

You can:
| Name | Neurotensin receptor type 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | NTSR2 |
| Synonym | levocabastine-sensitive neurotensin receptor neurotensin receptor type 2 high-affinity levocabastine-sensitive neurotensin receptor NT2R NTR2 [ Show all ] |
| Disease | N/A |
| Length | 410 |
| Amino acid sequence | METSSPRPPRPSSNPGLSLDARLGVDTRLWAKVLFTALYALIWALGAAGNALSAHVVLKARAGRAGRLRHHVLSLALAGLLLLLVGVPVELYSFVWFHYPWVFGDLGCRGYYFVHELCAYATVLSVAGLSAERCLAVCQPLRARSLLTPRRTRWLVALSWAASLGLALPMAVIMGQKHELETADGEPEPASRVCTVLVSRTALQVFIQVNVLVSFVLPLALTAFLNGVTVSHLLALCSQVPSTSTPGSSTPSRLELLSEEGLLSFIVWKKTFIQGGQVSLVRHKDVRRIRSLQRSVQVLRAIVVMYVICWLPYHARRLMYCYVPDDAWTDPLYNFYHYFYMVTNTLFYVSSAVTPLLYNAVSSSFRKLFLEAVSSLCGEHHPMKRLPPKPQSPTLMDTASGFGDPPETRT |
| UniProt | O95665 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | O95665 |
| 3D structure model | This predicted structure model is from GPCR-EXP O95665. |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2514 |
| IUPHAR | 310 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 3810 | 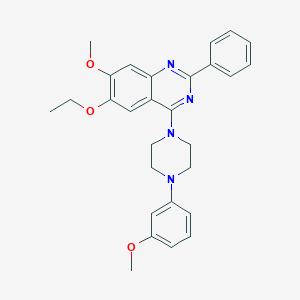 CHEMBL2431236 CHEMBL2431236 | C28H30N4O3 | 470.573 | 7 / 0 | 5.5 | No |
| 25851 | 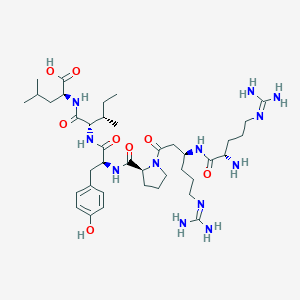 CHEMBL1766936 CHEMBL1766936 | C39H66N12O8 | 831.033 | 11 / 11 | -2.5 | No |
| 32320 | 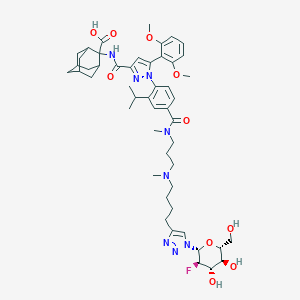 CHEMBL3086356 CHEMBL3086356 | C50H67FN8O10 | 959.13 | 15 / 5 | 2.9 | No |
| 548293 | 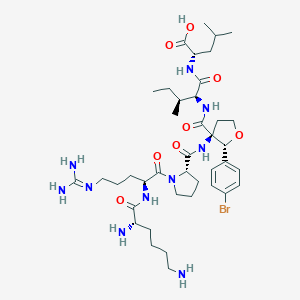 CHEMBL3951982 CHEMBL3951982 | C40H65BrN10O8 | 893.926 | 11 / 9 | -1.1 | No |
