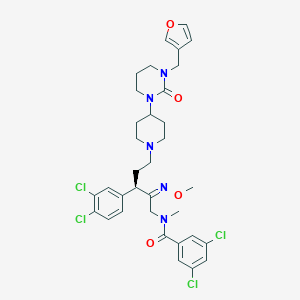 CHEMBL307733 CHEMBL307733 | C34H39Cl4N5O4 | 723.517 | 6 / 0 | 7.0 | No |
| 21939 | 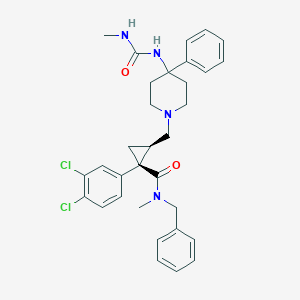 CHEMBL1682670 CHEMBL1682670 | C32H36Cl2N4O2 | 579.566 | 3 / 2 | 5.3 | No |
| 553345 | 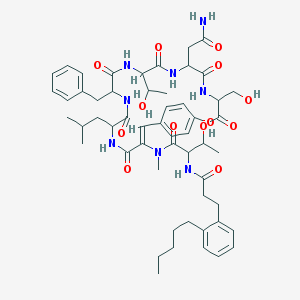 FK 224 FK 224 | C54H72N8O13 | 1041.21 | 13 / 10 | 4.9 | No |
| 522138 | 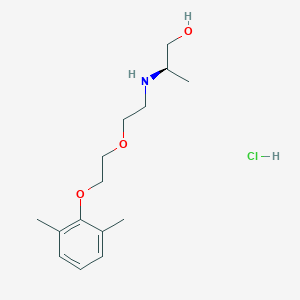 CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522198 | 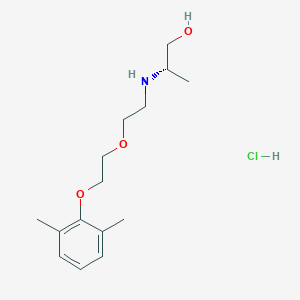 CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 536568 | 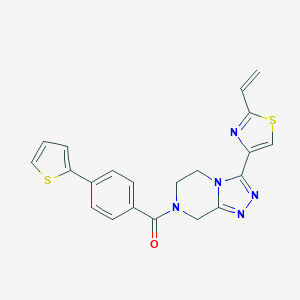 CHEMBL3964222 CHEMBL3964222 | C21H17N5OS2 | 419.521 | 6 / 0 | 3.0 | Yes |
| 558001 | 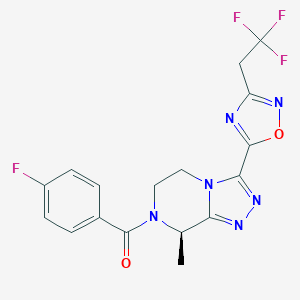 SCHEMBL16115678 SCHEMBL16115678 | C17H14F4N6O2 | 410.333 | 10 / 0 | 1.9 | Yes |
| 548154 | 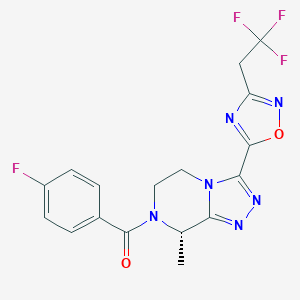 CHEMBL3898651 CHEMBL3898651 | C17H14F4N6O2 | 410.333 | 10 / 0 | 1.9 | Yes |
| 23707 | 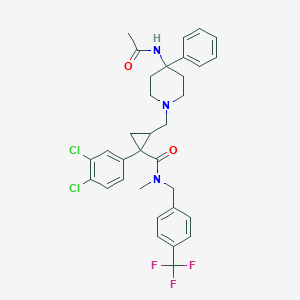 CHEMBL1682637 CHEMBL1682637 | C33H34Cl2F3N3O2 | 632.549 | 6 / 1 | 6.4 | No |