| 38862 | 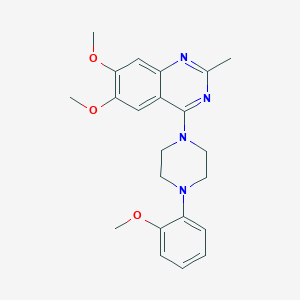 CHEMBL2431113 CHEMBL2431113 | C22H26N4O3 | 394.475 | 7 / 0 | 3.9 | Yes |
| 40577 | 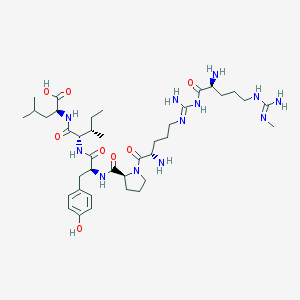 CHEMBL434227 CHEMBL434227 | C39H66N12O8 | 831.033 | 12 / 11 | -1.5 | No |
| 548363 | 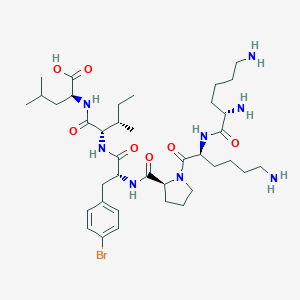 CHEMBL3950695 CHEMBL3950695 | C38H63BrN8O7 | 823.875 | 10 / 8 | -1.0 | No |
| 548365 | 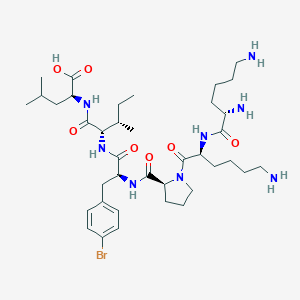 CHEMBL3979668 CHEMBL3979668 | C38H63BrN8O7 | 823.875 | 10 / 8 | -1.0 | No |
| 43026 | 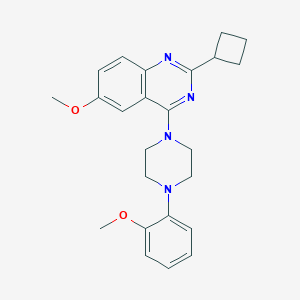 CHEMBL2431125 CHEMBL2431125 | C24H28N4O2 | 404.514 | 6 / 0 | 4.6 | Yes |
| 548418 | 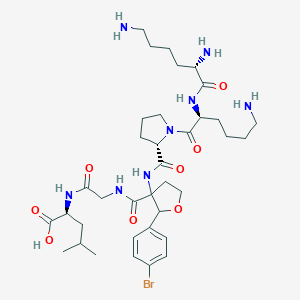 CHEMBL3899699 CHEMBL3899699 | C36H57BrN8O8 | 809.804 | 11 / 8 | -1.9 | No |
| 61622 | 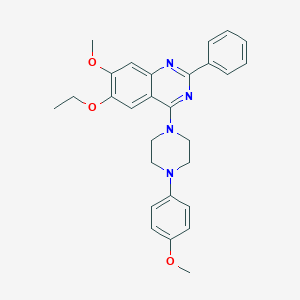 CHEMBL2431237 CHEMBL2431237 | C28H30N4O3 | 470.573 | 7 / 0 | 5.5 | No |
| 548624 | 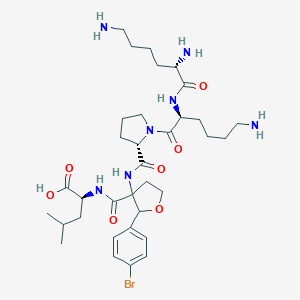 CHEMBL3890335 CHEMBL3890335 | C34H54BrN7O7 | 752.752 | 10 / 7 | -1.2 | No |
| 62112 |  CHEMBL1766945 CHEMBL1766945 | C39H66N10O8 | 803.019 | 11 / 9 | -1.4 | No |
| 64700 |  CHEMBL2431110 CHEMBL2431110 | C17H22N4O2 | 314.389 | 6 / 1 | 1.9 | Yes |
| 66200 |  CHEMBL1766929 CHEMBL1766929 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66202 |  CHEMBL1766932 CHEMBL1766932 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66207 |  CHEMBL342252 CHEMBL342252 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66208 |  CHEMBL1766934 CHEMBL1766934 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 471091 |  neurotensin (8-13) neurotensin (8-13) | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66211 |  CHEMBL1766931 CHEMBL1766931 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66212 | 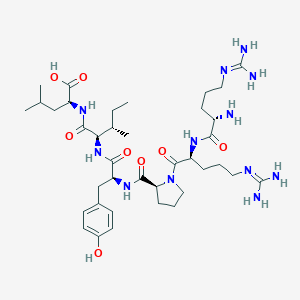 CHEMBL1766933 CHEMBL1766933 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 66215 | 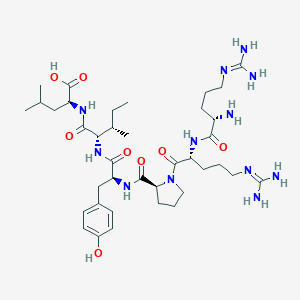 CHEMBL1766930 CHEMBL1766930 | C38H64N12O8 | 817.006 | 11 / 11 | -4.0 | No |
| 68134 | 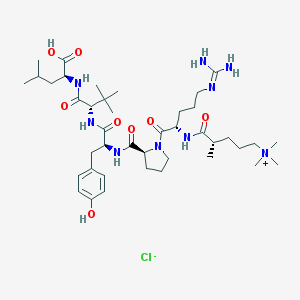 CHEMBL1172376 CHEMBL1172376 | C41H70ClN9O8 | 852.516 | 10 / 8 | N/A | No |
| 71472 | 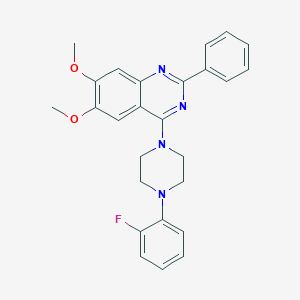 CHEMBL2431135 CHEMBL2431135 | C26H25FN4O2 | 444.51 | 7 / 0 | 5.3 | No |
| 72071 | 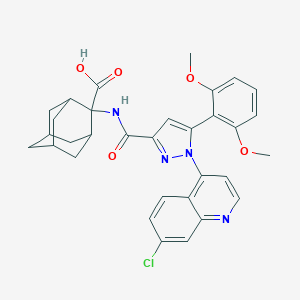 Meclinertant Meclinertant | C32H31ClN4O5 | 587.073 | 7 / 2 | 5.8 | No |
| 72888 | 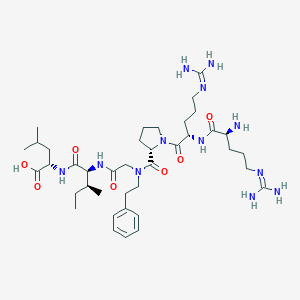 CHEMBL1766942 CHEMBL1766942 | C39H66N12O7 | 815.034 | 10 / 9 | -2.0 | No |
| 75436 | 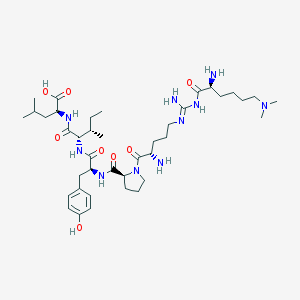 CHEMBL132507 CHEMBL132507 | C40H68N10O8 | 817.046 | 12 / 9 | -0.1 | No |
| 88582 | 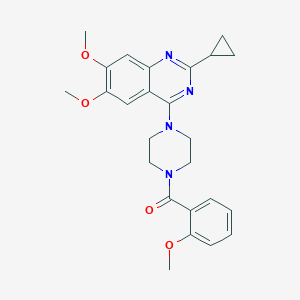 CHEMBL2431109 CHEMBL2431109 | C25H28N4O4 | 448.523 | 7 / 0 | 3.4 | Yes |
| 93316 | 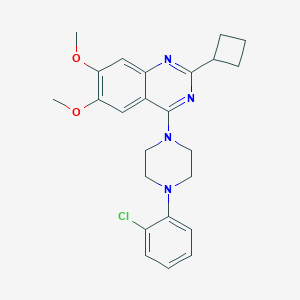 CHEMBL2431234 CHEMBL2431234 | C24H27ClN4O2 | 438.956 | 6 / 0 | 5.3 | No |
| 474854 | 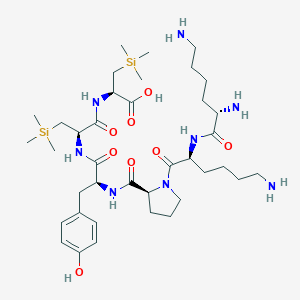 CHEMBL3622803 CHEMBL3622803 | C38H68N8O8Si2 | 821.18 | 11 / 9 | N/A | No |