| 24267 | 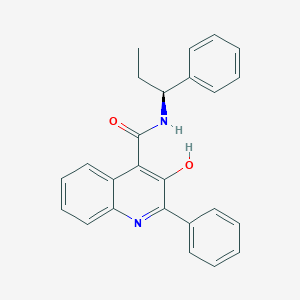 Talnetant Talnetant | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24274 | 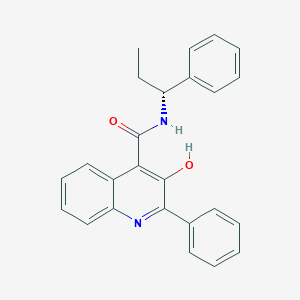 CHEMBL430118 CHEMBL430118 | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24276 | 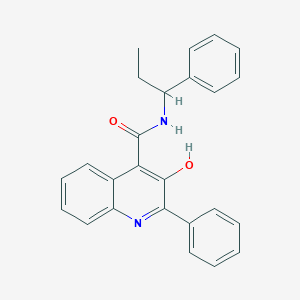 SB 223412 SB 223412 | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24551 | 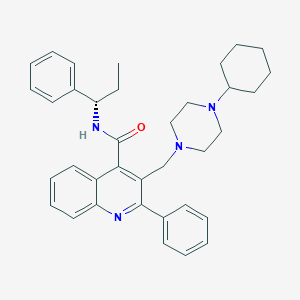 CHEMBL33868 CHEMBL33868 | C36H42N4O | 546.759 | 4 / 1 | 7.0 | No |
| 24565 | 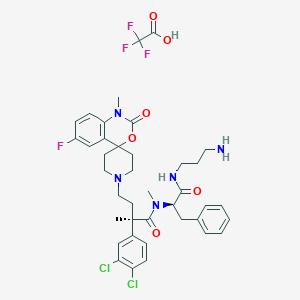 CHEMBL3288170 CHEMBL3288170 | C39H45Cl2F4N5O6 | 826.712 | 12 / 3 | N/A | No |
| 25437 | 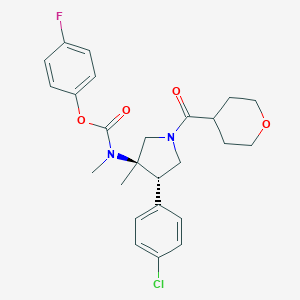 CHEMBL3680182 CHEMBL3680182 | C25H28ClFN2O4 | 474.957 | 5 / 0 | 4.0 | Yes |
| 536650 | 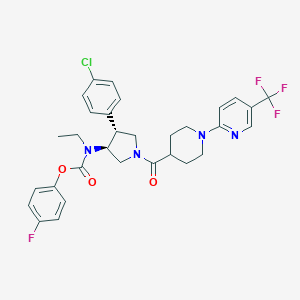 SCHEMBL1985493 SCHEMBL1985493 | C31H31ClF4N4O3 | 619.058 | 9 / 0 | 6.3 | No |
| 558056 | 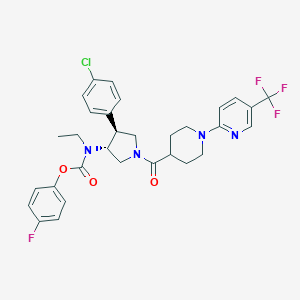 SCHEMBL1987661 SCHEMBL1987661 | C31H31ClF4N4O3 | 619.058 | 9 / 0 | 6.3 | No |
| 25702 | 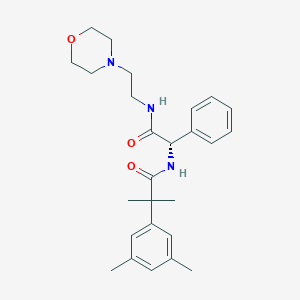 CHEMBL3104771 CHEMBL3104771 | C26H35N3O3 | 437.584 | 4 / 2 | 3.5 | Yes |