| 97573 | 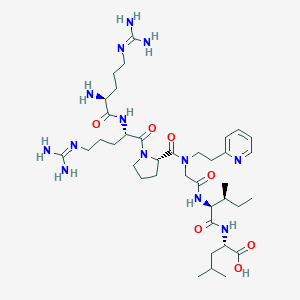 CHEMBL1766943 CHEMBL1766943 | C38H65N13O7 | 816.022 | 11 / 9 | -3.0 | No |
| 99455 | 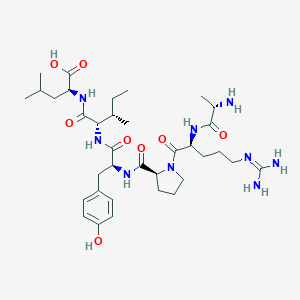 CHEMBL1766923 CHEMBL1766923 | C35H57N9O8 | 731.896 | 10 / 9 | -3.0 | No |
| 99924 |  CHEMBL1766937 CHEMBL1766937 | C39H66N12O8 | 831.033 | 11 / 11 | -2.5 | No |
| 100509 |  CHEMBL2431233 CHEMBL2431233 | C24H27FN4O2 | 422.504 | 7 / 0 | 4.8 | Yes |
| 102078 |  CHEMBL1417160 CHEMBL1417160 | C23H27N5O2 | 405.502 | 7 / 0 | 3.9 | Yes |
| 477516 |  CHEMBL3622802 CHEMBL3622802 | C38H66N8O8Si | 791.079 | 11 / 9 | N/A | No |
| 119639 |  CHEMBL1766944 CHEMBL1766944 | C39H66N10O8 | 803.019 | 11 / 9 | -1.4 | No |
| 119991 |  CHEMBL2431127 CHEMBL2431127 | C23H26N4O | 374.488 | 5 / 0 | 4.7 | Yes |
| 539578 |  CHEMBL3942339 CHEMBL3942339 | C36H58N12O9 | 802.935 | 12 / 11 | -4.8 | No |
| 549678 |  CHEMBL3971946 CHEMBL3971946 | C36H58N12O9 | 802.935 | 12 / 11 | -4.8 | No |
| 148523 |  CHEMBL258221 CHEMBL258221 | C38H64N8O8 | 760.978 | 11 / 9 | -2.1 | No |
| 481837 |  CHEMBL3622804 CHEMBL3622804 | C40H72N8O8Si2 | 849.234 | 11 / 9 | N/A | No |
| 154200 |  CHEMBL2431118 CHEMBL2431118 | C25H32N4O3 | 436.556 | 7 / 0 | 5.1 | No |
| 157462 |  CHEMBL336836 CHEMBL336836 | C40H68N12O8 | 845.06 | 12 / 11 | -1.2 | No |
| 482996 |  CHEMBL3622805 CHEMBL3622805 | C39H68N10O8Si | 833.12 | 11 / 10 | N/A | No |
| 483383 |  CHEMBL3622806 CHEMBL3622806 | C39H68N8O8Si | 805.106 | 11 / 9 | N/A | No |
| 166218 |  CHEMBL435199 CHEMBL435199 | C41H67N11O7 | 826.057 | 11 / 10 | -0.2 | No |
| 166850 |  CHEMBL1766939 CHEMBL1766939 | C39H66N12O8 | 831.033 | 11 / 11 | -2.5 | No |
| 172914 |  CHEMBL2431131 CHEMBL2431131 | C23H25ClN4O2 | 424.929 | 6 / 0 | 4.7 | Yes |
| 173314 |  CHEMBL2369509 CHEMBL2369509 | C38H64N8O8 | 760.978 | 11 / 8 | -0.8 | No |
| 174034 |  CHEMBL132409 CHEMBL132409 | C38H64N12O8 | 817.006 | 12 / 11 | -3.7 | No |
| 554156 |  JMV458 JMV458 | C45H73N13O9 | 940.161 | 11 / 11 | 1.9 | No |
| 182599 |  CHEMBL2431133 CHEMBL2431133 | C27H28N4O3 | 456.546 | 7 / 0 | 5.2 | No |
| 182891 |  CHEMBL2431108 CHEMBL2431108 | C24H26N4O3 | 418.497 | 6 / 0 | 3.4 | Yes |
| 186857 |  CHEMBL1766941 CHEMBL1766941 | C40H68N12O8 | 845.06 | 11 / 9 | -2.0 | No |
| 190500 |  CHEMBL133340 CHEMBL133340 | C38H64N10O8 | 788.992 | 12 / 10 | -0.9 | No |
| 192162 |  CHEMBL132524 CHEMBL132524 | C40H68N12O8 | 845.06 | 14 / 12 | -1.7 | No |
| 550322 |  CHEMBL3954824 CHEMBL3954824 | C34H54BrN11O7 | 808.78 | 10 / 9 | -3.1 | No |
| 196206 |  CHEMBL2431111 CHEMBL2431111 | C29H32N4O2 | 468.601 | 6 / 0 | 6.3 | No |
| 196975 |  CHEMBL270332 CHEMBL270332 | C76H124N16O14 | 1485.93 | 18 / 16 | 4.4 | No |
| 197835 | 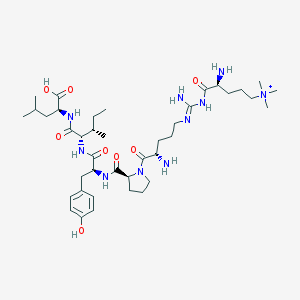 CHEMBL129953 CHEMBL129953 | C40H69N10O8+ | 818.054 | 11 / 9 | -0.4 | No |
| 198398 | 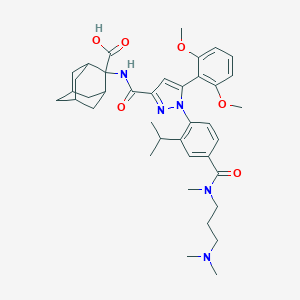 SR142948A SR142948A | C39H51N5O6 | 685.866 | 8 / 2 | 3.6 | No |