| 25737 | 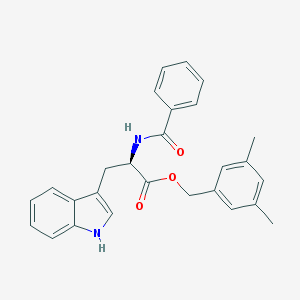 CHEMBL2114308 CHEMBL2114308 | C27H26N2O3 | 426.516 | 3 / 2 | 5.3 | No |
| 25738 | 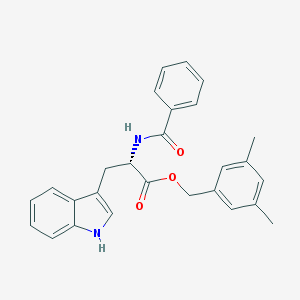 CHEMBL278351 CHEMBL278351 | C27H26N2O3 | 426.516 | 3 / 2 | 5.3 | No |
| 548173 | 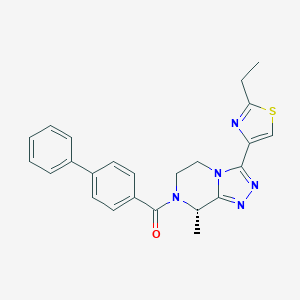 CHEMBL3984131 CHEMBL3984131 | C24H23N5OS | 429.542 | 5 / 0 | 3.7 | Yes |
| 558070 | 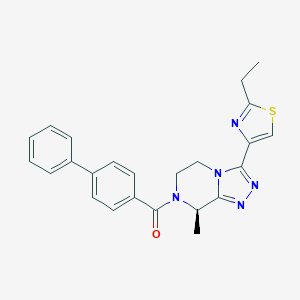 SCHEMBL14850857 SCHEMBL14850857 | C24H23N5OS | 429.542 | 5 / 0 | 3.7 | Yes |
| 26279 | 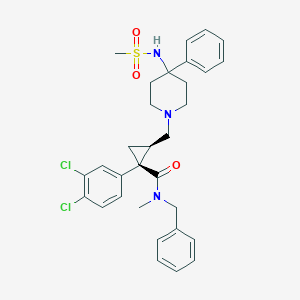 CHEMBL1682673 CHEMBL1682673 | C31H35Cl2N3O3S | 600.599 | 5 / 1 | 5.3 | No |
| 442719 | 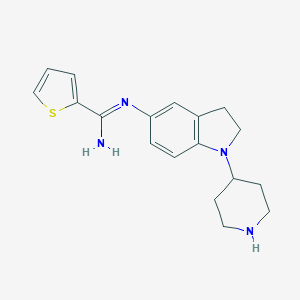 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27828 | 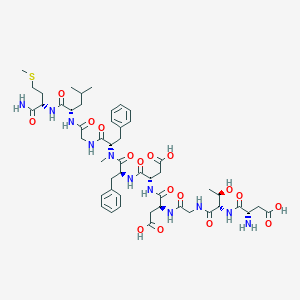 CHEMBL2347501 CHEMBL2347501 | C50H71N11O17S | 1130.24 | 19 / 14 | -4.6 | No |
| 28460 | 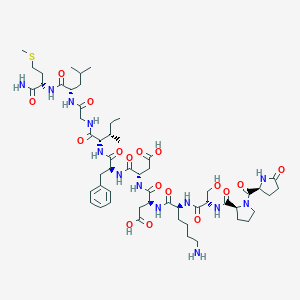 CHEMBL2347505 CHEMBL2347505 | C55H85N13O17S | 1232.42 | 19 / 15 | -4.5 | No |
| 442796 | 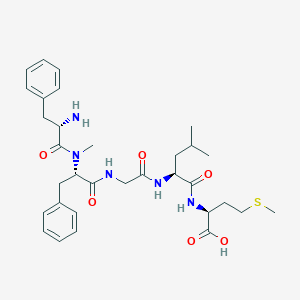 CHEMBL3361402 CHEMBL3361402 | C32H45N5O6S | 627.801 | 8 / 5 | 0.9 | No |
| 536738 | 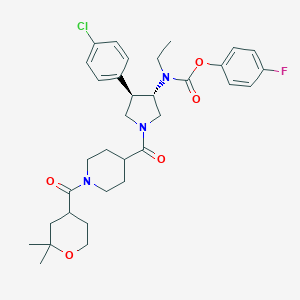 SCHEMBL1986618 SCHEMBL1986618 | C33H41ClFN3O5 | 614.155 | 6 / 0 | 4.7 | No |
| 30721 | 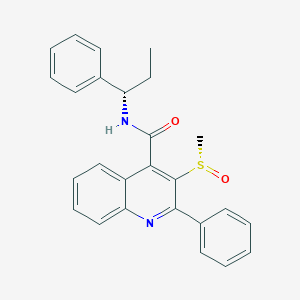 CHEMBL1682939 CHEMBL1682939 | C26H24N2O2S | 428.55 | 4 / 1 | 4.7 | Yes |
| 30722 | 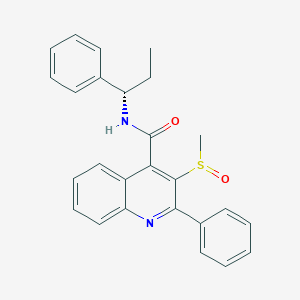 CHEMBL1682938 CHEMBL1682938 | C26H24N2O2S | 428.55 | 4 / 1 | 4.7 | Yes |
| 558242 | 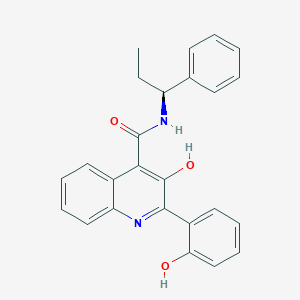 CHEMBL3813719 CHEMBL3813719 | C25H22N2O3 | 398.462 | 4 / 3 | 5.4 | No |
| 32464 | 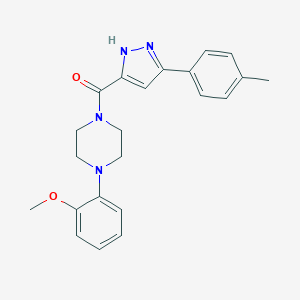 CHEMBL1760210 CHEMBL1760210 | C22H24N4O2 | 376.46 | 4 / 1 | 3.6 | Yes |
| 32769 | 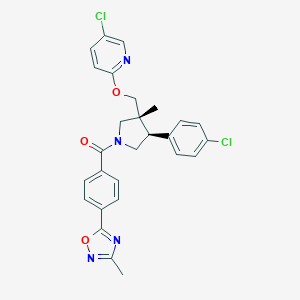 SCHEMBL6837110 SCHEMBL6837110 | C27H24Cl2N4O3 | 523.414 | 6 / 0 | 6.0 | No |
| 536854 | 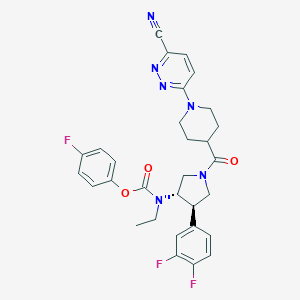 SCHEMBL1990795 SCHEMBL1990795 | C30H29F3N6O3 | 578.596 | 10 / 0 | 4.0 | No |
| 442959 | 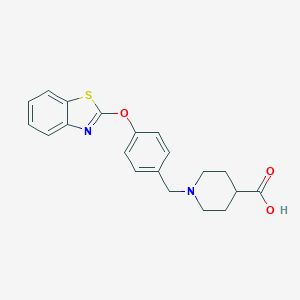 UNII-7W7511NPF8 UNII-7W7511NPF8 | C20H20N2O3S | 368.451 | 6 / 1 | 1.8 | Yes |
| 34095 | 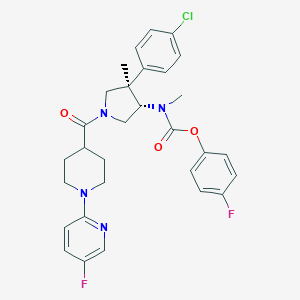 CHEMBL3680200 CHEMBL3680200 | C30H31ClF2N4O3 | 569.05 | 7 / 0 | 5.7 | No |