| 205387 | 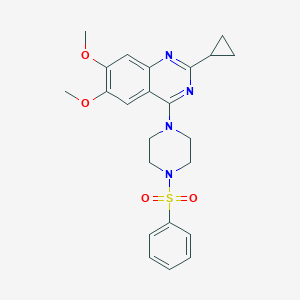 CHEMBL2431132 CHEMBL2431132 | C23H26N4O4S | 454.545 | 8 / 0 | 3.1 | Yes |
| 461049 | 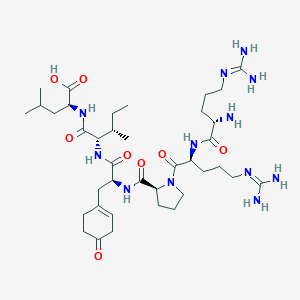 BDBM50342239 BDBM50342239 | C38H66N12O8 | 819.022 | 11 / 10 | -3.7 | No |
| 209944 | 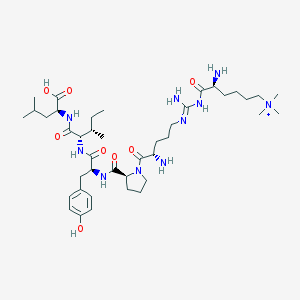 CHEMBL133798 CHEMBL133798 | C41H71N10O8+ | 832.081 | 11 / 9 | 0.0 | No |
| 211034 | 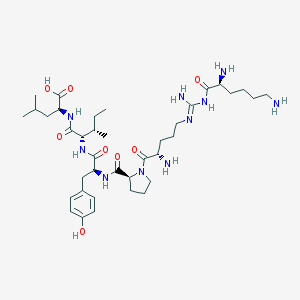 CHEMBL430910 CHEMBL430910 | C38H64N10O8 | 788.992 | 12 / 10 | -1.0 | No |
| 550544 | 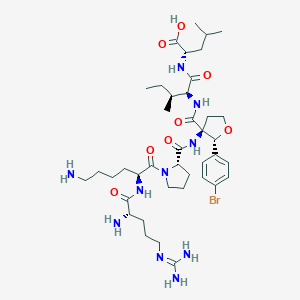 CHEMBL3915284 CHEMBL3915284 | C40H65BrN10O8 | 893.926 | 11 / 9 | -1.1 | No |
| 214680 | 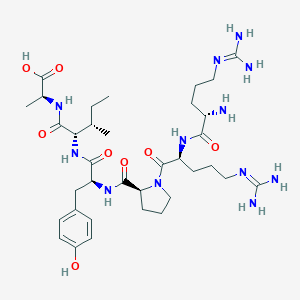 CHEMBL1766927 CHEMBL1766927 | C35H58N12O8 | 774.925 | 11 / 11 | -4.2 | No |
| 219625 | 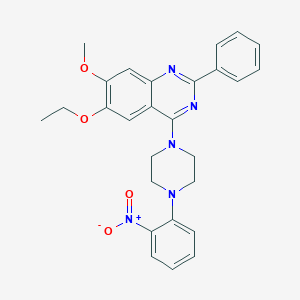 CHEMBL2431235 CHEMBL2431235 | C27H27N5O4 | 485.544 | 8 / 0 | 5.4 | No |
| 231750 | 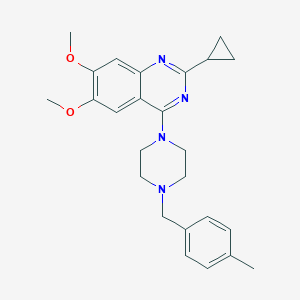 CHEMBL2431106 CHEMBL2431106 | C25H30N4O2 | 418.541 | 6 / 0 | 4.2 | Yes |
| 232249 | 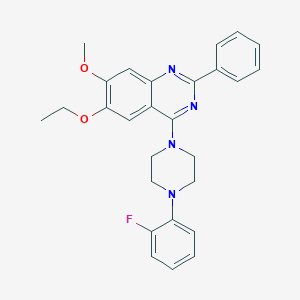 CHEMBL2431140 CHEMBL2431140 | C27H27FN4O2 | 458.537 | 7 / 0 | 5.7 | No |
| 233713 | 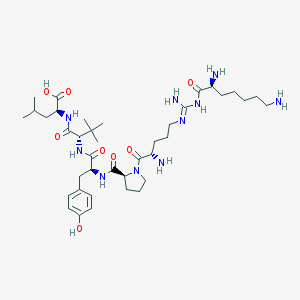 CHEMBL335846 CHEMBL335846 | C39H66N10O8 | 803.019 | 12 / 10 | -0.6 | No |
| 461240 | 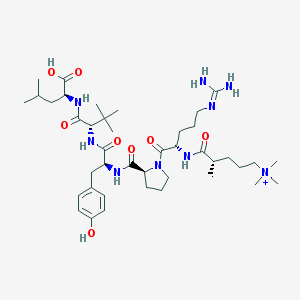 CHEMBL1172376 CHEMBL1172376 | C41H70N9O8+ | 817.066 | 9 / 8 | 2.8 | No |
| 234477 | 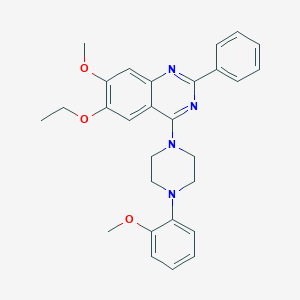 CHEMBL2431138 CHEMBL2431138 | C28H30N4O3 | 470.573 | 7 / 0 | 5.5 | No |
| 243562 | 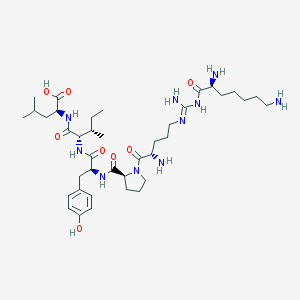 CHEMBL337644 CHEMBL337644 | C39H66N10O8 | 803.019 | 12 / 10 | -0.7 | No |
| 247284 | 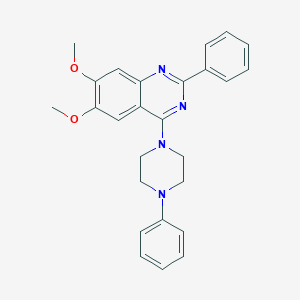 CHEMBL2431134 CHEMBL2431134 | C26H26N4O2 | 426.52 | 6 / 0 | 5.2 | No |
| 249829 |  CHEMBL133850 CHEMBL133850 | C41H70N10O8 | 831.073 | 12 / 9 | 0.3 | No |
| 250148 |  CHEMBL2431112 CHEMBL2431112 | C21H24N4O3 | 380.448 | 7 / 0 | 3.5 | Yes |
| 250156 |  CHEMBL1766825 CHEMBL1766825 | C39H66N12O8 | 831.033 | 11 / 10 | -2.3 | No |
| 250879 |  CHEMBL254770 CHEMBL254770 | C38H64N8O8 | 760.978 | 11 / 8 | -0.9 | No |
| 251007 | 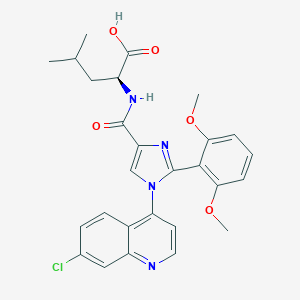 CHEMBL3099773 CHEMBL3099773 | C27H27ClN4O5 | 522.986 | 7 / 2 | 5.1 | No |
| 251577 | 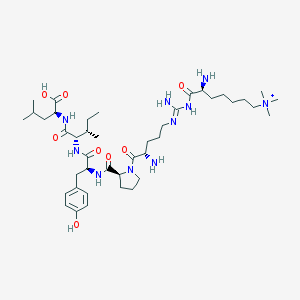 CHEMBL132794 CHEMBL132794 | C42H73N10O8+ | 846.108 | 11 / 9 | 0.3 | No |
| 256140 | 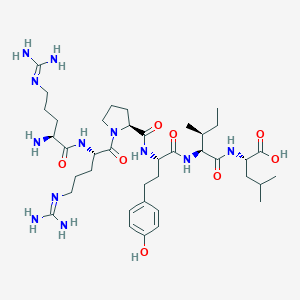 CHEMBL253527 CHEMBL253527 | C39H66N12O8 | 831.033 | 11 / 11 | -3.6 | No |
| 256347 | 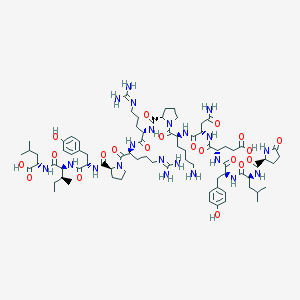 NEUROTENSIN NEUROTENSIN | C78H121N21O20 | 1672.95 | 23 / 21 | -3.7 | No |
| 256846 | 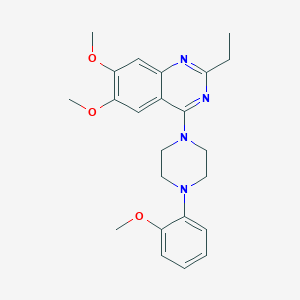 CHEMBL2431115 CHEMBL2431115 | C23H28N4O3 | 408.502 | 7 / 0 | 4.3 | Yes |
| 257994 | 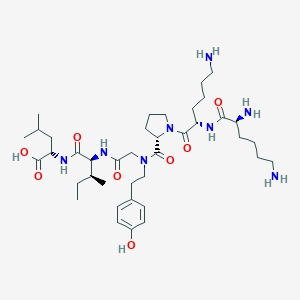 CHEMBL1766946 CHEMBL1766946 | C39H66N8O8 | 775.005 | 11 / 8 | -0.4 | No |
| 258605 | 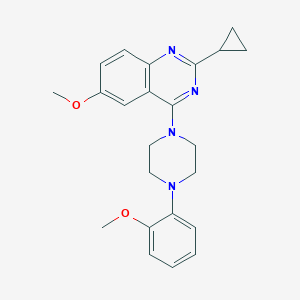 CHEMBL2431129 CHEMBL2431129 | C23H26N4O2 | 390.487 | 6 / 0 | 4.1 | Yes |