| 536875 | 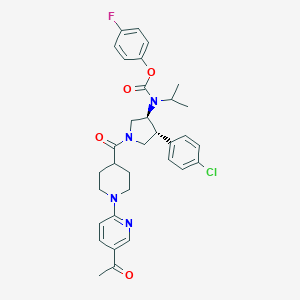 SCHEMBL1990051 SCHEMBL1990051 | C33H36ClFN4O4 | 607.123 | 7 / 0 | 5.5 | No |
| 443043 | 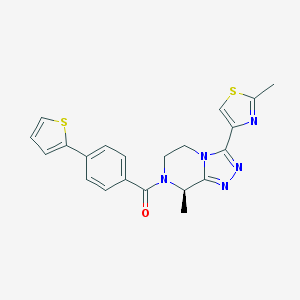 CHEMBL3422010 CHEMBL3422010 | C21H19N5OS2 | 421.537 | 6 / 0 | 3.0 | Yes |
| 443050 | 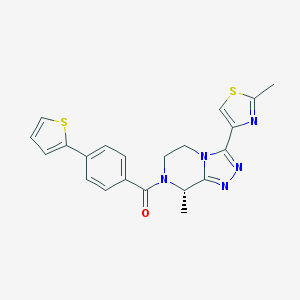 CHEMBL3422011 CHEMBL3422011 | C21H19N5OS2 | 421.537 | 6 / 0 | 3.0 | Yes |
| 34909 | 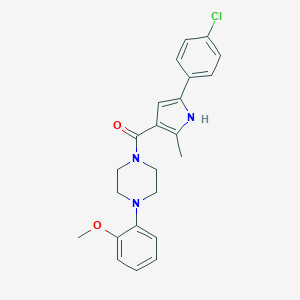 CHEMBL1760207 CHEMBL1760207 | C23H24ClN3O2 | 409.914 | 3 / 1 | 4.4 | Yes |
| 35008 | 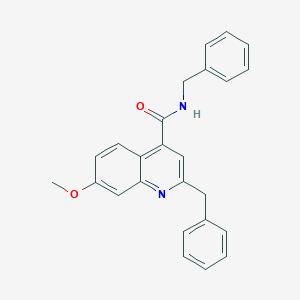 CHEMBL50905 CHEMBL50905 | C25H22N2O2 | 382.463 | 3 / 1 | 4.9 | Yes |
| 36008 | 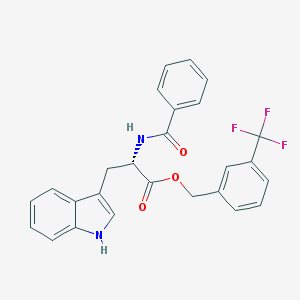 CHEMBL26336 CHEMBL26336 | C26H21F3N2O3 | 466.46 | 6 / 2 | 5.4 | No |
| 548309 | 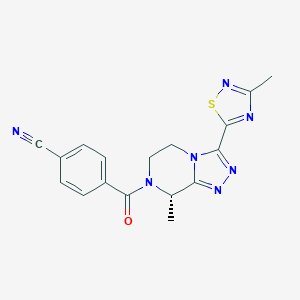 CHEMBL3896219 CHEMBL3896219 | C17H15N7OS | 365.415 | 7 / 0 | 1.2 | Yes |
| 558380 | 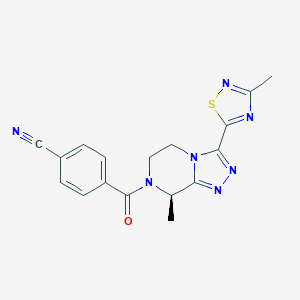 SCHEMBL17005219 SCHEMBL17005219 | C17H15N7OS | 365.415 | 7 / 0 | 1.2 | Yes |
| 36138 | 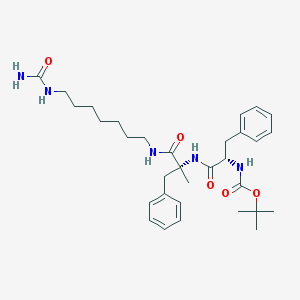 PD157672 PD157672 | C32H47N5O5 | 581.758 | 5 / 5 | 4.5 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417