| 259932 | 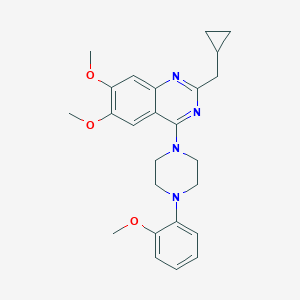 CHEMBL2431122 CHEMBL2431122 | C25H30N4O3 | 434.54 | 7 / 0 | 4.9 | Yes |
| 260434 |  CHEMBL439494 CHEMBL439494 | C69H108N20O18 | 1505.74 | 22 / 20 | -7.4 | No |
| 551174 |  CHEMBL3941970 CHEMBL3941970 | C34H54BrN9O7 | 780.766 | 10 / 8 | -2.2 | No |
| 270516 |  CHEMBL2431128 CHEMBL2431128 | C24H26N4O3 | 418.497 | 7 / 0 | 4.5 | Yes |
| 273365 |  CHEMBL2431232 CHEMBL2431232 | C25H30N4O2 | 418.541 | 6 / 0 | 5.0 | Yes |
| 279995 | 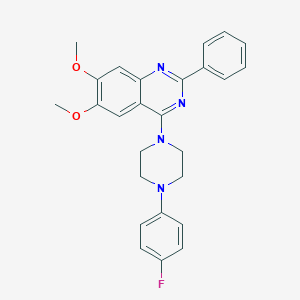 CHEMBL2431136 CHEMBL2431136 | C26H25FN4O2 | 444.51 | 7 / 0 | 5.3 | No |
| 551411 | 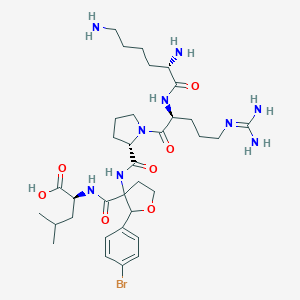 CHEMBL3932989 CHEMBL3932989 | C34H54BrN9O7 | 780.766 | 10 / 8 | -2.2 | No |
| 284031 | 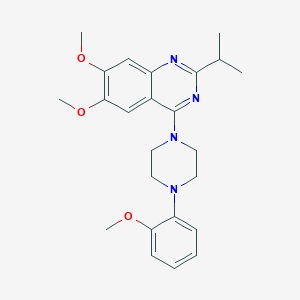 CHEMBL2431117 CHEMBL2431117 | C24H30N4O3 | 422.529 | 7 / 0 | 4.7 | Yes |
| 291026 | 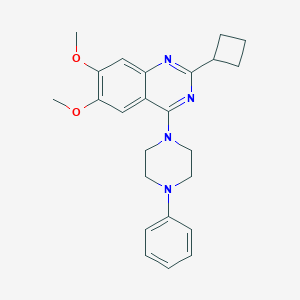 CHEMBL2431231 CHEMBL2431231 | C24H28N4O2 | 404.514 | 6 / 0 | 4.6 | Yes |
| 291914 | 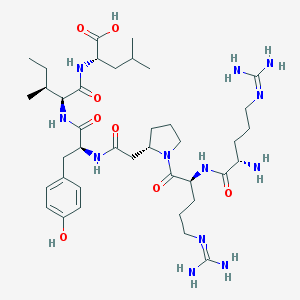 CHEMBL266230 CHEMBL266230 | C39H66N12O8 | 831.033 | 11 / 11 | -2.5 | No |
| 299167 | 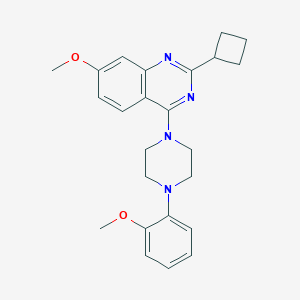 CHEMBL2431126 CHEMBL2431126 | C24H28N4O2 | 404.514 | 6 / 0 | 4.6 | Yes |
| 303446 | 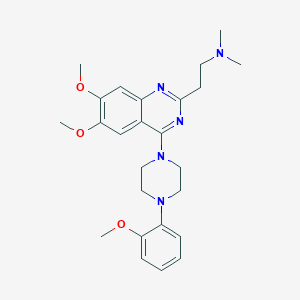 CHEMBL2431123 CHEMBL2431123 | C25H33N5O3 | 451.571 | 8 / 0 | 3.8 | Yes |
| 305033 | 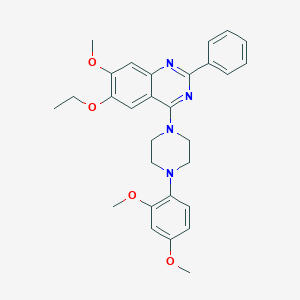 CHEMBL2431238 CHEMBL2431238 | C29H32N4O4 | 500.599 | 8 / 0 | 5.5 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417